Abstract
Unloaded shortening speeds, V, of muscle are thought to be limited by actin-bound myosin heads that resist shortening, or V = a·d·τon−1 where τon−1 is the rate at which myosin detaches from actin and d is myosin's step size. The a-term describes the efficiency of force transmission between myosin heads, and has been shown to become less than one at low myosin densities in a motility assay. Molecules such as inorganic phosphate, Pi, and blebbistatin inhibit both V and actin-myosin strong binding kinetics suggesting a link between V and attachment kinetics. To determine whether these small molecules slow V by increasing resistance to actin sliding or by decreasing the efficiency of force transmission, a, we determine how inhibition of V by Pi and blebbistatin changes the force exerted on actin filaments during an in vitro sliding assay, measured from changes in the rate, τbreak−1, at which actin filaments break. Upon addition of 30 mM Pi to a low (30 μM) [ATP] motility buffer V decreased from 1.8 to 1.3 μm·sec−1 and τbreak−1 from 0.029 to 0.018 sec−1. Upon addition of 50 μM blebbistatin to a low [ATP] motility buffer, V decreased from 1.0 to 0.7 μm·sec−1 and τbreak−1 from 0.059 to 0.022 sec−1. These results imply that blebbistatin and Pi slow V by decreasing force transmission, a, not by increasing resistive forces, implying that actin-myosin attachment kinetics influence V.
Introduction
Muscle contracts when myosin heads cyclically interact with actin filaments through their ATPase reaction (1-4). With the weak-to-strong actin-binding intermediate step in the ATPase reaction, a single myosin head undergoes a discrete lever arm rotation that displaces an actin filament a distance d(5-11). A single myosin head takes one of these displacement steps every time it goes around its ATPase cycle and thus in theory can move an actin filament at a speed V = d/(τon + τoff), where τon and τoff are lifetimes of the A-M strong- and weak-binding states respectively.
In theory, V would increase with increasing numbers, N, of myosin heads as V = N·d/(τon + τoff), but in the context of a muscle system (Fig. 1A) where many myosin heads interact with the same actin filament, the lever arm rotation of a given myosin head becomes damped by other actin-bound myosin heads that oppose actin movement (12,13), making the discrete lever arm rotation more of a force-generating than a motion-generating step. With enough myosin heads strongly bound to an actin filament, “the actin filament cannot move any faster than the heads can let go” (12), and V becomes limited by strongly-bound myosin heads that resist actin movement. It is commonly assumed that V reaches a maximum value when N = (τon + τoff)/τon, or when
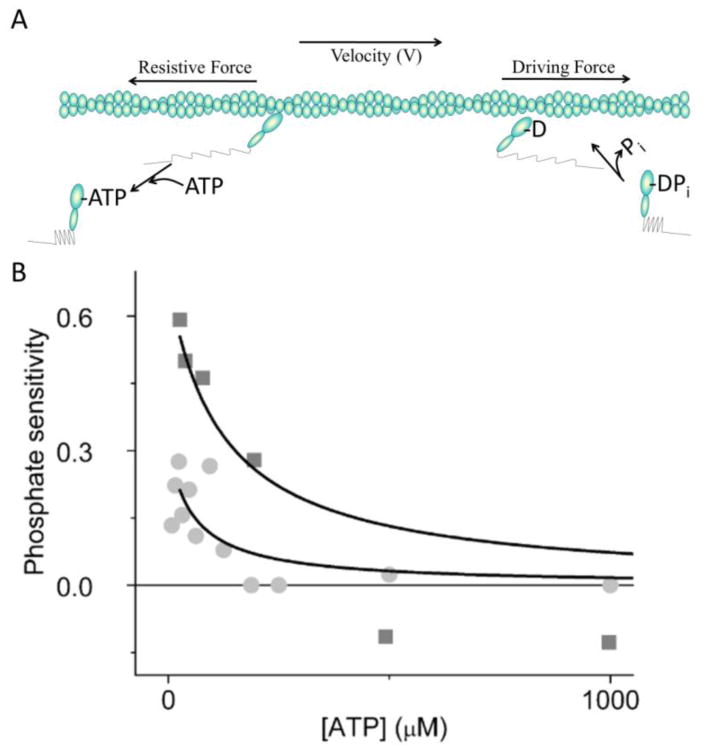
Figure 1.
Model of actin sliding velocities. (A) Driving forces (springs on right) are generated in the direction of actin movement by myosin weak-to-strong binding transitions (heads on right). These forces are opposed by resistive forces (springs on left) exerted by actin-bound myosin heads that have become negatively strained by actin sliding (heads on left). Here, the spatial separation of positively and negatively strained heads is used for illustrative purposes only. (B) The [ATP]-dependence of phosphate sensitivity (1 – VPi/V°) calculated from data in Amrute-Nayak et al. (20) (squares) and Hooft et al. (17) (circles), where V° is the actin sliding velocity in the absence of added Pi and VPi is the actin sliding velocity with added Pi.
According to this model, τon is the sole biochemical determinant of Vmax, and thus velocities are said to be detachment limited. An additional factor, a, is sometimes included to describe the observation that Vmax decreases at low myosin densities due to inefficient force transmission (39).
| (Eq. 1) |
The goal of the present study is to determine whether inhibition of actin-myosin strong binding kinetics results in a similar decrease in a.
A myosin head can transmit forces over a relatively limited length of actin. Harris and Warshaw showed (16) that when the average spacing between myosin heads increases beyond 130 nm, the efficiency, a, with which one head transmits forces to another decreases significantly, slowing Vmax (Eq. 1). Their analysis raises an important question. To achieve a maximum velocity, Vmax, does the force generated by one head need to be efficiently transmitted to any actin-associated myosin head (including weakly-associated heads) or does it need to be efficiently transmitted to another strongly bound myosin head? Harris and Warshaw assume the former, but the limited ability of weak-binding heads to maintain force suggests that force transmission probably occurs between strongly bound heads. In this case, the data in Harris and Warshaw would indicate that the distance over which a myosin head can efficiently transmit force to another strongly bound myosin head is ∼130 nm·(τon + τoff)/τon (∼2600 nm for a 5% duty ratio). Because the distance between strongly bound myosin heads increases when the rate, katt, of actin-myosin strong binding decreases (τoff increases), the efficiency of force transmission, a, could decrease with a decrease in katt, resulting in a decrease in Vmax and a corresponding decrease in the interhead forces generated in a motility assay.
Recent studies suggest that both τon and τoff influence V (17). Specifically, Hooft et al. demonstrated that at low [ATP], in vitro actin sliding velocities, V, decrease with the addition of 30 mM phosphate, Pi, consistent with a Pi-induced increase in τoff (per cycle) through the reversal of myosin's force-generating, weak-to-strong binding transition (18, 19). However, Brenner and colleagues argue that the inhibition of V by Pi at low [ATP] results from a Pi–induced increase in τon (consistent with the detachment-limited model) caused by Pi competing with ATP for the myosin active site.
To determine if Pi slows V by increasing resistance to actin sliding (i.e., increasing τon) or if it slows V by decreasing the driving force of weak-to-strong binding transitions (i.e., increasing τoff), an in vitro motility assay can be used to determine the effects of Pi on the forces exerted by myosin heads along actin filaments during actin sliding (20). If Pi slows V by decreasing driving forces, then it should decrease the rate of the force-induced actin filament breaking during motility (Fig. 1A). On the other hand, if Pi slows V by increasing τon, Pi should increase the rate of force-induced actin filament breaking (Fig. 1A). To estimate actin filament breaking rates, Amrute-Nayak et al. (20) attempted to make these measurements by imaging fluorescently-labeled actin filaments at two time points during a motility assay. They showed that after 100 seconds of motility, filaments were on average shorter in the presence of 5 mM Pi than in the absence of added Pi. The authors inferred that Pi increases the rate of actin filament breaking, concluding that Pi slows V by increasing τon.
There are two potential problems with these previous experiments. First, as shown below, the rate of actin filament breaking varies with filament length, and the authors did not control for this in their study. Second, the visible pool of actin filaments changes during a motility assay due to actin filaments moving in and out of the field of view, and so actin filaments analyzed at time t = 100 sec are not the same as those analyzed at t = 0 sec. A more rigorous approach for determining the rate (τbreak−1) at which actin filaments break in a motility assay is to perform a frame-by-frame analysis that detects actual actin filament breaking events. Below, we use this approach, controlling for actin filament length, to determine the effects of Pi on the rate of actin filament breaking. In contrast to the results of Amrute-Nayak et al. (20), we show that at low [ATP] the addition of Pi to a motility assay significantly decreases τbreak−1. Our data imply that inhibition of V by Pi results from decreased driving forces rather than increased τon, supporting the hypothesis that inhibiting the forces generated in a motility assay slows actin sliding velocities.
To further test this hypothesis we examine the effects of the small molecule inhibitor blebbistatin on both V and the rate of actin filament breaking. Blebbistatin is known to slow both V and the weak-to-strong binding transition without significantly influencing actin-myosin detachment kinetics (21, 22), and it is often assumed that blebbistatin inhibits V by sequestering a non-cycling pool of myosin heads (23) that resists actin filament movement through weak actin-myosin interactions (Fig. 1A). If this were the primary mechanism by which blebbistatin slows V, we would expect blebbistatin to increase the rate of actin filament breaking. However, here we show that like with Pi, 50 μM blebbistatin slows both V and the rate at which actin filaments break, suggesting that blebbistatin slows V by decreasing force transmission in a motility assay. These results provide additional support for the hypothesis that, in addition to actin-myosin detachment kinetics, the driving force of actin-myosin binding influences actin sliding velocities. This novel perspective on muscle contraction provides potential new insights into how muscle mechanics are altered by small molecule inhibitors such as BDM and BTS (24-27), by mutations associated with muscle myopathies (28), and by regulatory proteins such as troponin and tropomyosin (29).
Experimental Procedures
Protein Purification
Fast skeletal muscle myosin was prepared from rabbit psoas as previously described (30) and stored in glycerol at -20° C. Actin was isolated from rabbit psoas (31) and stored on ice at 4° C. For in vitro motility assays, actin was incubated with tetramethylrhodamine isothiocyanate (TRITC) phalloidin overnight.
Buffers
For actin-based motility assays, myosin buffer (300 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT), actin buffer (25 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT), motility buffer (25 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, 0.03 – 1 mM ATP, 0.5% methylcellulose, 5.8 mg/ml glucose, 0.033 mg/ml glucose oxidase, and 0.045 mg/ml catalase) were prepared. For blebbistatin experiments, we added 50 μM blebbistatin (Sigma-Aldrich, St. Louis, MO) to the motility buffer. Phosphate buffers contained a 3:2 ratio of K2HPO4 and KH2PO4 (Sigma-Aldrich, St. Louis, MO) as previously described (32). In Pi experiments ionic strength was maintained by adjusting KCl according to Fabiato and Fabiato (33).
Activity Assays
The velocity of fluorescently labeled actin filaments sliding over a bed of myosin molecules was measured using an in vitro motility assay (34, 35) at 25° C. Flow cells were prepared by attaching a nitrocellulose-coated coverslip to a microscope slide with shim spacers. Flow cells for the motility assay were treated as follows: 2 × 50 μL of 100 μg·mL-1 myosin with a 1 min. incubation period, 2 × 50 μL of 0.5 mg·mL-1 BSA, 2 × 50 μL of 10 μM actin with a 1 min. incubation period, 2 × 50 μL of actin buffer, and 1 × 80 μL of motility buffer. Actin filaments bound to the coverslip surface were visualized prior to addition of motility buffer so that the time elapsed between adding motility buffer and recording actin filament movement was less than 10 sec. For control experiments, actin was not immobilized specifically. The actin filaments in the control experiments were free to thermally fluctuate. For blebbistatin experiments, light exposure to blebbistatin buffer was minimized by making buffers, storing buffers, and performing experiments with minimal light (36).
Motility assays were performed using a Nikon TE2000 epifluorescence microscope with fluorescence images digitally acquired at 7.3 frames per second with a Roper Cascade 512B (Princeton Instruments, Trenton, NJ) CCD camera. We prepared at least three flow cells for each experimental condition, and for each flow cell we recorded one three-minute image sequence. For each image sequence, we analyzed actin movement using Simple PCI tracking software (Compix, Sewickley, PA) to obtain actin-sliding velocities, V. Objects were defined by applying an exclusionary area threshold to minimize background noise. Intersect filters were applied to exclude intersecting filaments. The velocities of the moving actin filaments were plotted as a histogram and fitted to a Gaussian distribution. The average velocity, V, for the field was taken from the mean of the Gaussian fit. All errors reported are standard deviation (SD).
Analysis of Actin-filament Breaking
Actin filament breaking was analyzed using Image J (NIH, Bethesda, MD). For each actin filament, the filament length and the time, τbreak, it took to break were recorded. τbreak was calculated relative to the frame of origin for a filament, defined as i) the initial frame of the image sequence for filaments fully in the field of view, ii) the frame at which the filament entered the field of view; or iii) the frame at which the filament broke from a longer filament. τbreak values for all actin filaments within a given length range were plotted in a histogram and fitted to a single exponential to determine the lifetime (τbreak) and break rate (τbreak−1) using Origin (Origin Lab Corporation, North Hampton, MA). Approximately 50 – 100 filaments were analyzed per condition. Non-moving filaments were omitted from our analysis. Actin filaments appeared to break through bending rather than through a tensile pull consistent with previous studies (37).
Results
According to conventional models of muscle contraction (V = d·τon−1), Pi and blebbistatin (both known effectors of weak-to-strong binding kinetics) slow V by altering chemical forces that resist myosin-based actin motility (Fig. 1A). Specifically, Pi has been proposed (18,20,32) to increase τon by competing with ATP for myosin binding (Fig. 1A), whereas blebbistatin is assumed to introduce resistive forces through weak interactions between actin and blebbistatin-sequestered, non-cycling myosin heads (Fig. 1A). An alternative hypothesis is that both Pi and blebbistatin slow V by diminishing the driving forces of strong actin-myosin binding (Fig. 1A). Here, to determine if V is slowed by altered resistive forces or altered driving forces (Fig. 1A), we measure the effects of 30 mM Pi and 50 μM blebbistatin on the rate (τbreak−1) of actin filament breaking during a motility assay. An increase in τbreak−1 is indicative of increased resistance to movement whereas a decrease in τbreak−1 suggests a decrease in driving force (Fig. 1A).
Actin breaking is myosin-dependent
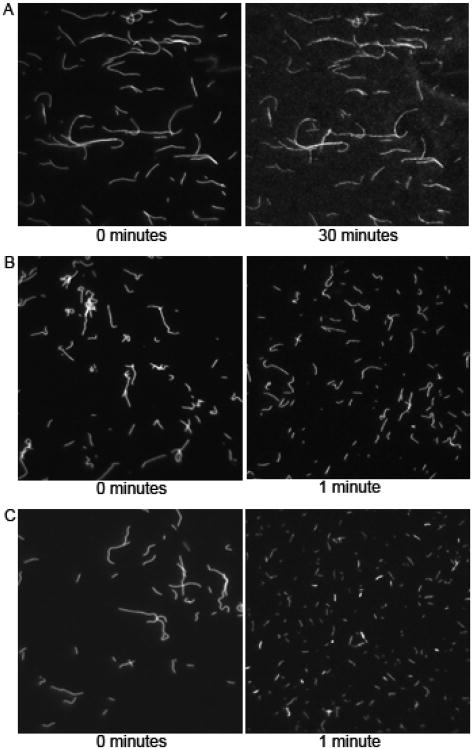
In the absence of myosin, actin filaments suspended over a nitrocellulose-coated coverslip in motility buffer (1 mM ATP) did not noticeably break over a period of 30 minutes (Fig. 2A), whereas during a myosin-based actin motility assay (1mM ATP) we observed significant actin filament fragmentation in less than one minute (Fig. 2B) showing that breaking is in fact myosin based. Actin filaments broke even more over this time period when [ATP] was decreased to 30 μM (Fig. 2C). We observed little or no [ATP]-dependence of actin filament fragmentation in the absence of myosin (data not shown). These data indicate that actin filament breaking observed in a motility assay is caused by forces generated between myosin heads.
Figure 2.
Actin breaking is myosin dependent. (A) Images of fluorescently labeled actin filaments in a flow cell with no myosin and 30 μM ATP obtained at 0 (left) and 30 minutes (right) after addition of actin. (B) Images of actin filaments during a myosin-based in vitro motility assay performed at 1 mM ATP obtained at 0 (left) and 1 minute (right) after addition of motility buffer. (C) Images of actin filaments during a myosin-based in vitro motility assay performed at 0.03 mM ATP obtained at 0 (left) and 1 (right) minute following addition of motility buffer.
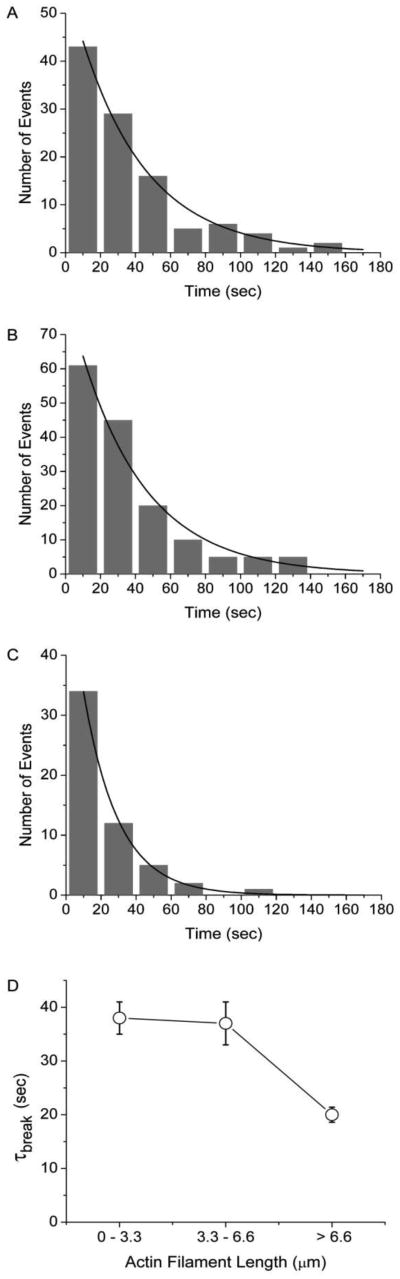
To determine the rate at which actin filaments break, we timed breaking events for individual actin filaments through a frame-by-frame analysis of actin filaments moving in a motility assay. For each actin filament we counted the number of frames elapsed before that filament broke, obtaining a time to break, τbreak, for that filament. We plotted these times in a histogram and fitted the data to a single exponential to obtain a mean actin filament lifetime (τbreak), or break rate (τbreak−1). To test and control for potential actin-length dependences of τbreak, we grouped individual break times (τbreak) by actin filament length. Figure 3 shows histograms of τbreak values obtained at 30 μM ATP for actin filaments with lengths < 3.3 μm (Fig. 3A), 3.3 – 6.6 μm (Fig. 3B), and > 6.6 μm (Fig. 3C). The average time to break (τbreak) for each range of actin filament length was obtained by fitting each histogram to a single exponential and was plotted in Fig. 3D. These data show a small actin length-dependence of τbreak for filaments between 0 and 6.6 μm, but a significant decrease in τbreak values for filaments > 6.6 μm, demonstrating that actin length is a variable that must be controlled when assessing Pi-dependent changes in τbreak.
Figure 3.
Length dependence of the rate of actin filament breaking. (A) Histogram of actin filament break times, τbreak, measured during an in motility assay performed at 30 μM ATP for actin filaments with lengths (A) between 0 and 3.3 μm (n = 106), (B) between 3.3 and 6.6 μm (n = 151), and (C) greater than 6.6 μm (n = 54). Each histogram is fitted to a single exponential (solid line) to obtain the actin filament lifetime (τbreak). (D) Actin lifetimes for 0 – 3.3 μm, 3.3 – 6.6 μm, and > 6.6 μm length actin filaments are plotted.
Relatively high interhead forces are generated in an in vitro motility assay at low [ATP]
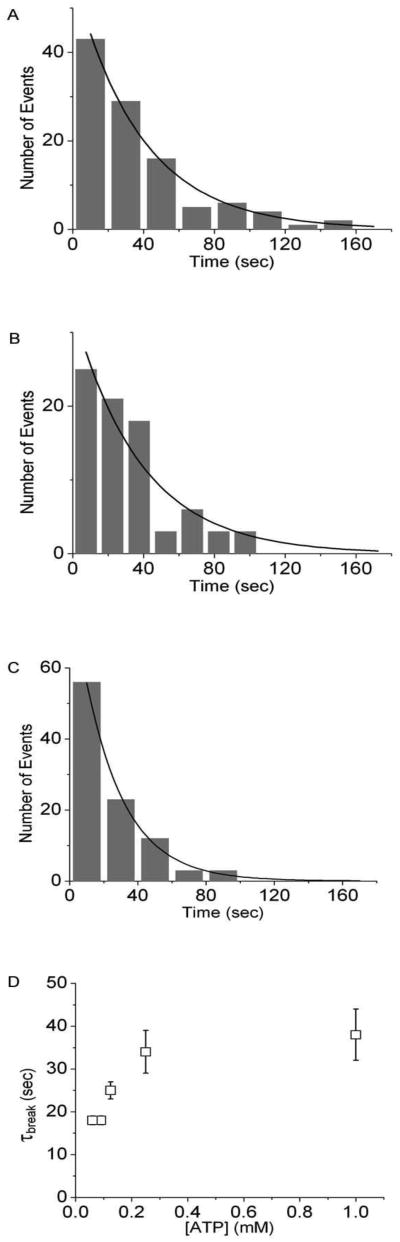
In an in vitro motility assay, actin sliding velocities measured at [ATP] > ∼100 μM exceed velocities predicted by a Michaelis-Menten analysis of velocities obtained at [ATP] < 100 μM (13,17,40). Interestingly, above this same critical ATP concentration, V becomes independent of Pi (Fig. 1A) (17). Both observations are consistent with a model in which at low [ATP], high forces are transmitted along actin filaments, whereas at high [ATP] less force is transmitted due to a smaller a, resulting either from a larger distance between heads at high [ATP] or from the mechanical acceleration of ADP release kinetics (15,17). To further test this hypothesis, we determined the [ATP]-dependence of the rate of actin filament breaking. Figures 4A – 4C show histograms of actin break times (τbreak) obtained at 1.0, 0.125, and 0.06 mM ATP. The ATP-dependence of the average actin filament lifetimes (τbreak) obtained from single exponential fits to the histograms shows a transition from slow break rates at high [ATP] (> 100 μM) to more rapid break rates at low [ATP] (< 100 μM), demonstrating that at high [ATP] force transmission, a, decreases. The data in Fig. 4 were acquired for actin filaments with lengths between 3.3 and 6.6 μm, and a similar [ATP]-dependence was observed for actin filament lengths > 6.6 μm (data not shown).
Figure 4.
ATP-dependence of the rate of actin filament breaking. Actin filament break times, τbreak, were measured at (A) 1.0 mM (n = 151), (B) 0.125 mM (n = 80), and (C) 0.06 mM (n = 76) ATP and plotted in histograms. Each histogram was fitted with a single exponential (solid line) to obtain actin filament lifetime (τbreak). (D) Actin filament lifetimes (τbreak) for 1.0, 0.250, 0.125, 0.06 and 0.03 mM ATP are plotted.
Pi and blebbistatin decrease force transmission along actin filaments
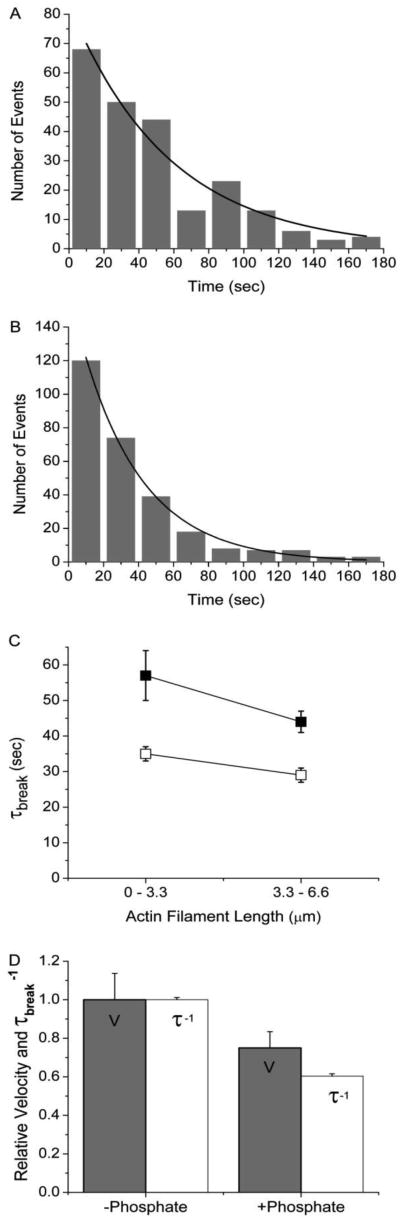
Hooft et al. argue that inhibition of V by Pi results from a decrease in the chemical forces that drive myosin-based actin motility (Fig. 1A). This model predicts that inhibition of V by Pi will be associated with a decrease in forces generated along actin filaments in an in vitro motility assay, resulting in a decrease in the rate at which actin filaments break. In contrast, a model in which Pi inhibits V by binding actin-bound myosin rigor heads predicts that the inhibition of V by Pi will increase forces exerted on actin (Fig. 1A), increasing the rate at which actin filaments break (20). To test these model predictions, we determined the effects of Pi on the rate of actin filament breaking (τbreak−1) in a motility assay performed at 30 μM ATP. Figure 5 shows representative histograms of τbreak values obtained both in the presence (Fig. 5A) and absence (Fig. 5B) of 30 mM Pi for actin filaments < 3.3 μm. These and other histograms were fitted to single exponentials to obtain rates (τbreak−1) of actin filament breaking (Fig. 5C). Figure 5D shows that the inhibition of V by Pi correlates with a decrease in the rate of actin filament breaking, indicating that Pi inhibits V by decreasing the driving forces of motility. These data support our hypothesis that driving forces associated with weak-to-strong binding kinetics influence actin sliding velocities.
Figure 5.
[Pi] dependence of actin sliding velocities and the rate of actin filament breaking. (A) Actin filament break times, τbreak, were measured in the presence of 30 mM Pi at 30 μM ATP for filaments < 3.3 μm and plotted in a histogram (n = 224). The data were fitted to a single exponential (solid line) to obtain the actin filament lifetime (τbreak). (B) Actin filament break times, τbreak, were measured in the presence 30 μM ATP with no added Pi for filaments < 3.3 μm and plotted in a histogram (n = 279). The data were fitted with a single exponential (solid line) to obtain the average time to break (τbreak). (C) Values for the actin filament lifetime (τbreak) obtained with (dark squares) and without (white squares) added Pi at 30 μM ATP are plotted both for actin filament lengths < 3.3 μm and for actin filament lengths between 3.3 and 6.6 μm. (D) For all actin filament lengths, mean actin sliding velocities (grey bars), normalized to 1.8 μm·sec−1, and the rates of actin breaking (white bars), normalized to 0.029 sec−1, obtained in the presence and absence of 30 mM Pi at 30 μM ATP are plotted, showing that the inhibition of V by Pi corresponds to a decrease in interhead forces.
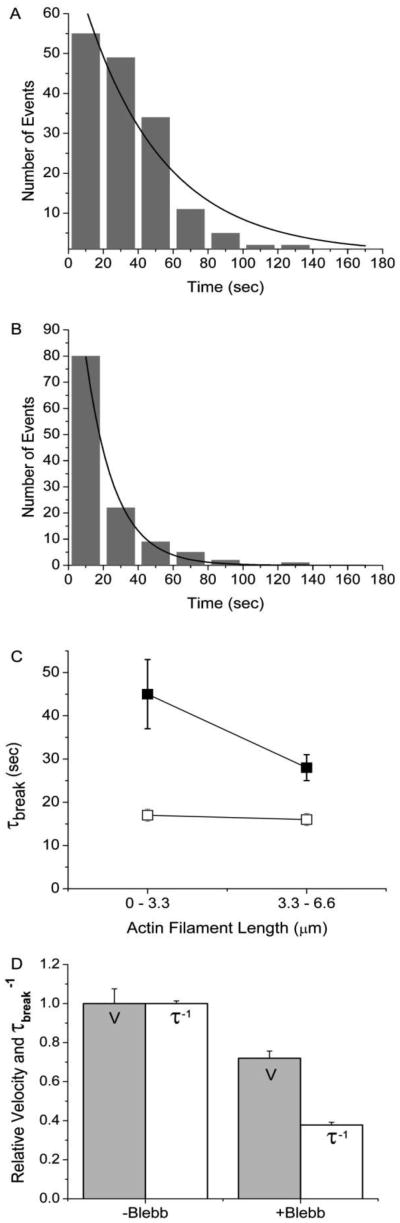
To further test this hypothesis, we studied the effects of blebbistatin on the rate of actin filament breaking. Blebbistatin is a small molecule inhibitor of actin-myosin ATPase activity, actin sliding velocities, and isometric muscle force (22,34). The kinetic basis for this effect is inhibition of myosin's force-generating, weak-to-strong binding transition (22). According to a model in which blebbistatin slows V by increasing resistive forces, inhibition of V by blebbistatin should increase the rate of actin filament breaking (Fig. 1A). In contrast, according to a model in which blebbistatin slows V by decreasing driving forces, inhibition of V by blebbistatin should decrease the rate of actin filament breaking (Fig. 1A). To test these models, we determined the effects of blebbistatin on both V and the rate (τbreak−1) of actin filament breaking using a motility assay performed at 30 μM ATP. Figure 6 shows histograms of τbreak values obtained in the presence (Fig. 6A) and absence (Fig. 6B) of 50 μM blebbistatin. High concentrations (50 μM) of blebbistatin relative to those used in previous studies (45) were needed to inhibit V presumably because at the low [ATP] used in this study, ATPase activity becomes limited by ATP-induced actin-myosin detachment kinetics and thus inhibiting weak-to-strong binding is not rate limiting and thus its inhibition has less of an effect on V. Actin filament lifetimes (τbreak) were obtained from single exponential fits to each histogram and plotted in Fig. 6C. Figure 6D shows that blebbistatin decreased both V and τbreak−1, implying that blebbistatin inhibits V by decreasing the driving force of the weak-to-strong binding transition.
Figure 6.
Blebbistatin dependence of actin sliding velocities and the rate of actin filament breaking. (A) Actin filament break times, τbreak, were measured in the presence (A, n = 158) and absence (B, n = 119) of 50 μm blebbistatin at 30 μM ATP for filaments < 3.3 μm and plotted in a histogram. These data were fitted to a single exponential (solid line) to obtain an actin filament lifetime, τbreak. (C) Actin filament lifetimes obtained at 30 μm ATP both with (dark squares) and without (white squares) 50 μM blebbistatin for actin filament lengths < 3.3 μm and between 3.3 and 6.6 μm. (D) For all actin filament lengths, mean actin sliding velocities (grey bars), normalized to 1.0 μm·sec−1, and rates of actin filament breaking (white bars), normalized to 0.059 sec−1, obtained in the presence and absence of 50 μM blebbistatin at 30 μM ATP are plotted, showing the inhibition of V by blebbistatin correlates with a decrease in interhead forces.
Inhibition of V with Blebbistatin cannot be recovered by increasing myosin density
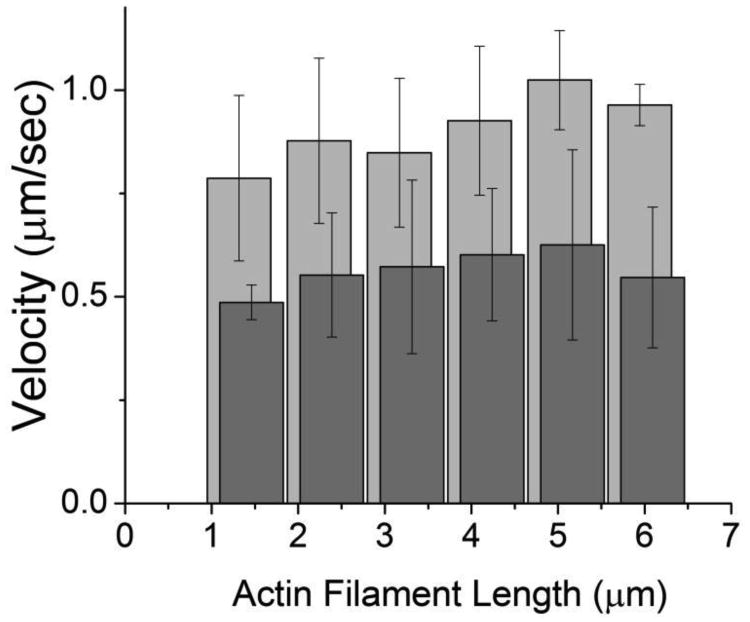
One alternative interpretation for the effects of blebbistatin on V and τbreak−1 in Fig. 6 is that blebbistatin sequesters a non-cycling pool of myosin heads, leaving an insufficient number of myosin heads available for maximal velocity. To test this hypothesis, we determined whether inhibition of V by blebbistatin can be reversed by increasing the total number of myosin heads available to bind an actin filament. We measured the actin length dependence of V at 30 μM ATP both in the presence and absence of 50 μM blebbistatin and calculated the number of myosin heads available to bind an actin filament of a given length as previously described (16). Figure 7 shows that inhibition of V by blebbistatin is not reversed by increasing the number of myosin heads (number of myosin heads can be calculated based off of actin filament length) available to bind an actin filament, ruling out sequestering of non-cycling crossbridges as the primary mechanism by which blebbistatin inhibits V.
Figure 7.
The effects of increasing myosin density on blebbistatin-inhibited V. Actin filament velocities (V) are plotted as a function of actin filament length (n = 120), where longer actin filaments interact with a higher number of myosin heads (N). N can be calculated from actin filament length as previously described (16). Points are shown in the absence (light gray) and presence (dark gray) of 50 μM blebbistatin. These data show that the inhibition of V by blebbistatin cannot be reversed by increasing myosin density.
Discussion
During unloaded muscle shortening and in vitro myosin-based actin sliding, myosin heads generate force in the direction of actin movement (Fig. 1A) against a population of actin-bound myosin that resists actin movement (41). The conventional view of actin sliding (12,41) is that the driving force of a myosin head is well-defined by a molecular mechanical constant, d, and that the sole biochemical determinant of actin sliding velocities is the rate (τon−1) at which resistive heads detach from actin (V = d·τon−1).
Hooft et al. showed that, although τon influences actin sliding velocities, weak-to-strong binding kinetics also contribute to V(17). Specifically we showed that at low [ATP], actin sliding velocities decrease with added Pi, indicating a link between V, actin-myosin affinity, and the force generating capacity of myosin (13,18,42,43). This interpretation differs from the conventional view that Pi slows V by competing with ATP for myosin binding thereby increasing τon. Amrute-Nayak et al. (20) correctly argue that the two models can be tested by determining the effects of Pi on the forces exerted along actin by myosin heads in a motility assay, where changes in actin forces can be estimated from changes in the rate at which actin filaments break. In the present study, we further validate this assertion, showing in Fig. 2 that actin breaking in a motility assay is associated with the actin-myosin ATPase reaction.
Because the visible pool of actin filaments changes over time in a motility assay, it is important to measure filament breaking events rather than before-and-after snapshots of filaments when determining the rate of filament breaking. Moreover, because the starting length of actin filaments in a motility assay can be variable, the length-dependence of the rate of actin filament breaking demonstrated in Fig. 3 implies that actin filament lengths must be controlled when measuring the rate of filament breaking. We believe the above factors contribute to the discrepancies between our results and those reported by Amrute-Nayak et al.
Whereas Amrute-Nayak et al. inferred an increase in the rate of filament breaking upon addition of Pi, in Fig. 5 we show that τbreak−1 decreases significantly upon addition of 30 mM Pi. This result implies that, as argued by Hooft et al., Pi slows V by decreasing the driving force of weak-to-strong binding, τoff. These studies were performed at low [ATP] because the weak-to-strong binding transition is made reversible by Pi only under the relatively high forces (13,18,42) generated in a motility assay at low [ATP] (15,17).
To further test the hypothesis that V is influenced by τoff, we studied the effects of blebbistatin on V and τbreak−1. Blebbistatin is a small molecule inhibitor of actin-myosin ATPase activity, which inhibits isometric muscle force, muscle shortening velocities, and actin sliding velocities in vitro (22,36). The kinetic mechanism for its inhibitory effect is that blebbistatin slows the rate of weak-to-strong binding (21,22,44) with no significant effect on ADP release, ATP binding, or τon. These kinetic studies suggest that blebbistatin inhibits V through its effects on the weak-to-strong binding kinetics. Figure 6 shows that the decrease in actin sliding velocities induced by blebbistatin corresponds to a decrease in the rate of actin filament breaking, consistent with blebbistatin inhibiting V through a decrease in the forces transmitted along an actin filament with the weak-to-strong binding transition.
The data reported herein and in a previous paper (17) support a model in which actin sliding velocities depend not only on detachment kinetics but also on attachment kinetics and energetics. Interestingly, both blebbistatin and Pi inhibit actin-myosin affinity, isometric muscle force, actin sliding velocities, and driving forces generated during an in vitro motility assay. Our observations that Pi and blebbistatin similarly decrease both the forces transmitted along an actin filament and actin sliding velocities are consistent with a model in which Pi and blebbistatin decrease the force transmission term, a, in Eq. 1.
We describe a simple expression for a as follows. First, we assume that inefficient force transmission results when the mechanical step of a myosin head pulls out “slack” in a flexible actin filament rather than straining an adjacent myosin head, and we estimate this slack length by treating an actin filament as a worm-like chain. For simplicity, we assume that the worm-like chain stiffness of actin is negligible relative to that of actin-bound myosin heads. (Incorporating worm-like-chain stiffness into a model of muscle shortening results in attachment kinetics contributing directly to actin sliding velocities – an analysis developed elsewhere). According to a worm-like-chain model (47), the average slack, s, in the average length, L, of an actin filament spanning two myosin heads is
where P is the persistence length of actin. The efficiency with which one myosin head can transmit forces a distance, L, along an actin filament is then a = (d – s)/d.
The effects of attachment kinetics on V can be accounted for by considering that the average distance, L, between two strongly bound heads on an actin filament is simply r/ρ, where ρ is the linear myosin head density along an actin filament and r = τon/(τon + τoff) is the fraction of those myosin heads that are strongly bound to actin, or
| (Eq. 2) |
Thus τoff can influence V (Eq. 1) via the effects of duty ratio on force transmission, a. Equation 2 also provides a qualitative description of the internal forces generated during unloaded shortening. When the distance between myosin heads is large (i.e., ρ is small), the slack, s, exceeds myosin's step size, which means no force is transmitted, a = 0, and no internal forces are generated. When the distance between myosin heads is small (i.e., ρ is large), a approaches one and maximal internal forces are generated.
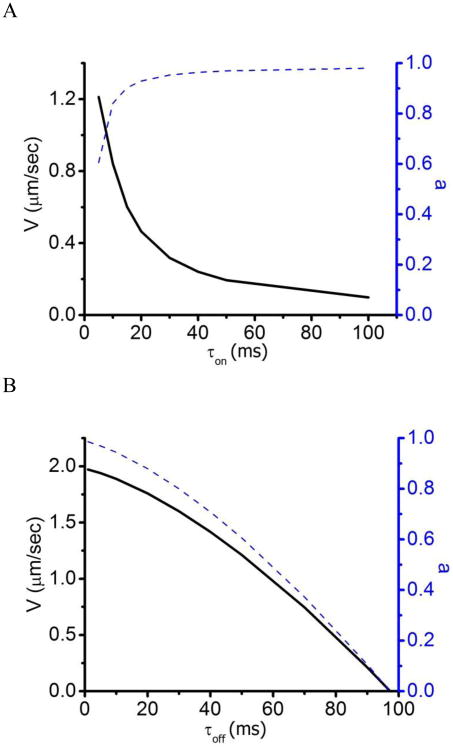
This model is consistent with the data reported herein. In Fig. 8, Eq. 1 (substituting a from Eq. 2) and Eq. 2 are plotted at different τon (Fig. 8A) and τoff (Fig. 8B) values. Figure 8A shows that when V is inhibited through slowing actin-myosin detachment kinetics (i.e., increasing τon) a increases, resulting in an increase in the internal forces generated. This is consistent with the data in Fig. 4. In contrast, when V is inhibited through slowing actin-myosin attachment kinetics (i.e., increasing τoff) a decreases, resulting in a decrease in internal forces generated. This is consistent with the data in Figs. 5 and 6. A more quantitative analysis will require developing a formal connection between force transmission, force generation, and the effects of force on the actin filament breaking rate, which is beyond the scope of this paper.
Figure 8.
The τoff and τon dependence of Equations 1 and 2. (A) Plots of both V (Eq. 1, substituting a from Eq. 2, solid line) and a (Eq. 2, dashed line) at different τon values show that increasing τon inhibits V and increases internal forces, consistent with the data in Fig. 4 [τoff = 50 ms; P = 2 mm; ρ = 1/(36 nm)]. (B) Plots of both V (Eq. 1, substituting a from Eq. 2, solid line) and a (Eq. 2, dashed line) at different τoff values show that increasing τoff inhibits V and decreases internal forces, consistent with the data in Figs. 5 and 6 [τon = 5 ms; P = 2 mm; ρ = 1/(36 nm)].
The above analysis suggests that, analogous to the effects of decreasing myosin density on V, blebbistatin and Pi inhibit V by decreasing the number of strongly bound myosin heads. It is also possible that Pi and blebbistatin inhibit V in the loaded (low [ATP]) motility assay by decreasing the average myosin step size, d, much like Pi inhibits the average force generated per myosin head in isometric muscle (46). Many other effectors of actin-myosin ATPase activity exhibit similar inhibition of both actin sliding velocities and isometric muscle force. The data presented above provide a potentially new perspective on the effects of many modulators of muscle contraction, including the small molecule inhibitors BDM and BTS (24-27), mutations associated with familial hypertrophic cardiomyopathy (28), and thin filament regulatory proteins troponin and tropomyosin (29).
Acknowledgments
We thank Kevin Facemyer for his helpful suggestions and Olivia John for purifying proteins. This study was funded by NIH 1R01HL090938.
This research was funded by NIH Grant 1R01HL090938
Abbreviations and Textual Footnotes
- Pi
Inorganic Phosphate
References
- 1.Lymn RW, Taylor EW. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 2.Goldman YE. Annu Rev Physiol. 1987;49:637–654. doi: 10.1146/annurev.ph.49.030187.003225. [DOI] [PubMed] [Google Scholar]
- 3.Cooke R. Physiol Rev. 1997;77:671–697. doi: 10.1152/physrev.1997.77.3.671. [DOI] [PubMed] [Google Scholar]
- 4.Sellers JR. Myosins. 2nd. Oxford University Press; Oxford UK: 1999. [Google Scholar]
- 5.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 6.Finer JT, Simmons RM, Spudich JA. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 7.Guilford WH, Dupuis DE, Kennedy G, Patlak JB, Warshaw DM. Biophys J. 1997;72:1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker JE, Brust-Mascher I, Ramachandran S, LaConte LE, Thomas DD. Proc Natl Acad Sci U S A. 1998;95:2944–2949. doi: 10.1073/pnas.95.6.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reedy MK, Holmes KC, Tregear RT. Nature. 1965;207:1276–1280. doi: 10.1038/2071276a0. [DOI] [PubMed] [Google Scholar]
- 10.Huxley HE. Science. 1969;164:1356–1365. [Google Scholar]
- 11.Eisenberg E, Hill TL. Science. 1985;227:999–1006. doi: 10.1126/science.3156404. [DOI] [PubMed] [Google Scholar]
- 12.Spudich JA. Nature. 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 13.Baker JE, Brosseau C, Joel PB, Warshaw DM. Biophys J. 2002;82:2134–2147. doi: 10.1016/S0006-3495(02)75560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaya M, Higuchi H. Science. 2010;3219:686–689. doi: 10.1126/science.1191484. [DOI] [PubMed] [Google Scholar]
- 15.Jackson DRJ, Baker JE. Phys Chem Chem Phys. 2009;11:4808–4814. doi: 10.1039/b900998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris DE, Warshaw DM. J Biol Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]
- 17.Hooft AM, Maki EJ, Cox KK, Baker JE. Biochemistry. 2007;46:3513–3520. doi: 10.1021/bi0614840. [DOI] [PubMed] [Google Scholar]
- 18.Cooke R, Pate E. Biophys J. 1985;48:789–798. doi: 10.1016/S0006-3495(85)83837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pate E, Cooke R. J Muscle Res Cell Motil. 1989;10:181–196. doi: 10.1007/BF01739809. [DOI] [PubMed] [Google Scholar]
- 20.Amrute-Nayak M, Antognozzi M, Scholz T, Kojima H, Brenner B. J Biol Chem. 2008;283:3773–3781. doi: 10.1074/jbc.M706779200. [DOI] [PubMed] [Google Scholar]
- 21.Limouze J, Straight AF, Mitchison T, Sellers JR. J Muscle Res Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 22.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. J Biol Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 23.Farman GP, Tachampa K, Mateja R, Cazorla O, Lacampagne A, de Tombe PP. Pflügers Archiv European J of Physiol. 2008;4552:995–1005. doi: 10.1007/s00424-007-0375-3. [DOI] [PubMed] [Google Scholar]
- 24.Shaw MA, Ostap EM, Goldman YE. Biochemistry. 2003;42:6128–6135. doi: 10.1021/bi026964f. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann C, Wray J, Travers F, Barman T. Biochemistry. 1992;31:12227–12232. doi: 10.1021/bi00163a036. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Pate E, Baker AJ, Cooke R. Biophys J. 1995;69:994–999. doi: 10.1016/S0006-3495(95)79974-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regnier M, Chase PB, Martyn DA. J Muscle Res Cell Motil. 1999;20:425–432. doi: 10.1023/a:1005594001334. [DOI] [PubMed] [Google Scholar]
- 28.Debold EP, Schmitt JP, Moore JR, Patlak JB, Beck SE, Seidman JG, Seidman C, Warshaw DM. Am J Physiol Heart Circ Physiol. 2007 doi: 10.1152/ajpheart.00128.2007. [DOI] [PubMed] [Google Scholar]
- 29.Lehrer SS, Geeves MA. J Mol Biol. 1998;277:1081–1089. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]
- 30.Warshaw DM, Desrosiers JM, Work SS, Trybus KM. J Cell Biol. 1990;111:453–463. doi: 10.1083/jcb.111.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardee JD, Spudich JA. Methods Enzymol. 1982;85 Pt B:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 32.Warshaw DM, Desrosiers JM, Work SS, Trybus KM. J Biol Chem. 1991;266:24339–24343. [PubMed] [Google Scholar]
- 33.Fabiato A, Fabiato F. J Physiol (Paris) 1979;75:463–505. [PubMed] [Google Scholar]
- 34.Kron SJ, Spudich JA. Proc Natl Acad Sci U S A. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harada Y, Noguchi A, Kishino A, Yanagida T. Nature. 1987;326:805–808. doi: 10.1038/326805a0. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto T, Limouze J, Combs CA, Straight AF, Sellers JR. Biochemistry. 2005;44:584–588. doi: 10.1021/bi0483357. [DOI] [PubMed] [Google Scholar]
- 37.Yuri Tsuda HY‡, Akihiko Ishijima†, Toshio Yanagida†‡§. Proc Natl Acad Sci U S A. 1996;93:12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T. J Mol Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- 39.Uyeda TQ, Kron SJ, Spudich JA. J Mol Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 40.Baker JE, Brosseau C, Fagnant P, Warshaw DM. J Biol Chem. 2003;278:28533–28539. doi: 10.1074/jbc.M303583200. [DOI] [PubMed] [Google Scholar]
- 41.Huxley AF. Prog Biophys. 1957;7:255–315. [PubMed] [Google Scholar]
- 42.Dantzig JA, Goldman YE, Millar NC, Lacktis J, Homsher E. J Physiol. 1992;451:247–278. doi: 10.1113/jphysiol.1992.sp019163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takagi Y, Homsher EE, Goldman YE, Shuman H. Biophys J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramamurthy B, Yengo CM, Straight AF, Mitchison TJ, Sweeney HL. Biochemistry. 2004;43:14832–14839. doi: 10.1021/bi0490284. [DOI] [PubMed] [Google Scholar]
- 45.Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A, Sellers JR. J Biol Chem. 2004;279(34):35557–63. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 46.Baker JE, LaConte EW, Brust-Mascher I, Thomas D. Biophysical Journal. 1999;77(5):2657–2665. doi: 10.1016/S0006-3495(99)77100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doi M, Edwards SF. The Theory of Polymer Dynamics. Oxford University Inc; NY: p. 1986. [Google Scholar]