Abstract
Problem
Intraamniotic pathogens and byproducts activate innate immune responses encompassing multitudes of signaling molecules and pathways that can result in spontaneous preterm birth (PTB). This study investigates fetal membrane response to bacterial stimulation using a bioinformatics approach.
Method of Study
Dysregulated biomarker (IL1-β, IL-2, IL-8, IL-10 and TNF-α) data from fetal membranes at term stimulated with Ureaplasma urealyticum, Ureaplasma parvum, Mycoplasma hominis, E. coli, Group B Streptococci, Polyporhans gingivalis, or Gardnerella vaginalis with 50% (v/v) amniotic fluid (AF) were analyzed by Ingenuity Pathway Analysis.
Results
In racially stratified analysis, networks representing late stage immune inflammation were seen in African Americans in AF absence. Inflammation was dominant in AF presence as well. In Caucasians, late stage immune response was dominant with AF but not in its absence.
Conclusions
Fetal membrane biofunctions in response to bacteria reflect early and late stage innate immune defense that vary based on presence of AF and subject race.
Keywords: cytokines, fetal membranes, inflammation, intraamniotic infection, prematurity
Introduction
Intraamniotic infections (IAI) are associated with ∼ 50 % of all spontaneous preterm births (PTB) and 75% of preterm premature rupture of the membranes (pPROM)1;2 cases. IAI is most common during early gestational periods (< 34 weeks), where incidence of neonatal mortality and morbidity are highest1;2. Intraamniotic pathogens and their byproducts activate an innate immune response that encompasses multitudes of signaling molecules and pathways, resulting in a predominantly cytokine and chemokine mediated inflammatory response that can lead to PTB3. Establishing infection in a sterile intraamniotic cavity requires overcoming antimicrobial properties contained in amniotic fluid such as defensins, bactericidal/permeability-increasing protein, cathelicidin, lysozyme, calprotectin, hyaluronan, and heat shock protein-containing exosomes. Compromise of the amniotic fluid (AF) may be necessary for the establishment of IAI;however, the contribution of AF to the host-immune response to IAI is not well understood4-7.
African-Americans are at a greater risk for infections during pregnancy as well as consequences of pPROM and preterm birth compared to Caucasians and Hispanics8;9. Pathophysiologic manifestations of these infections may differ between African-Americans and other racial groups despite similar environmental conditions and exposure to risk factors9. We theorize that IAI associated PTB or pPROM in a given woman depends on several factors including, but not limited to: pathogen type, anti-microbicidal/immunomodulatory activities of AF, and patient race.
Using an in vitro model system, we recently demonstrated that the innate immune response to each pathogen has its own unique immune/inflammatory signature. We also noted racial disparity amongst biomarkers and pathways of PTB in response to different pathogens. African-American women have immune responses to reproductive pathogens that may enhance their risk for PTB10. Autologous AF exposure of fetal membranes stimulated with IAI pathogens demonstrated that the immunomodulatory properties of AF vary between different pathogens. AF may inhibit or enhance inflammatory signals depending on the type of pathogen10. Although findings that distinct fetal membrane immune response to pathogens in individuals from different races are important and informative, these reports are limited to the biomarkers tested. Recent advances in bioinformatics have led to the development of computational methods that can identify downstream molecular pathways from assayed biomarkers.
We recently used Ingenuity Pathway Analysis (IPA) to analyze data from a multiplex analysis to identify potential pathways associated with disease function and other biomarkers that network with tested biomarkers11;12. In this study, we used IPA to analyze data from an ongoing study to: 1) characterize the networks affected by dysregulated biomarkers in terms of specific aspects of the host response to the bacterial pathogens; 2) determine how downstream pathways may differ between African-American and Caucasian women and 3) to characterize how downstream mediators of the host response to pathogens may be influenced by AF.
Methods
The tissue culture experiments used to generate the data used in this report were performed at The Perinatal Research Center, The Centennial Women's Hospital Nashville, TN, and IPA analysis of the data were performed at The University of Texas Medical Branch in Galveston, TX. All tissue collections, cultures and sample analyses were in compliance with Institutional Review Board-approved protocols from both institutions.
Source Data
This project is a secondary analysis of data published elsewhere, and details of the study design can be found in the original report10. Briefly, placental tissues were harvested from Caucasian and African-American women undergoing elective repeat Cesarean sections at term (≥37 weeks of gestation) prior to onset of labor. Fetal membranes were transported to the laboratory and placed in an organ explant system. Cultures were incubated overnight and then stimulated with 10 colony forming units (CFU)/color changing units (CCU) of Ureaplasma urealyticum (UU), Ureaplasma parvum (UP), Mycoplasma hominis (MH), Gardnerella vaginalis (GV), Polyporhorans gingivalis (PG), Group B Streptococci (GBS), or Escherichia coli (EC) in the presence or absence of 50% autologous AF for an additional 24 hours. Cytokine (TNF-α IL-6, IL-8, IL-10, and IL-2) concentrations in conditioned medium were quantified using the Bioplex™ Bead-Array system and analyzed using linear mixed effects models in R. Results were summarized as least-squares means ± SEM.
Bioinformatics analysis
Bioinformatics analysis was performed using Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems, http://www.ingenuity.com)13 to understand network interaction between dysregulated markers in our study and associated markers11;12. IPA uses information from its knowledgebase to create cellular and molecular networks that depict binding direct and indirect relationships between molecules. The statistical algorithm identifies networks involving our focus biomarkers based on their selective interconnectivity with each other and additional molecules stored in the knowledgebase. These networks are illustrated and ranked by the software for significance of focus gene enrichment. There are 27 higher-order disease and disorder categories in the IPA knowledgebase. In addition, there are several lower level and specific functions. Molecules in top ranking networks can be associated with one or more of these disease/disorder functions and are termed network functions. IPA constructed a series of connectivity maps, or network diagrams, derived from millions of molecular interactions and regulatory processes in the IPA knowledgebase. IPA network diagrams were generated based on their score, which is calculated by taking the negative log of the p-value for the likelihood these molecules would be found together by chance alone. A higher score indicates greater statistical significance that molecules depicted in the network are interconnected. Each connection is supported by at least one reference from the literature, a textbook, or canonical information stored in the IPA knowledgebase. Each network includes several partner molecules that were assigned to the network by IPA, but these are not among the input focus genes13.
Least square mean (LSM) ratios were calculated for each bacterial stimulation by dividing the least square mean of a bacterial stimulation by the least square mean value for the control for each of our focus biomarkers (IL-1β, TNF-α, IL-10, IL-8, and IL-2). LSM ratios and corresponding p-values were used to create separate spreadsheets for each racial group, both and with and without amniotic fluid, resulting in six spreadsheets total (Combined race -AF, combined race +AF, African-Americans -AF, African-Americans +AF, Caucasians -AF, Caucasians +AF). These spreadsheets were then uploaded separately into IPA.
Results
Characteristics of the source data
Details of the primary analysis which produced the source data for this project have been previously reported. In brief, the seven IAI pathogens (UU, UP, MH, GV, PG, GBS, and EC) tested10 produced different cytokine responses by fetal membranes. All bacterial species increased IL-8 production, and no change in IL-2 was seen with any pathogen. IL-1β and TNF-α production were stimulated by EC and GV compared to control, but responses to GBS and PG were limited to IL-1β and TNF-α, respectively. Genital mycoplasmas stimulated TNF-α and IL-10 but had no detectible effect on IL-1β production. African-Americans had twice the IL-1β response to EC as Caucasians (p=0.031). Conversely, Caucasians produced more IL-8 in response to LPS than African-Americans (p=0.026). AF had both pro- and antiinflammatory properties that varied between races and pathogens. Table I summarizes these findings.
Table I. Bacterial Stimulation of Fetal Membranes in Presence and Absence of Amniotic Fluid.
Effect of different bacterial species on cytokine production in fetal membranes in a racially stratified analysis [African American (AA) and Caucasian (C). Arrows indicate a statistically significant (p<0.05) fold change in cytokines compared to unstimulated control) and effect of AF on cytokine production for a given race-pathogen combination compared to controls stimulated with AF + Medium. (↑) - Up regulation compared to unstimulated controls; (↓) down regulation compared to unstimulated controls; - no change compared to unstimulated controls
| IL-1β | IL-8 | IL-10 | TNF-α | |||||
|---|---|---|---|---|---|---|---|---|
| Pathogen | AA | C | AA | C | AA | C | AA | C |
| AF + Medium | - | - | - | - | - | - | - | - |
| E. coli | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| E. coli +AF | ↓ | - | - | - | ↓ | ↓ | - | - |
| GBS | ↑ | - | ↑ | ↑ | - | - | - | - |
| GBS +AF | - | - | - | - | - | - | - | - |
| P. gingivalis | - | - | ↑ | ↑ | - | ↑ | ↑ | ↑ |
| P. gingivalis +AF | ↑ | - | - | - | - | - | - | - |
| G. vaginalis | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| G. vaginalis +AF | ↑ | - | - | - | ↑ | ↑ | ↑ | ↑ |
| M. hominis | - | - | ↑ | ↑ | ↑ | ↑ | ↑ | - |
| M. hominis +AF | - | - | - | - | ↑ | - | - | - |
| U. urealyticum | - | - | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| U. urealyticum +AF | - | ↓ | - | - | - | ↑ | - | - |
| U. parvum | - | - | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ |
| U. parvum +AF | - | - | - | - | ↓ | - | - | ↑ |
Tables II and Table III show shared functions in the presence and absence of AF for each bacterial stimulation in combined and racially stratified analyses. These are the top functions associated with each network that were mapped by IPA based on our input dataset. Scores for these networks were highly significant, ranging from 3 to 6 (p-value: 0.001 to 0.000001). Network functions are derived from a finite number of categories in the IPA knowledgebase. Figure I shows examples of IPA generated network diagrams for bacterial stimulation by EC. The remaining network diagrams and a list of molecules in each network can be found in the Appendix S1
Table II. IPA Network Biofunctions for Combined Dataset.
Ingenuity Pathway Analysis Network Functions for pathogens stimulated with either E. coli, Group B Streptococci, G. vaginalis, M. hominis, P. gingivalis, U. parvum, or U. urealyticum in the presence (+AF) or absence (−AF) of amniotic fluid. The ‘shared functions’ column indicates network functions that were the same both in the presence and absence of amniotic fluid. Abbreviations: Cell-to-Cell Signaling and Interaction (Cell Interaction), Hematological System Development and Function (Hem Function), Small Molecule Biochemistry (SMB)
| Combined Race | |||
|---|---|---|---|
| Pathogen | Shared Functions | −AF | +AF |
| E. coli |
|
|
|
| Group B . Streptococci |
|
|
|
| G. vaginalis |
|
||
| M. hominis |
|
||
| P. gingivalis |
|
|
|
| U. parvum |
|
||
| U. urealyticum |
|
||
Table III. IPA Network Biofunctions stratified by Race.
Ingenuity Pathway Analysis Network Functions for pathogens stimulated with either E. coli, Group B Streptococci, G. vaginalis, M. hominis, P. gingivalis, U. parvum, or U. urealyticum in the presence (+AF) or absence (−AF) of amniotic fluid. The ‘shared functions’ column indicates network functions that were the same both in the presence and absence of amniotic fluid. Abbreviations: Cell-to-Cell Signaling and Interaction (Cell Interaction), Hematological System Development and Function (Hem Function), Small Molecule Biochemistry (SMB)
| African Americans | Caucasians | |||||
|---|---|---|---|---|---|---|
| Pathogen | Shared Functions | −AF | +AF | Shared Functions | -AF | +AF |
| E. coli |
|
|
|
|
|
|
| Group B . Streptococci |
|
|
|
|
|
|
| G. vaginalis |
|
|
|
|
|
|
| M. hominis |
|
|
|
|
||
| P. gingivalis |
|
|
|
|
||
| U. parvum |
|
|
|
|
|
|
| U. urealyticum |
|
|
|
|
||
Figure I. Selected Examples of IPA generated Network Diagrams.

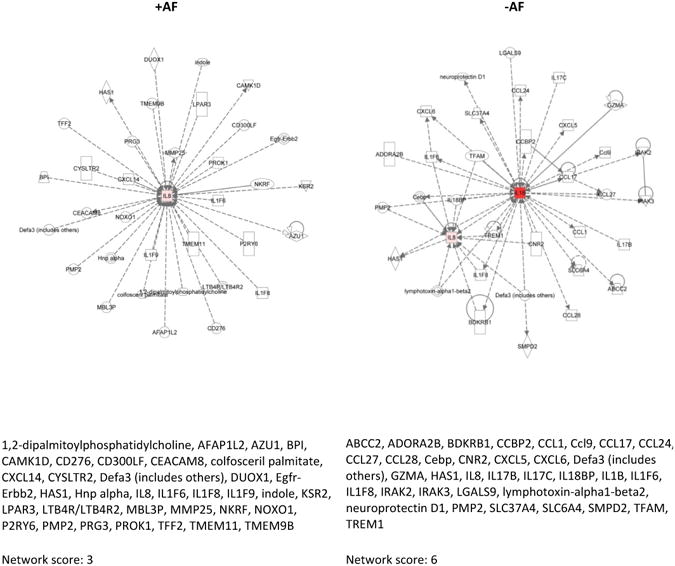

Our results showed that many network functions overlapped, while some were exclusive to the presence or absence of AF. Networks resulting from bacterial stimulations without the presence of AF varied across pathogen and race in several instances.
Analysis of combined race
We initially performed IPA analysis using combined data irrespective of race. Network associated biological functions are indicative of underlying pathologic processes when membranes were treated with IAI bacteria compared to unstimulated controls. When data were combined by race, treatment with AF did not have an impact on the network associated functions of several pathogens, as indicated by the number of shared functions for pathogen stimulations with and without AF treatment. The functions identified by biomarker networks indicate different types of immune mechanisms that are expected in response to bacterial infections. The type of immune functions associated with each pathogen-stimulated network differed based on dysregulation of the biomarkers, their significance, and network interactions. Except EC, all pathogens shared network functions with and without AF treatment. These functions included well defined inflammatory response (GBS, MH, PG, UP, UU) and metabolic derangements (GV), a likely precursor of early stage inflammatory response.
Dominant functions differed for EC, GBS, and PG in the presence and absence of AF. In the absence of AF, stimulation by EC produced functions indicative of metabolic derangement. Immune response related functions such as antigen presentation and cellular movement were top network-associated functions for GBS and PG respectively in the absence of AF. Stimulations by GV and the genital mycoplasmas (MH, UP, and UU) did not have any network-associated functions exclusive to AF absence. EC stimulation in the presence of AF produced antigen presentation and cellular movement (suggesting chemotaxis) as top network functions, which were different than the functions noted without AF treatment. The immune response functions were reversed for GBS and PG stimulations with AF compared to stimulation without AF, where cellular movement and antigen presentation were top network associated biofunctions for GBS and PG respectively. Stimulations by GV and genital mycoplasmas did not have network associated functions that were exclusive to only the AF treated compartment. All networks were significant (GBS, MH, PG: Score=3, p=0.001; UP, UU: Score=6, p=0.000001; EC, GV: Score=5, p=0.00001).
Racially stratified analysis of the data
In African-Americans, several network functions were common and overlapping in response to bacterial stimulations, irrespective of AF presence. Inflammatory response was a common function across most stimulation. To note, as indicated by the network diagrams (Appendix S1), the type of biofunction represented by inflammatory mediators are not the same in all bacteria. EC, GBS, GV, and UP all produced distinct forms of inflammatory response that are denoted by IPA as inflammatory response, antigen presentation, changes associated with tissue morphology (adaptation to immune stimuli or phagocytosis), cellular movement, and immune cell trafficking (associated with chemotaxis and diapedesis). No functions were common to both AF treatment and absence for stimulations by PG and UU.
Immune responses were more pronounced for African Americans in the absence of AF in culture media. All bacteria produced inflammation associated fetal membrane responses (cellular movement, immune cell trafficking, hematologic systems related functions, tissue morphologic changes, and antigen presentation). Presence of AF in culture eliminated the inflammatory response induced by PG and UU in African Americans, but some form of inflammatory response remained with other bacteria. All networks that were found were significant, and network p-values for each bacteria with and without AF treatment were the same (EC, UP: Score=6, p=0.000001; GBS, MH, PG, UU: Score=3, 0.001; GV: Score=5, 0.00001). No significant networks were found for PG and UU in the presence of AF.
Network functions for Caucasian membranes were markedly different from African American membranes. Presence or absence of AF produced common network associated functions for all bacterial stimulations. These functions, all forms of immune response, included hematologic systems related functions (EC, UP and UU), lipid metabolism (GV) and inflammation associated antigen presentation (GBS, MH, PG). In the absence of AF, immune response was not well represented as a network associated biofunction, but metabolic changes associated with early stage immune response, hematologic systems development, antigen presentation, and cell to cell signaling and interactions were observed for all bacterial stimulations. The presence of AF modified these functions to have more defined inflammatory response. All networks were significant, and network p-values for each bacteria with and without AF treatment were the same (EC, GBS, GV, MH, PG, UP, UU: Score=5, p=0.00001).
Discussion
Host inflammatory response is a major contributor of adverse pregnancy outcomes14-16. The inflammatory process is a complex system dependent on multitudes of factors associated with both an infectious agent and a host's innate and acquired immune defense system16. Pathogen related factors that influence inflammation may include the type of pathogen, its dose, site of localization, antigenicity, and presence of polymicrobial etiology. Factors that affect pregnancy may also include various host specific factors such as race, genetic and environmental factors and their interactions16. In order to tailor successful interventions to prevent infection associated PTB, it is necessary to better understand the infectious agent, host specific immune response, and related biofunctions (pathophysiologic changes) resulting in host affected pregnancy complications. In this study, a bioinformatics approach was employed to provide 1) in depth information on changes in biomarkers profile associated with seven IAI pathogens 2) pathogen-specific modification of host response pathways by AF and 3) information on possible racial disparities in molecular pathways stimulated by IAI pathogens and the influence of AF factors.
We found that individual pathogens mount distinct inflammatory responses, representing different biofunctions likely to influence pregnancy outcome. Using IPA, network maps including dysregulated molecules from our study in addition to markers that were not a part of this study (derived from the IPA knowledgebase) were generated. This allowed us to better categorize inflammatory stages induced in fetal membranes as early or late stage. IPA function labels such as “antigen presentation, cellular movement, cell to cell interaction, metabolic derangement, immune cell trafficking, hematologic system development and function” are suggestive of early-stage host innate immune response. Conversely, the late-stage immune response that IPA identified as “inflammatory response” represents a very well established inflammatory condition that likely represents the end stage of a fully developed host immune response. The former condition is presented by most pathogens, as fetal membranes were likely adapting to immune challenges after twenty four hour stimulations. To note, terminologies identified by the IPA application are more suitable to represent classical immune cell functions (neutrophils, T cells, macrophages antigen presentation, chemotaxis, diapedesis, etc.) and do not necessarily indicate that fetal membranes perfrom those exact functions. Based on the data, we provide the best explanation for the bioinformatics data in context of infection, fetal membrane inflammatory response and adverse pregnancy outcome. Network interaction diagrams and biofunctions suggest similar mechanistic properties exhibited by the membranes in response to different pathogens.
Our earlier reports demonstrated the need for racial stratification of data prior to analysis as race is a risk modifier. As expected, data stratified by race show vastly different response compared to combined data analysis, and therefore a discussion of combined data is not attempted here. Late-stage inflammation is presented by African American derived fetal membranes for most pathogens (see shared response column irrespective ofAF presence, Table II). However, the late stage was limited to GV + AF stimulation and Ureaplasma species when AF was absent. This suggests that AF likely plays a role in controlling Ureaplasma mediated pathologic complications in African Americans. Shared functions (functions shared in presence and absence of AF) showed early-stage immune response by Caucasian derived membranes in response to all pathogens, compared to shared functions by African Americans that demonstrated late stage inflammatory response. Absence of AF showed early stage immune response whereas presence of AF enhanced the response to late stage inflammation. In conclusion, the presence of AF produced marked inflammatory networks and biofunctions in Caucasians (Table III).
IPA allows data and knowledge driven construction of fetal membrane immune response that is not otherwise possible with limited biomarker data. IPA studies are not without limitations. Like all computational techniques, IPA generates information that is based on current knowledge, therefore mistakes in the literature could lead to erroneous evaluations and misidentification of pathways beyond the control of IPA users. As more knowledge about the functional human molecular markers are accumulated, new information may lead to identification of pathways that were not detected in this study. Although this can limit the reproducibility of the results, having the database be built on well-established relationships between proteins suggests that we will be more at risk of not identifying pathways than misidentifying them17.
The current management of preterm labor is based on the logical but overly simplistic approach of inhibiting uterine contractions. This strategy has not been effective, as seen by the increasing rate of preterm birth in recent decades. Proper identification of risk factors, understanding of specific initiators and effectors of labor process, or knowledge of pathophysiologic pathways is critical to the development of improved interventions. The information provided here demonstrates that immunologic pathways and biomarkers associated with different infections agents are unique, and their manifestation resulting in PTB or pPROM likely will depend on the type of innate immune defense mounted against them. Understanding the unique biofunctions exerted by each pathogen is critical for intervention design.
Supplementary Material
Acknowledgments
This study is supported by NIH #1R03HD067446-01 to R Menon.
Details of Ethics Approval: This study was approved by the Western Institutional Review Board (protocol ID no. 20101328; 9/29/2010).
Footnotes
Disclosure of Interests: None of the authors have any conflicts of interest to report.
Contribution to Authorship: GB analyzed the data and wrote the article. MP provided bacterial cultures and helped with manuscript preparation. TAS helped with tissue culture. COD recruited subjects, and collected and processed samples. GS assisted with data interpretation and manuscript preparation. RM conceived and designed the experiments, recruited subjects, collected samples, analyzed the data and mentored GB in writing this article.
Supporting Information: Additional supporting information may be found in the online version of this article. This version will include network diagrams produced by each bacterium in fetal membranes with and without AF in combined race, African Americans and Caucasians.
Disclosure: The authors report no conflict of interest
Reference List
- 1.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastek JA, Gomez LM, Elovitz MA. The role of inflammation and infection in preterm birth. Clin Perinatol. 2011;38:385–406. doi: 10.1016/j.clp.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Sezen D, Perni U, Herway C, Bongiovanni AM, Skupski D, Witkin SS. Hyaluronan modulates pro-inflammatory immune activity in the mid-trimester amniotic cavity. J Reprod Immunol. 2009;82:89–93. doi: 10.1016/j.jri.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Sezen D, Bongiovanni AM, Jean-Pierre C, Linhares IM, Skupski D, Witkin SS. Ex vivo cytokine production by whole mid-trimester amniotic fluid. J Reprod Immunol. 2008;78:22–27. doi: 10.1016/j.jri.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Perni SC, Kalish RB, Hutson JM, et al. Differential expression of immune system-related components in midtrimester amniotic fluid from singleton and twin pregnancies. Am J Obstet Gynecol. 2005;193:942–946. doi: 10.1016/j.ajog.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 7.Asea A, Jean-Pierre C, Kaur P, et al. Heat shock protein-containing exosomes in mid-trimester amniotic fluids. J Reprod Immunol. 2008;79:12–17. doi: 10.1016/j.jri.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Fiscella K. Racial disparities in preterm births. The role of urogenital infections. Public Health Rep. 1996;111:104–113. [PMC free article] [PubMed] [Google Scholar]
- 9.Menon R. Spontaneous preterm birth. Race and Genetics in Understanding the Complexities of Preterm Birth. Expert Rev Obstet Gynecol. 2009;4:695–704. [Google Scholar]
- 10.Peltier MR, Drobek CO, Bhat G, et al. Amniotic Fluid and Maternal Race Influence Responsiveness of Fetal Membranes to Bacteria. J Reprod Immunol. 2012 doi: 10.1016/j.jri.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brou L, Almli LM, Pearce BD, et al. Dysregulated biomarkers induce distinct pathways in preterm birth. BJOG. 2012;119:458–473. doi: 10.1111/j.1471-0528.2011.03266.x. [DOI] [PubMed] [Google Scholar]
- 12.Menon R, Pearce B, Velez DR, et al. Racial disparity in pathophysiologic pathways of preterm birth based on genetic variants. Reprod Biol Endocrinol. 2009;7:62. doi: 10.1186/1477-7827-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingenuity Systems. 2012 Ref Type: Online Source ( http://www.ingenuity.com)
- 14.Romero R, Gotsch F, Pineles B, Kusanovic JP. Inflammation in pregnancy: its roles in reproductive physiology, obstetrical complications, and fetal injury. Nutr Rev. 2007;65:S194–S202. doi: 10.1111/j.1753-4887.2007.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 15.Gotsch F, Romero R, Kusanovic JP, et al. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- 16.Romero R, Espinoza J, Goncalves LF, Kusanovic JP, Friel L, Hassan S. The role of inflammation and infection in preterm birth. Semin Reprod Med. 2007;25:21–39. doi: 10.1055/s-2006-956773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer-Mehren A, Furlong LI, Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol Syst Biol. 2009;5:290. doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



