Summary
Both p53 and Mdmx are ubiquitinated and degraded by the same E3 ligase Mdm2; interestingly, however, while p53 is rapidly degraded by Mdm2, Mdmx is a stable protein in most of cancer cells. Thus, the mechanism by which Mdmx is degraded by Mdm2 needs further elucidation. Here, we identified the noncoding 5S rRNA as a major component of Mdmx-associated complexes from human cells. We show that 5S rRNA acts as a natural inhibitor of Mdmx degradation by Mdm2. RNAi-mediated knockdown of endogenous 5S rRNA, while not affecting p53 levels, significantly induces Mdmx degradation and subsequently, activates p53-dependent growth arrest. Notably, 5S rRNA binds the RING domain of Mdmx and blocks its ubiquitination by Mdm2 whereas Mdm2-mediated p53 ubiquitination remains intact. These results provide insights into the differential effects on p53 and Mdmx by Mdm2 in vivo and reveal an critical role of noncoding 5S rRNA in modulating the p53-Mdmx axis.
Keywords: 5S rRNA, Non-coding small RNA, Mdm2, Mdmx, p53, HAUSP, Ubiquitination, Deubiquitination
Introduction
The p53 tumor suppressor acts as the major sensor for a regulatory circuit that monitors signaling pathways from diverse sources, including DNA damage, oncogenic events, ribosomal stress and others abnormal cellular processes (Kruse and Gu, 2009; Vousden and Prives, 2009). While p53 mutations have been documented in more than half of human tumors, deregulation in other key components of the p53 pathway, such as overexpression of its inhibitors Mdm2 and Mdmx, are frequently observed in tumor cells that retain wild type p53 (Gilkes and Chen, 2007; Marine et al., 2007; Zhang and Lu, 2009) . It is well accepted that Mdm2 and its related protein Mdmx (also called Mdm4) play a major part in the scope of specifically inhibiting p53 activities in cancer cells . Mdm2, a RING finger oncoprotein, acts as a specific E3 ubiquitin ligase in p53 degradation. The critical role of Mdm2 in regulating p53 is best illustrated by studies carried out in mice where inactivation of p53 was shown to completely rescue the embryonic lethality caused by the loss of Mdm2 function (Marine and Lozano, 2010). Despite its high sequence homology with Mdm2 and the presence of a RING domain, Mdmx does not have intrinsic E3-ligase activity for p53 but was instead shown to inhibit p53-induced transcription via their interactions; nevertheless, Mdmx knockout mice die even in the presence of Mdm2 and this lethality is also rescued by inactivation of p53 (Marine et al., 2007). Accumulating evidence indicates that degradation-independent mechanisms are crucial for both Mdm2 and Mdmx in controlling p53 activities by forming a protein complex with p53 on the promoters of specific p53-responsive genes (Chen et al., 2010; Minsky and Oren, 2004; Tang et al., 2008). Thus, the role of Mdmx in repressing p53 function is as critical as that of Mdm2 and physiological levels of Mdmx are required in a non-redundant manner to regulate p53 activity in vivo. Indeed, Mdmx overexpression is found in several types of human tumors retaining wild type p53 (Danovi et al., 2004; Marine et al., 2007). Notably, like p53, Mdmx is ubiquitinated and degraded by Mdm2 (de Graaf et al., 2003; Kawai et al., 2003; Pan and Chen, 2003); several studies indicate that degradation of Mdmx is crucial for optimal activation of p53 function (Jin et al., 2006; LeBron et al., 2006; Lopez-Pajares et al., 2008; Meulmeester et al., 2005; Okamoto et al., 2005; Pereg et al., 2005). However, unlike p53, Mdmx is a very stable protein in most human cancer cells and the molecular mechanism by which Mdmx is regulated in vivo needs to be further elucidated.
Results
5S rRNA is a major component of Mdmx associated complexes
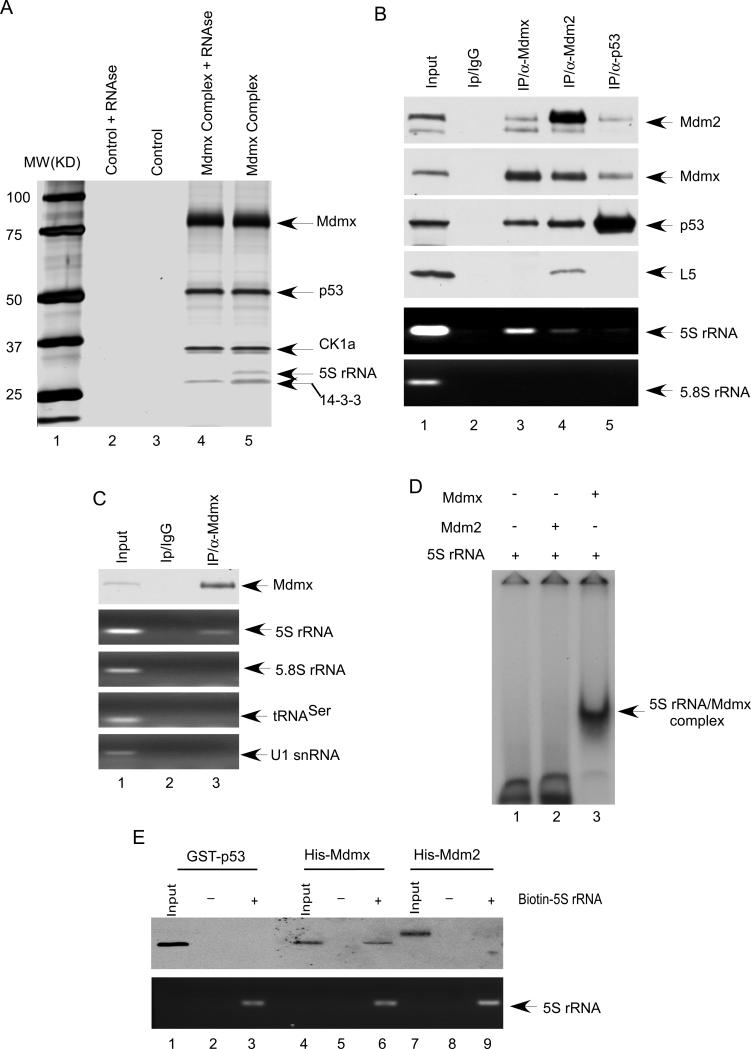
To understand the mechanism by which Mdmx stability is regulated in vivo, we sought to identify specific cofactors from physiologically-formed Mdmx protein complexes in human cells. To this end, we generated a derivative of human osterosarcoma U2OS cell line that stably expresses an exogenous Mdmx polypeptide containing N-terminal FLAG and C-terminal HA epitopes (Flag-Mdmx-HA). To isolate Mdmx-associated protein complexes, cell extracts from the stably transformed U2OS cells were sequentially subjected to affinity chromatography on M2 (Flag antibody) agarose beads and an HA-affinity column. The bound proteins were then fractionated by SDS-PAGE and visualized by silver staining (Figue 1A). Both Flag-HA tagged Mdmx and endogenous p53 polypeptides were recovered as major components of the complexes, indicating that ectopic Mdmx can form a stable complex with endogenous p53. Consistent with previous studies, mass spectrometry analysis of the major bands of the complex further identified CK1a and 14-3-3 as Mdmx-associated proteins (Chen et al., 2005; Jin et al., 2006; LeBron et al., 2006; Lopez-Pajares et al., 2008). Interestingly, the Mdmx complexes also contained a band with a relative molecular mass of 30,000 (Mr 30K) (lane 5). Mass spectrometry analysis of this band failed to produce any peptide identity. However, treatment of the Mdmx complexes with RNase A completely eliminated the 30K band (lane 4), suggesting that it may contain a small RNA molecule associated with Mdmx. The RNA band was extracted and cloned; sequencing analysis identified this band as 5S rRNA, comprising 121 nucleotides.
Figure 1. 5S rRNA directly interacts with Mdmx both in vitro and in vivo.
(A) The Mdmx complex affinity-purified from the whole extract of the U2OS/Flag-Mdmx-HA stable cell line (lane 4 and lane 5) were incubated with RNase A (lane 4) or the buffer alone (lane 5), and analyzed by SDS-PAGE and silver attaining. The parental U2OS whole cell extract were used in a parallel procedure for control (lane 2 and lane 3).
(B) Coimmunoprecipitation of 5S rRNA with Mdmx from U2OS cells. The whole-cell extract (lane 1) or immunoprecipitates with a control IgG (lane 2), anti-Mdmx antibody (lane 3), anti-Mdm2 antibody (lane 4) or anti-p53 antibody (lane 5) were subjected to western blot for Mdm2, Mdmx, p53, and L5. The 5S rRNA, and 5.8s rRNA were analyzed by RT-PCR
(C) Mdmx specifically interacts with 5S rRNA. The whole cell extract (lane 1) or immunoprecipitates with a control IgG (lane 2) or anti-Mdmx antibody (lane 3) were analyzed by RT-PCR with primers for 5S rRNA, 5.8s rRNA, tRNAser and U1 snRNA.
(D) Mdmx directly interacts with 5S rRNA. [32P] UTP- labeled 5S rRNA was incubated with 75 ng of His-Mdm2 (lane 2), or 60 ng of His-Mdmx (lane 3) for 45 min at room temperature, and analyzed by native gel electrophoresis and autoradiography.
(E) Mdmx, but not p53 or Mdm2 interacts with 5S rRNA. Biotin labeled 5S rRNA was incubated with GST-p53 (lane 3), His-Mdmx (lane 6) or His-Mmd2 (lane 9). As the control, nonlabeled 5S rRNA was incubated with GST-p53 (lane 2), His-Mdmx (lane 5) or His-Mdm2 (lane 8). The precipitates with Avidin Agarose beads were analyzed by Western bolt with anti-body against p53, Mdmx or Mdm2; or by RT-PCR with primer for 5S rRNA.
5S rRNA directly interacts with Mdmx both in vitro and in vivo
To elucidate the physiological role of this Mdmx-5S rRNA interaction, we first analyzed the specificity of this interaction at the endogenous levels. To this end, cell extracts from native U2OS cells were immunoprecipitated with α-Mdmx, α-Mdm2 and α-p53 or with control IgG. As seen in Figure 1B, high levels of 5S rRNA were readily detected in the immunoprecipitates obtained with the α-Mdmx antibody but not with control IgG (lane 3 vs. 2). As expected, both p53 and Mdm2 were also present in the Mdmx-associated protein complexes. Notably, only a very small amount of 5SRNA was detected in Mdm2-associated complexes while 5S rRNA was barely detected in the p53-complexes. Since 5S rRNA is always found in the complexes with ribosomal proteins, it is possible that Mdmx may interact with 5S rRNA indirectly through ribosomal proteins. Thus, we monitored the levels of ribosomal proteins in these complexes. As expected, endogenous ribosome proteins such as L5 were present in Mdm2-associated protein complexes (Dai and Lu, 2004; Horn and Vousden, 2008; Lindstrom et al., 2007); however, we failed to detect any ribosomal proteins in Mdmx associated complexes (lane 4 vs 3, and Figure S1A). Moreover, we also examined several other noncoding RNAs such as 5.8S rRNA, tRNA and U1 snRNA. As shown in Figure 1C, none of these RNA was detectable in Mdmx-associated complexes. These data demonstrate the specific interaction between endogenous Mdmx and 5S rRNA in human cells and also indicate that 5S rRNA is a major component of Mdmx associated complexes compared to either p53- or Mdm2-associated complexes.
To determine whether Mdmx directly binds to 5S rRNA, we test this interaction in a purified system. 5S rRNA was in vitro transcribed and radiolabeled. The purified radiolabeled 5S rRNA was incubated with or without highly purified, bacterial expressed Mdmx or Mdm2 protein (Figure S1B) and then analyzed by electrophoresis mobility shift assay (EMSA). Although previous studies suggest that Mdm2 may directly interact with RNA in vitro (Elenbaas et al., 1996), the binding affinity and specificity are not well understood. EMSA analysis revealed that the binding between 5S rRNA and Mdm2 apparently, was extremely weak in the presence of non-specific competitors such as tRNA (lane 2, Figure 1D). However, under the same conditions, the mobility of the vast majority of the labeled 5S rRNA was shifted by Mdmx (lane 3), suggesting that Mdmx is a much stronger binding protein for 5S rRNA. To corroborate with these data, we also tested the binding specificity with biotin-labeled 5S rRNA. To this end, biotin-labeled 5S rRNA was first incubated with highly purified recombinant proteins (p53, Mdm2 and Mdmx), respectively, and then the RNA-protein complexes were precipitated with Avidin Agarose beads. As shown in Figure 1E, western blot analysis revealed that Mdmx was readily detected in 5S rRNA complexes (lane 6). Neither Mdm2 nor p53 was detectable in 5S rRNA-associated complexes under the same conditions. Taken together, these data indicate that Mdmx, but not p53 or Mdm2 directly interacts with 5S rRNA in vitro and in vivo.
RNAi-mediated knockdown of 5S rRNA does not affect the formation of the large ribosomal RNA-protein complex, but induces Mdmx degradation
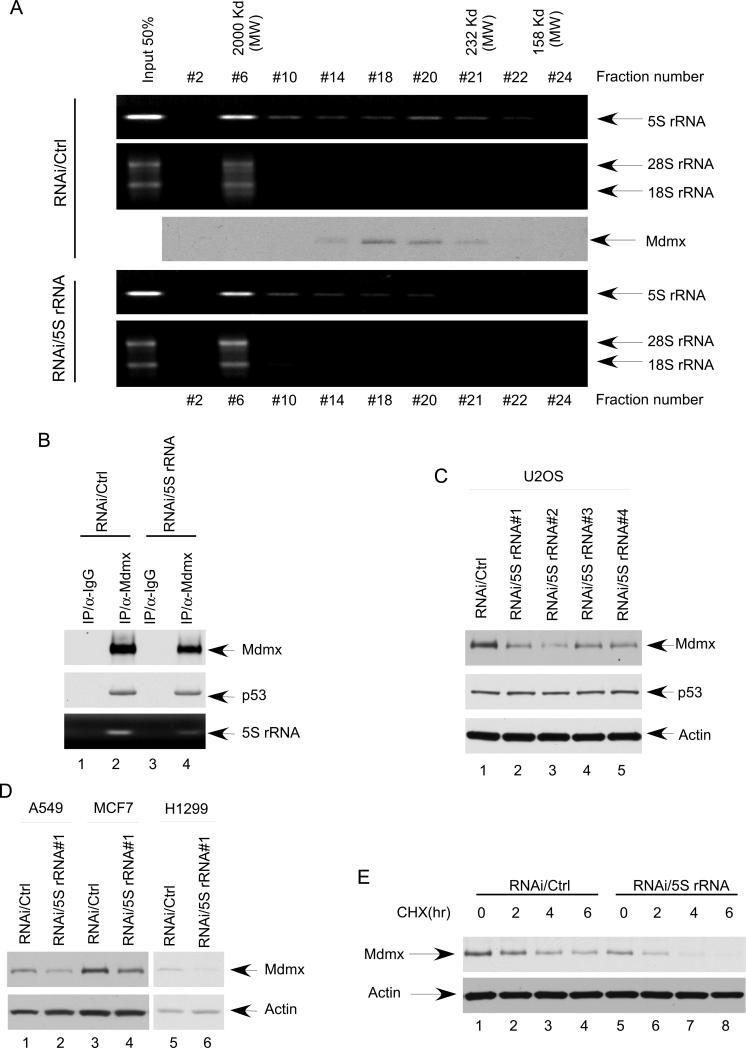
5S rRNA is a small non-coding RNA component of the large ribosomal RNA-protein complex, that is critically involved in protein translation process (Szymanski et al., 2003). Indeed, when the 5S rRNA complexes were subjected to gel-filtration chromatography on Superose 6 (SMART system), the majority of 5S rRNA was present in a large RNA-protein ribosomal complex (Fraction #6) together with other components of ribosomal RNA such as 28S rRNA and 18S rRNA, with the molecular weight of about 2 MDa (Figure 2A, upper panel). Interestingly, a fraction of 5S rRNA was co-eluted with the Mdmx polypeptide in fraction #18-21 with much smaller molecular weight sizes, suggesting that 5S rRNA can form smaller complexes with Mdmx, independent of the large ribosomal complex. To understand the physiological role of 5S rRNA in regulating Mdmx, we examined whether knockdown of endogenous 5S rRNA has any effect on the Mdmx- 5S rRNA complex. To this end, U2OS cells were transfected with either a 5S rRNA-specific (RNAi/5S rRNA) or a control (RNAi/control) siRNA. As shown in Figure 2A (lower panel), the amount of endogenous 5S rRNA in the large ribosomal complexes (fraction 6) was essentially the same after transfections with 5S rRNA-RNAi and no obvious effect on protein translation was observed in these cells (data not shown). Conversely, the levels of 5S rRNA in the smaller complexes (fraction #18 to #21) were significantly reduced. Moreover, the Mdmx-associated complexes from 5S rRNA knockdown cells, as well as from control cells were immunoprecipitated with the α-Mdmx antibody in the presence of proteasome inhibitor treatment. Indeed, the levels of 5S rRNA in the Mdmx complex were reduced upon 5S rRNA knockdown whereas the Mdmx-p53 interaction remained intact (lane 4 vs. lane 2, Figure 2B). Taken together, these data indicate that RNAi-mediated knockdown of 5S rRNA described above does not affect the formation of the large ribosomal RNA-protein complex, but significantly decreases the levels of 5S rRNA present in the Mdmx- associated complexes.
Figure 2. RNAi-mediated knockdown of 5S rRNA does not affect the formation of the large ribosomal RNA-protein complex, but induces Mdmx degradation.
(A) Minimal reduction of 5S rRNA does not affect the integrity of ribosome. U2OS cells were treated with either control siRNA or siRNA for 5S rRNA. Whole cell extracts were fractionated on a Superpose 6 gel filtration column. The fractions were analyzed by RT-PCR with primer for 5S rRNA, and by Western blot with antibody against Mdmx. 18s rRNA and 28s rRNA were used to indicate the fraction contains the integrated ribosome.
(B) Minimal reduction of 5S rRNA reduces Mdmx bound 5S rRNA. U2OS cells were treated with either control siRNA (lane 1 and 2) or siRNA for 5S rRNA (lane 3 and 4), and then the whole cell extracts were prepared after 6 hours treatment with proteasome inhibitor. The immunoprecipitates with a control IgG (lane 1 and 3) or anti-Mdmx antibody (lane 2 and 4) were subjected to western blot for Mdmx and p53, and to RT-PCR for 5S rRNA
(C) 5S rRNA knocking down reduces Mdmx levels. The cell extracts from the U2OS treated with a control RNAi (lane 1), or four different siRNA for 5S rRNA (lane2, 3, 4, and 5) were subjected to western blot for Mdmx, p53, and Actin.
(D) 5S rRNA was knocked down by RNAi in A549, MCF7, and H1299 cells. The cell extracts from the cells treated with a control siRNA (lane 1, 3, and 5), or siRNA for 5S rRNA (lane2, 4 and 6) were analyzed by western bolt using the antibodies against Mdmx, and Actin.
(E) Knocking down 5S rRNA decreases the half-life of Mdmx protein. U2OS cells treated with either control siRNA (lane 1, 2, 3 and 4) or siRNA for 5S rRNA (lane 5, 6, 7 and 8) were harvested at indicated time points (hour) after cyclohexamide (CHX) treatment. The cell extracts were subjected to western blot for Mdmx and Actin.
To examine the role of 5S rRNA in regulating Mdmx function, we tested the effect on Mdmx stability by RNAi-mediated knockdown. As shown in Figure 2C, although the levels of p53 were unaffected by 5S rRNA knockdown, Mdmx protein levels were significantly reduced (lane 2 vs. lane 1). To exclude off-target effects, we also treated cells with three additional 5S rRNA siRNAs (RNAi/5S rRNA #2, RNAi/5S rRNA #3 and RNAi/5S rRNA #4) that recognize different regions of the 5S rRNA sequence. Again, the endogenous levels of Mdmx protein were all reduced by 5S rRNA knockdown, although the mRNA levels of Mdmx remained unchanged (Figure S2A). Similar effects on Mdmx levels by 5S rRNA knockdown were also obtained in other human cancer cell lines such as human lung cancer cell line A549, human breast cancer cell line MCF-7 and human lung cancer cell line p53-null H1299 (Figure 2D). Moreover, the half-life of endogenous Mdmx was significantly decreased from about 4 hrs to less than 2 hrs by knockdown of 5S rRNA (Figure 2E; Figure S2B). These data demonstrate that 5S rRNA plays an important role in regulating Mdmx stability in human cancer cells.
5S rRNA knockdown induces p53 activation and triggers p53-dependent cell growth arrest
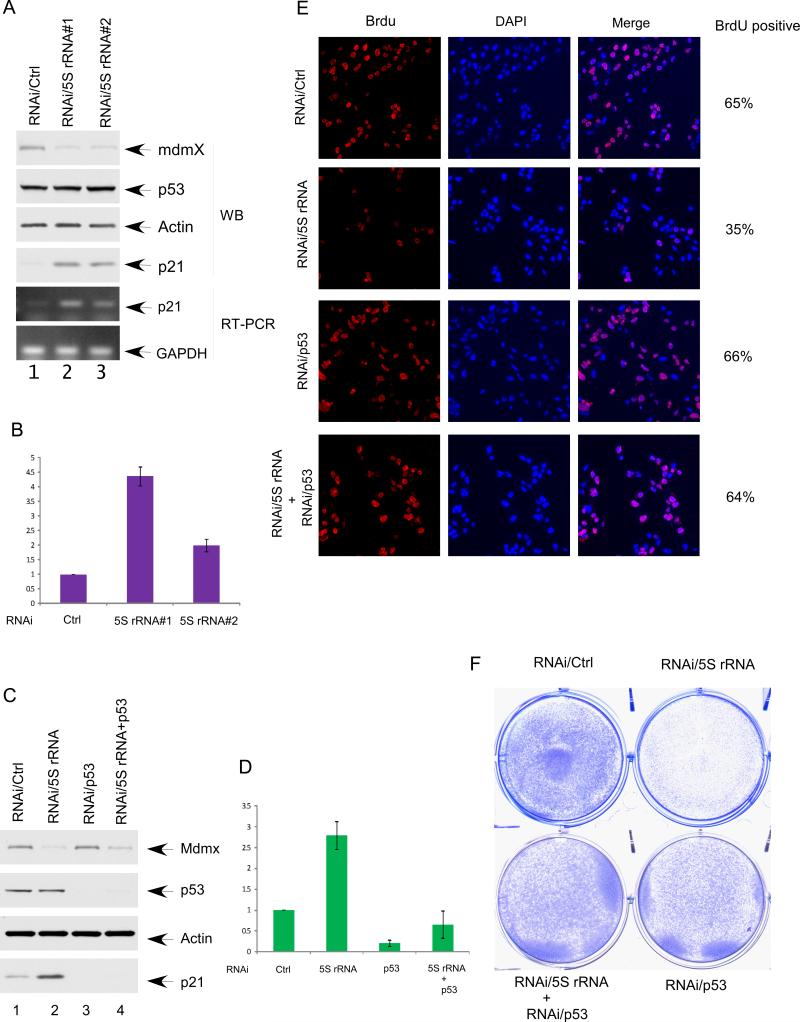
Although Mdmx may also play a role in p53 degradation by associating with Mdm2, accumulating evidence suggests that the key function of Mdmx is to repress p53-mediated transcriptional activation (Kruse and Gu, 2009; Marine et al., 2007). Thus, we evaluated the role of 5S rRNA in regulating p53-dependent transcriptional activity. As shown in Figure 3A, RNAi-mediated knockdown of 5S rRNA did not elevate the p53 levels but significantly increased protein and mRNA levels of p21, a well established target of p53 affected by Mdmx (also see Figure 3B). To ascertain whether the 5S rRNA-mediated effect on p21is p53-dependent, we tested the consequences of 5S rRNA knockdown in the absence of p53. Indeed, p21 activation was markedly diminished in these cells upon siRNA-mediated co-depletion of p53 (Figures 3C and 3D), suggesting that the activation of p21 induced by 5S rRNA knockdown is solely p53-dependent. To examine whether 5S rRNA knockdown affects cell growth, we used BrdU staining for ly synthesized DNA to monitor cell proliferation. 5S rRNA knockdown significantly decreased the fraction of BrdU-positive cells (from 65% to 35%), but this effect was reversed by concomitant knockdown of p53 (66% vs. 64%) (Figures 3E and Figure S3). Moreover, cell proliferation was also visibly inhibited upon 5S rRNA knockdown after three days of culture (Figure 3F). Again, these effects were dramatically reversed by concomitant knockdown of endogenous p53. These experiments indicate that 5S rRNA knockdown induces Mdmx degradation and triggers p53-dependent cell growth arrest.
Figure 3. 5S rRNA knockdown induces p53 activation and triggers p53-dependent cell growth arrest.
(A) 5S rRNA was knocked down by RNAi in U2OS cells. The cell extracts from the U2OS treated with a control RNAi (lane 1), or two different siRNA for 5S rRNA (lane2 and lane3) were analyzed by western bolt using the antibodies against Mdmx, p53, p21, and Actin. The RNA levels of p21 and GAPDH were determined by RT-PCR.
(B) RNA levels of p21 in the experiment described in (A) was quantitative analyzed by RT-qPCR. Error bars represent standard errors calculated from 3 different experiments.
(C) 5S rRNA knocking down induces p21 in a p53 dependent manner. U2OS cells treated with control siRNA (lane 1), siRNA for 5S rRNA (lane 2), p53 siRNA (lane 3) or combination of p53 siRNA and siRNA for 5S rRNA (lane 4). The cell extracts were analyzed by western blot using the antibodies against Mdmx, p53, p21, and Actin.
(D) RNA levels of p21 in the experiment described in (C) was quantitative analyzed by RT-qPCR. Error bars represent standard errors calculated from 3 different experiments.
(E) The BrdU incorporation of the U2OS cells treated with a control RNAi, siRNA for 5S rRNA, p53 siRNA, or combination of p53 siRNA and siRNA for 5S rRNA. The cells were labeled with BrdU for 1 hr and immunostained with anti-BrdU antibody. The nuclei are in blue (DAPI), and the BrdU positive nuclei are shown in red.
(F) U2OS cells were treated as (B), the cells were stained with crystal violet three days after siRNA treatment.
Mechanistic insights into 5S rRNA-mediated Mdmx regulation
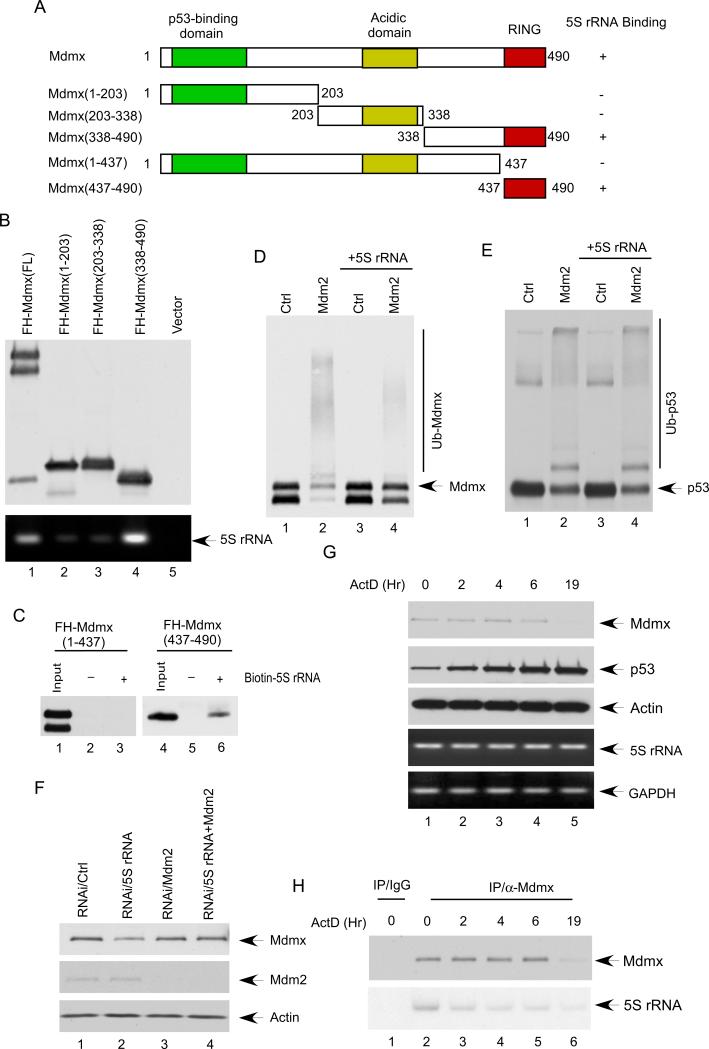
To elucidate the molecular mechanism by which 5S rRNA controls the stability of Mdmx, we further mapped the binding domain of Mdmx for 5S rRNA. By using co-immunoprecipitation assays, we found that 5S rRNA strongly interacts with the C-terminus (a.a.338-490) of Mdmx but has very low affinity for either the N-terminus (a.a. 1-203) or the middle region (a.a. 203-308) of Mdmx (Figures 4A and 4B). Furthermore, by using in vitro binding assays with biotin-labeled 5S rRNA, we found that the RING domain (a.a. 437-490) of Mdmx is the major binding domain for 5S rRNA (lane 6, Figure 4C). Although the RING domain of Mdmx does not contain intrinsic ubiquitin ligase activity for either p53 or itself, this domain is crucial for Mdmx degradation induced by Mdm2 (de Graaf et al., 2003; Kawai et al., 2003; Pan and Chen, 2003). Thus, it is very likely that 5S rRNA may modulate Mdm2-mediated ubiquitination of Mdmx through directly interacting with the RING domain of Mdmx. To validate this notion, we examined whether 5S rRNA has any specific effect on Mdmx ubiquitination in a purified system. As seen in Figure 4D, western blot analysis revealed that high levels of ubiquitinated Mdmx generated by Mdm2 (lane 2) were effectively abolished in the presence of 5S rRNA. In contrast, 5S rRNA had no significant effect on p53 ubiquitination under the same conditions (Figure 4E). To corroborate these findings, we tested whether 5S rRNA expression induces stabilization of Mdmx in cells. Western analysis revealed that Mdmx levels were induced in a dosage-dependent manner by 5S rRNA expression whereas the levels of p53 or Mdm2 were not affected (Figures S4A and S4B). To confirm the role of Mdm2 in 5S rRNA-mediated effect on Mdmx in vivo, we examined whether inactivation of Mdm2 could rescue Mdmx degradation induced by 5S rRNA knockdown. As shown in Figure 4F, as expected, the levels of Mdmx were reduced upon 5S rRNA knockdown (lane 2); however, in the cells in which endogenous Mdm2 was co-depleted by RNAi, the effect on Mdmx levels by 5S rRNA knockdown was completely reversed (lane 4). Taken together, these results demonstrate that 5S rRNA acts as a specific inhibitor for Mdmx ubiquitination and degradation induced by Mdm2.
Figure 4. Mechanistic insights into 5S rRNA-mediated Mdmx regulation.
(A) Schematic representation of the human Mdmx protein and various mutant used in this study. The p53 binding domain, the acidic domain, and the RING domain are indicated.
(B) C-terminal of Mdmx is involved in 5S rRNA interaction. U2OS cells were transfected with various plasmids, as indicated. The cell extracts were immunoprecipitated with Flag antibody and then analyzed by western bolt with HA anti-body. The associated 5S rRNA were analyzed by RT-PCR.
(C) RING domain of Mdmx directly interacts with 5S rRNA. Biotin labeled 5S rRNA was incubated with FH-Mdmx (1-437) (lane 3), or FH-Mdmx (437-490) (lane 6). As the control, nonlabeled 5S rRNA was incubated with FH-Mdmx (1-437) (lane 2), or FH-Mdmx (437-490) (lane 5). The precipitates with Avidin Agarose beads were analyzed by Western bolt with HA anti-body.
(D) 5S rRNA inhibits Mdm2-mediated ubiquitination of Mdmx. FH-Mdmx incubated with either buffer (lane 1 and 2) or 5S rRNA (lane 3 and 4) were subjected to in vitro ubiquitination reactions either in the absence of Mdm2 (lane1 and 3) or in the presence of Mdm2 (lane 2 and 4). The reactions were separated by SDS-PAGE and analyzed by Western bolt with HA anti-body.
(E) 5S rRNA has no effect on Mdm2-mediated ubiquitination of p53. F-p53 incubated with either buffer (lane 1 and 2) or 5S rRNA (lane 3 and 4) were subjected to in vitro ubiquitination reactions either in the absence of Mdm2 (lane1 and 3) or in the presence of Mdm2 (lane 2 and 4). The reactions were separated by SDS-PAGE and analyzed by Western bolt with anti-body against p53.
(F) U2OS cells treated with control siRNA (lane 1), siRNA for 5S rRNA (lane 2), Mdm2 siRNA (lane 3) or combination of Mdm2 siRNA and siRNA for 5S rRNA (lane 4). The cell extracts were analyzed by western blot using the antibodies against Mdm2, Mdmx, and Actin.
(G) Actinomycin D (ActD) treatment reduces the levels of mdmx protein. U2OS cells were harvested at indicated time points (hour) after Actinomycin D (ActD) treatment. The cell extracts were subjected to western blot for Mdmx, p53 and Actin. The RNA levels of 5S rRNA and GAPDH were determined by RT-PCR.
(H) U2OS cells were harvested at indicated time points (hour) after Actinomycin D (ActD) treatment. The whole cells extracts were immunoprecipitated with a control IgG (lane 1), or anti-Mdmx antibody (lanes 2 to 6), and then analyzed by western bolt with anti-body against Mdmx. The associated 5S rRNA were analyzed by radioactive RT-PCR.
To further elucidate the role of the Mdmx-5S rRNA interaction under physiological settings, we examined whether this interaction is regulated during the stress response. Several studies showed that Mdmx is phosphorylated by ATM and Chk2 upon DNA damage, which is crucial prerequisite for Mdmx degradation by Mdm2 during the DNA damage response (LeBron et al., 2006; Meulmeester et al., 2005; Wang et al., 2009). Moreover, phosphorylation of Mdmx also hinders its ability to interact with the deubiquitinase HAUSP and therefore, facilitates Mdmx degradation (Meulmeester et al., 2005). Interestingly, upon ribosomal stress, e.g. treatment with reagents such as Actinomycin D (ActD), p53 is activated in the absence of ATM/Chk2-mediated phosphorylation (Tang et al., 2008); likewise, phosphorylation of Mdmx is undetectable but still undergoes Mdm2-mediated degradation (Gilkes and Chen, 2007; Gilkes et al., 2006), suggesting a mechanism involved in destabilization of Mdmx upon ribosomal stress. To this end, we treated the cells with ActD and harvested the cells at different time points. As shown in Figure 4G, upon the ActD treatment, the levels of Mdmx were reduced slowly at the early time points but more drastic reduction was observed after 6 hrs. By co-immunoprecipitation assays, we found that the levels of 5S rRNA from Mdmx-associated complexes were significantly decreased upon ActD treatment at each time point even before the levels of Mdmx were dramatically reduced (Fig. 4H). Similar data were also obtained when the cells were treated with serum starvation (Figure S4C). Thus, these data suggest that inhibition of the 5S rRNA-Mdmx interaction at least, in part, contributes to Mdmx degradation during the cellular response to ribosomal stress.
Discussion
Despite the fact that Mdmx does not have intrinsic E3-ligase activity for p53, numerous studies show that the role of Mdmx in repressing p53 function is as critical as that of Mdm2 in tumorigenesis; accumulating evidence also indicates that regulation of Mdmx stability is crucial for its oncogenic effects in vivo (Gilkes and Chen, 2007; Marine et al., 2007; Vousden and Prives, 2009; Wang et al., 2009). Notably, both p53 and Mdmx are ubiquitinated and degraded by Mdm2 but the stabilities of p53 and Mdmx in cancer cells as well as the dynamic changes of both proteins during the stress response behave completely differently. For example, p53 is very unstable in cancer cells but stabilized upon stress; conversely, Mdmx is a stable protein under normal conditions but degraded rapidly when the cells are under stress. By identifying 5S rRNA as a specific regulator for Mdmx stability, our study implicates that 5S rRNA plays an important role in modulating Mdm2-mediated differential effects on p53 and Mdmx in vivo.
Numerous studies indicate that the p53 pathway is tightly regulated by ribosome biogenesis and ribosomal stress (Zhang and Lu, 2009). While several ribosomal proteins such as L5, L11 and L23 have been showed as important regulatory factors in regulating p53 function by interacting with Mdm2, none of these factors is found associated with Mdmx (Gilkes and Chen, 2007; Zhang and Lu, 2009). By identifying 5S rRNA as a critical regulator for Mdmx stability, our study provides insights into the regulation of the p53-Mdmx axis by ribosomal stress. Although earlier studies showed that the RING domain of Mdm2 can also interact with 5S rRNA (Elenbass et al., 1996; Marechal et al., 1994), our data indicate that the direct binding between Mdm2 and 5S rRNA is apparently extremely weak (Figure 1). Notably, by using co-immunoprecipitation assays, we found that 5S rRNA interacts indirectly with the middle part of Mdm2 (a.a. 205-339) in cells but had no obvious binding for either the N-terminus or the Ring domain containing C-terminal region of Mdm2 in human cells (Figures S5A and S5B). Since the middle part of Mdm2 (a.a. 205-339) is the region responsible for its direct interactions with ribosomal proteins (Zhang and Lu, 2009), it is very likely that Mdm2 can bind 5S rRNA indirectly through its association with ribosomal proteins such as L11 and L5 in human cells. Interestingly, the ability of L11 to bind the 5S rRNA is also critical for the cooperation with L5 to regulate Mdm2 activities (Horn and Vousden, 2008), suggesting that 5S rRNA may modulate the functions of both Mdm2 and Mdmx in vivo. Moreover, it was reported that p53 is covalently linked to 5.8S rRNA (Fontoura et al., 1992). Thus, the prescise roles of 5S rRNA and other noncoding rRNAs in regulating the p53 pathway are clearly very important issues that warrent further investigations. In this regard, it is noteworthy that 5S rRNA is commonly overexpressed in many types of human cancer and that 5S rRNA expression is tightly regulated by both oncogenic or tumor suppressor signals (Marshall et al., 2008; White, 2008). Finally, the mechanism by which the Mdmx -5S rRNA interaction is regulated during the stress response remains unclear. Future studies may be required to elucidate whether these ribosomal proteins such as L11 and L5, participate in modulating the Mdmx-5S rRNA interaction upon ribosomal stress (Gilkes et al., 2006). It will be interesting to examine whether protein modifications of Mdmx may play a role in modulating this interaction upon stress. The critical role of 5S rRNA in controlling Mdmx stability further indicates that non-coding RNAs have “protein-like” regulatory functions in many biological processes, which go beyond the traditional role defined for RNA.
Experimental Procedures
Electrophoresis mobility shift assay (EMSA)
5S rRNA was in vitro transcribed and radiolabeled. His-Mdmx and His-Mdm2 were purified from bacteria. The protein-RNA binding reactions (10 μl) contained 20 mM HEPES (pH 7.6), 35 mM KCl, 0.1 mM EDTA, 10% glycerol, 2.5 mM MgCl2, 0.1 mM spermidine, 1 mM DTT, 0.2% Triton X-100, 5ng tRNA, 1ng [32P] UTP- labeled 5S rRNA, 0.2 Unit SUPERase inhibitor [Ambion], 75 ng of His-Mdm2, or 60 ng of His-Mdmx. After 45 min of incubation at room temperature, the reactions were analyzed by native gel electrophoresis and autoradiography.
Biotin-Pull-down
5S rRNA was labeled with Biotin by in vitro transcribed. Biotin labeled 5S rRNA was incubated with GST-p53, His-Mdmx or His-Mmd2 in 20 μl of binding buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, 0.2% Triton X-100, 10% Glycerol, 0.2 Unit SUPERase inhibitor [Ambion] and protein inhibitor mixture [Sigma]). As the control, nonlabeled 5S rRNA was incubated with GST-p53, His-Mdmx or His-Mdm2 in the same buffer. The RNA was pulled down using Avidin Agarose beads and the beads were washed five times with binding buffer. The precipitates were analyzed by Western bolt
In Vitro Ubiquitination Assays
The in vitro ubiquitination assay was performed as described previously with some modifications. Mdmx or p53 was mixed with other components, including E1 (10 ng), E2 (His-UbcH5a, 100 ng), and 5 μg of His-ubiquitin (affinity) and Mdm2 in 10 μl of reaction buffer (40 mM Tris, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, pH 7.6). The reaction was stopped after 3 hrs at 37 °C by the addition of SDS sample buffer, and subsequently resolved by SDS-PAGE gels for Western blot analysis with anti-Mdmx antibody or anti-p53 antibody.
BrdU Labeling
The BrdU incorporation assay was performed as previously described. In brief, cells were grown in medium containing 20 μM BrdU (Calbiochem) for 2 h and then fixed in 70% ethanol. DNA was denatured, and cells were permeabilized in 2N HCl, 0.5% Triton X-100 (Sigma), neutralized in 0.1 M Na2B4O7 (pH 8.5), and then blocked with 1% BSA in PBS. Anti-BrdU was added following the manufacture's protocol (Amersham). After washing with 1% BSA/PBS, the cells were incubated with Alexa-488 conjugated anti-mouse IgG (Molecular Probes). Finally, cells were counterstained with DAPI to visualize the nuclei.
Supplementary Material
HIGHLIGHTS.
5S rRNA is a major component of Mdmx-associated complexes.
5S rRNA acts as a natural inhibitor of Mdmx degradation by Mdm2.
5S rRNA knockdown induces Mdmx degradation and p53 activation.
5S rRNA blocks Mdm2-mediated ubiquitination of Mdmx but not of p53.
Acknowledgement
We specifically thank Jun Qin and Yingming Zhao for validating some of the known components in Mdmx complexes by Mass spectrometry analysis, Shang-Jui Wang for carefully reading the manuscript. This study was supported by grants from NIH/NCI and the Leukemia and Lymphoma Society. W.G. is also supported by an Ellison Medical Foundation Senior Scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
More detailed procedures could be found in the Supplemental Information.
References
- Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 2005;25:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li Z, Zwolinska AK, Smith MA, Cross B, Koomen J, Yuan ZM, Jenuwein T, Marine JC, Wright KL, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J. 2010;29:2538–2552. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- Fontoura BM, Sorokina EA, David E, Carroll RB. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen J. Distinct roles of MDMX in the regulation of p53 response to ribosomal stress. Cell Cycle. 2007;6:151–155. doi: 10.4161/cc.6.2.3719. [DOI] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27:5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pajares V, Kim MM, Yuan ZM. Phosphorylation of MDMX mediated by Akt leads to stabilization and induces 14-3-3 binding. J Biol Chem. 2008;283:13707–13713. doi: 10.1074/jbc.M710030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Elenbaas B, Piette j., Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle. 2005;4:1166–1170. doi: 10.4161/cc.4.9.1981. [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol. 2005;25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5 S rRNA: structure and interactions. Biochem J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.