Abstract
Stem-cell-based therapies hold promise and potential to address many unmet clinical needs. Cell tracking with modern imaging modalities offers insight into the underlying biological process of the stem-cell-based therapies, with the goal to reveal cell survival, migration, homing, engraftment, differentiation, and functions. Adaptability, sensitivity, resolution, and non-invasiveness have contributed to the longstanding use of optical imaging for stem cell tracking and analysis. To identify transplanted stem cells from the host tissue, optically active probes are usually used to label stem cells before the administration. In comparison to the traditional fluorescent probes like fluorescent proteins and dyes, nanoparticle-based probes are advantageous in terms of the photo-stabilities and minimal changes to the cell phenotype. The main focus here is to overview the recent development of optically active nanoparticles for stem cells tracking. The related optical imaging modalities include fluorescence imaging, photoacoustic imaging, Raman and surface enhanced Raman spectroscopy imaging.
Keywords: Stem cell therapy, optical imaging, nanoparticles, fluorescence imaging, photoacoustic imaging, Raman and surface enhanced Raman spectroscopy imaging
Introduction
Stem-cell-based cell therapy holds great promise for patients living with serious and currently incurable diseases including cancer, Alzheimer disease, Parkinson disease, diabetes and etc [1-6]. These potentials of stem cells rely on their remarkable properties of self-renewal and differentiation into diverse specialized cells, offering hope for the regeneration of tissues/organs for replacing diseased and damaged areas in the body [7,8]. Since the first bone marrow transplant was performed to treat two siblings with severe combined immunodeficiency in 1968, scientists and clinicians have put tremendous effects to develop new stem-cell-based therapeutics. Promising approaches include bone marrow derived mesenchymal stem cell for graft-versus-host disease [9]. Currently, ~4000 clinical trials around the world involve some form of stem cell therapy, including therapies for cancer, cardiac disease, stroke (American’s top 3 causes of death), diabetes, bone repair and etc (Table 1) [10].
Table 1.
Clinical trials related with major stem-cell-based cell therapies (Results from www.clinicaltrials.gov)
| Types of diseases | Number of trials |
|---|---|
| Cancer | 3007 |
| Graft-versus-host disease | 631 |
| Cardiac diseases | 472 |
| Diabetes | 101 |
| Bone repair | 90 |
| Stroke | 66 |
While preclinical results are promising, few treatments have been translated to humans due to conflicting results [11]. In addition to the limitation of preclinical models (i.e. the different behavior of stem cells between preclinical models and human), it is in part due to the lack of a comprehensive understanding of the fate, distribution, and the function of transplanted stem cells in the local microenvironment [12]. Traditionally, transplanted stem cells are studied through the histological analysis, which is largely invasive at pre-determined time points after transplant [13]. Thus non-invasive imaging methods are highly needed to monitor transplanted stem cells qualitatively and quantitatively. This will facilitate the prediction of treatment efficacy, and reveal optimal transplantation conditions, allowing the cell dosage, delivery route, and timing of transplantations to be determined [14].
In comparison to other modalities like magnetic resonance imaging, positron emission tomography and computed tomography, optical imaging possesses distinctive advantages including low cost, easy accessibility, as well as high spatial and temporal sensitivity. Furthermore, the molecular fingerprints derived from the spectroscopy that were collected during the imaging process are promising contributions to reveal the viability and differentiation of transplanted stem cells. For example, Raman spectroscopy contains information of vibrational, rotational and other low-frequency modes of molecules, which make it a powerful technique for monitoring cellular processes (e.g. apoptosis or necrosis) [15,16]. To identify administered cells from the host tissue, contrast agents are usually required to label the cells. Current contrast agents for optical imaging include endogenous biomolecules, fluorescent proteins, organic dyes, and fluorescent lanthanide chelates, all of which suffer from photo-bleaching effects, as well as chemical and metabolic degradation in vivo [17]. These shortcomings hinder the efforts to track transplanted stem cell in vivo. More recently, the development of optically active nanoparticles (NPs) over the past two decades provides hope in addressing this challenge [18-20]. One promising agent is the semiconductor nanocrystals or quantum dots (QDs), which exhibits non-bleachable fluorescence with controllable wavelength ranging from visible to near infrared [21-24].
This review summarizes recent developments in the field of stem cell tracking with optically active NPs (Figure 1). We will start with the fluorescence imaging and photoacoustic imaging, and then explore the application of Raman and surface enhanced Raman spectroscopy (SERS) for stem cells tracking. Finally, we will discuss the strength and weakness of these three strategies aforementioned, and the challenges of NP-based contrast agents for optical tracking of stem cells.
Figure 1.
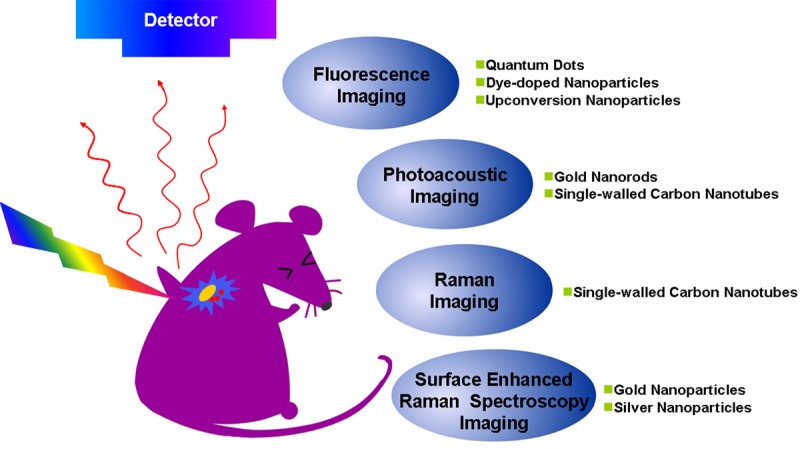
Optical imaging strategies for stem cells tracking with optically active NPs.
Stem cell tracking with nanoparticle-aided fluorescence imaging
Fluorescent imaging is attractive in terms of cost, sensitivity, and accessibility for the majority of researchers. Furthermore, it is the only modality so far that could indicate the activities and functions of transplanted cells tagged with reporter genes (genetic) [25] and endogenous [26] or exogenous fluorescent indicators (non-genetic) [27]. Normally, cells are pre-labeled with fluorescent NPs before their administration into the animals. Considering the quantum yield, brightness, and stability issues, popular fluorescent NPs for stem cell tracking include QDs, fluorescent silica NPs, and fluorescent polymer NPs.
Quantum dots
QDs are highly fluorescent semiconductor NPs with high extinction coefficients, tunable emissions, sharp emission bandwidths, and good photostability [28,29]. The tunable emission especially at the near infrared region (> ~800 nm) avoids the background signal of autofluorescence of the animal tissues (emissions are mainly at the visible region, ~300-550 nm). Good photostability allows QDs for the long-term tracking of stem cells.
Prior to the transplantation, stem cells have to be pre-labeled with QDs. There are at least six different ways to label cells including endocytosis through incubation, receptor-mediated uptake, lipid-based transduction, microinjection, electroporation, and peptide-mediated delivery [30-32]. Passive incubation and peptide-mediated delivery are the most commonly used labeling methods. Rosen and colleagues used passive incubation to load QDs into human mesenchymal stem cells (hMSCs), which was found more effective than electroporation and receptor-mediated uptake [33]. They found that QDs aggregated around the nucleus when electroporation and receptor-mediated uptake were used and the inefficiency of labeling might be correlated with the impairment of cell membrane. Furthermore, they demonstrated that QDs labeled hMSCs could be identified in histological sections of canine ventricle, and the fluorescence signals were visible for at least 8 weeks following the injection [33].
Besides passive incubation, cell-penetrating peptide is another favorable choice [34-36]. Yukawa et al introduced QDs into adipose tissue-derived stem cells (ASCs) with octa-arginine peptide (R8) [37]. When the concentration of R8 increased from 0.8 nM to 8 nM, the transduction efficiency of QDs increased from 81.4% to 90.8% [37]. In their stem cell tracking assay, QDs labeled ASCs were visualized in situ with Maestro in vivo imaging system following transplantation through the tail vein of a mouse (Figure 2) and almost all ASCs were trapped in the lung [37]. Interestingly, if QDs labeled ACSs were mixed with heparin before the tail-vein-injection into the acute liver failure mice, ASCs were accumulated not only in the lung, but also in the liver. The accumulation rate of ASCs in liver increased to about 30%, which suggested heparin was effective for increasing the accumulation of transplanted ASCs in the liver [38]. To our knowledge, it is more likely that heparin increases the ability of transplanted ASCs to bypass the pulmonary circulation which is the first capillary system that the cells will encounter after tail-vein-injection.
Figure 2.
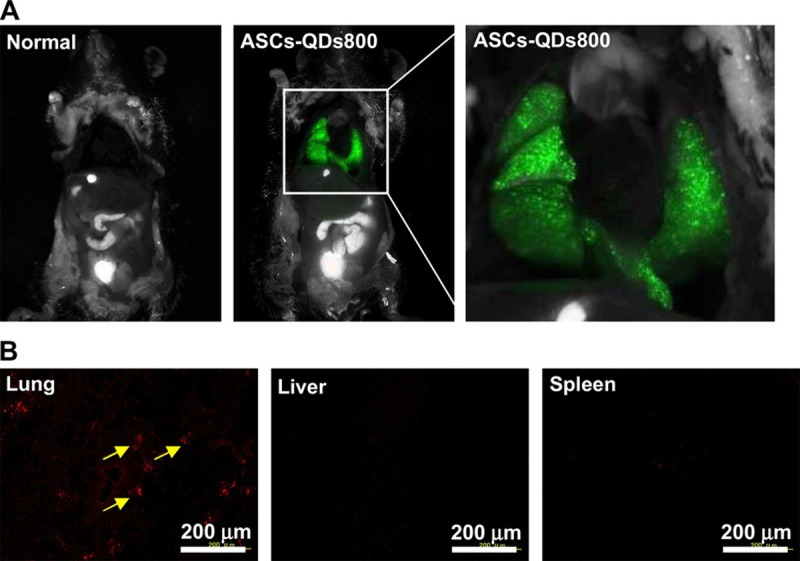
In vivo imaging of ASCs labeled with QDs after intravenous injection. A. ASCs (5.0 × 105 cells) labeled with QDs800 (0.8 nM) using R8 were transplanted through the tail vain into mouse. The images were taken 10 min after injection (excitation filter 575–605 nm, emission filter 645 nm long pass); B. The red fluorescence of QDs655 was detected in the lung only, with little or no QDs655 accumulation in the liver or spleen. Reprinted with permission from [37]. Copyright © 2010 Elsevier Ltd.
In contrast to the peptides that could improve the internalization of NPs by all types of cells, Lu et al recently developed a peptide which specifically targeted rhesus macaque embryonic stem cells (RM-ESCs) [39]. The peptide was identified by phage display and contains a sequence of APWHLSSQYSRT. Peptide was covalently conjugated on the QDs surface via N-Hydroxysuccinimide (NHS) and ethyl(dimethylaminopropyl) carbodiimide (EDC) chemistry. The final conjugates should be a promising contrast agent specifically for imaging embryonic stem cells in vivo [39].
One major concern of QDs for stem cell labeling is their cytotoxicity [40,41]. To address this issue, inert materials like silica have been used as the coating of QDs to decrease their cytotoxicity and to add extra functionalities. For example, silica coated QDs with cysteine (Cy) as capping ligands showed lower cytotoxicity to hMSCs without compromising the quantum yield [42]. When incubated with uncoated CdSe/ZnS-Cy at the concentration of 2.98 μM, the viability of hMSCs dropped down to 70%. In contrast, with the silica coated CdSe/ZnS-Cy at the same concentration, the cell viability was above 90% after 24 h incubation [42]. The confocal imaging illustrated efficient labeling and advised that no particles were located on the cell membrane or inside the nucleus.
Dye-doped nanoparticles
Despite their unique optical properties, QDs are not clinically applicable because of their potential cytotoxicity generated from the leakage of toxic metal ions [43]. As an alternative, researchers have designed biocompatible silica and polymeric NPs containing fluorescent dyes [44,45]. The biocompatible shell (e.g. polymer or silica) not only prevents organic dyes from oxidation or decomposition, but also enables the generation of strong fluorescence by concentrating the dyes.
Fluorescent silica NPs are mainly made through two approaches: sol-gel or reverse microemulsion [46,47]. One example is the fluorescent silica core-shell NPs, which were first named as Cornell dots or C-dots [48]. During the synthesis, organic dye molecules were covalently bound to a silica precursor to form adduct of the dye-rich core materials. Then silica sol-gel monomers were subsequently co-condensed with the core in specific order depending on the desired architecture to form a denser silica shell around the core [48]. C-dots possess enhanced brightness, photo-stability, biocompatibility, and versatile surface functionalities. Recently, the commercial version of C-dots, C•spec® from Hybrid Silica Technologies (HST) has already been regulated by FDA for tumor imaging in a phase-I clinical trials, which confirmed the safety of those silica-based NPs [49].
Besides C-dots, another type of silica NPs is cyanine dye-doped silica NPs (IRIS Dots), which were synthesized using a reverse microemulsion method [50]. Briefly, spherical silica NPs containing fluorescent trimethine indocyanine dyes were prepared using a water-in-oil microemulsion method with diameter 50 nm. Entrapment of dye molecules in the silica matrix stabilized the photoemission over several hours of continuous irradiation [51]. IRIS Dots did not affect the viability, proliferation and differentiation capability of hMSCs as well as C-dots [51]. More interestingly, IRIS Dots could allow the discrimination between live and early-stage apoptotic stem cells through the different surface distribution. Specifically, hMSCs were pretreated with apoptosis-inducing agent actinomycin D (ActD) to produce apoptotic cells. Then by incubating apoptotic and live hMSCs with IRIS Dots, they demonstrated that IRIS Dots were distributed in the cytoplasm of live cells (verified by stained with calcein-AM, Figure 3C), but on the outer cell surface of early apoptotic cells (stained with Annexin V-fluorescein isothiocyanate, Figure 3A and 3B) due to loss of active endocytosis.
Figure 3.
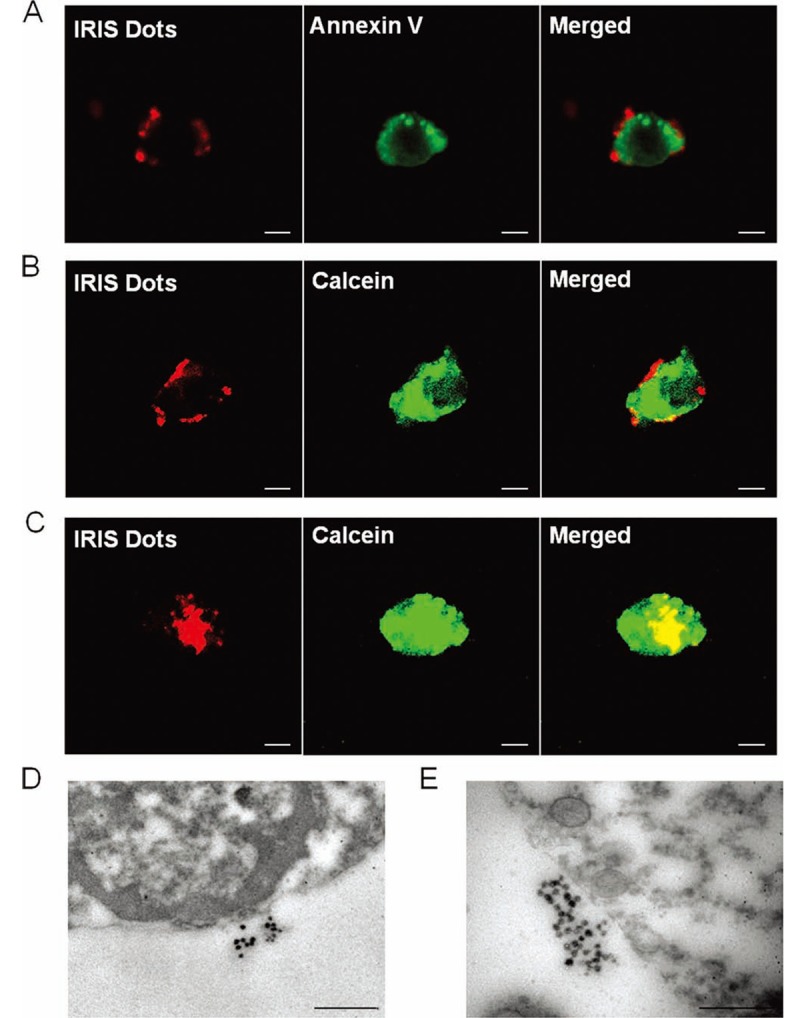
Detection of IRIS Dots uptake by hMSCs using confocal microscope and transmission electron microscope: A–C. hMSCs were pretreated for 48 h with 10 μg/mL ActD and then incubated with 20 μg/mL IRIS Dots for an additional 24 h. Both spontaneously detached and trypsinized cells were stained with either Annexin V–fluorescein isothiocyanate (A) or 2 μM calcein-AM (B, C) and then evaluated by confocal microscopy. The white scale bar represents 10 μm. One representative apoptotic cell co-stained with Annexin V–fluorescein isothiocyanate (A), one representative apoptotic cell co-labeled with calcein-AM (B), and one representative live cell co-labeled with calcein-AM (C) are shown. D, E. hMSCs were pretreated for 48 h with 10 μg/mL ActD, incubated with 20 μg/mL IRIS Dots for an additional 24 h, and then analyzed by TEM. The black scale bar represents 0.5 μm. Reprinted with permission from [51]. Copyright © 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Besides silica NPs, fluorescent polymeric NPs like polystyrene (PS) NPs are another popular choice likely due to their distribution by all the major biotechnology companies and the variety of functional groups including non-modified, sulfate-modified, aldehyde-modified, carboxylate-modified or amine-modified surface [44,52]. Recently, the uptake of the anionic PS NPs by hMSCs was investigated using spinning-disk confocal optical microscopy [53]. In this study, carboxyl-functionalized PS NPs were shown to be internalized mainly through the clathrin-mediated mechanism, which were more rapidly than the nonfunctionalized PS NPs.
Gold nanoparticles
With the rapid advancement of non-linear optics, fluorescence imaging with multiphoton excitation becomes a powerful technique for the high-resolution imaging. In this technique, noble metal NPs like gold NPs (Au NPs) were excited to a high energy state by two or more photons of red or near infrared (NIR) light simultaneously. Farrer et al. demonstrated that multiphoton-absorption-induced luminescence (MAIL) from Au NPs was generated efficiently with 800 nm laser and the luminescence spanned all the visible spectrum [54]. Compared with the traditional ultraviolet-visible (UV) excitation, NIR provided relatively higher depth of tissue penetration and minimized the interference of background fluorescence from the biological samples. In addition, Au NPs are generally considered biocompatible compared to other NPs [55], given that they have been used for human decorations for thousands of years. In another example, Nagesha et al. labeled mouse embryonic stem cells with Au NPs and visualized them through MAIL. They expected the further application of this method for the investigation of the molecular machinery of endocytosis, post-internalization vesicle trafficking, lineage tracking, and cellular motility assays [56]. Au NPs are also well known for their plasmonic properties as well as the applications in photoacoustic imaging and SERS, which we will discuss later in other sections.
Upconversion nanoparticles
Upconversion (UC) is a process in which the sequential absorption of two or more photons leads to the emission of light at shorter wavelength. It is a non-linear optical process, which refers to anti-Stokes type emission. The most efficient UC materials are formed by solid-state materials doped with rare-earth ions [57]. In the nanoscale, NPs made of UC materials (so called, UCNPs) benefit from this unique properties when they are utilized as contrast agents in molecular imaging. Imaging with UCNPs provides higher sensitivity (lack of autofluorescence background), less toxic components (in comparison to QDs), high penetration depths (excitation with NIR light), and good photostability (no photobleaching) [58,59].
A number of groups have labeled progenitor cells with UCNPs for both in vitro and in vivo fluorescence tracking [60-62]. For example, Wang et al labeled and tracked mouse MSCs (mMSCs) with UCNPs [63], which were conjugated with R8 to facilitate cellular uptake. Little exocytosis of UCNPs from labeled mMSCs was found in the transwell assay after 10 days incubation, suggesting its potential for long-term cell tracking. Even after two weeks, the labeling with UCNPs did not show any influence over the survivability, proliferation, and differentiation of mMSCs. To determine the detection sensitivity of UCNP-labeled mMSCs in vivo, various amount of mMSCs (10-104) labeled with UCNPs were subcutaneously injected beneath the skin of a nude mouse. It was then imaged by a modified Maestro in vivo imaging system with a 980 nm laser as excitation source. The sensitivity of fluorescence imaging with UCNPs was as few as 10 cells, nearly at the single cell level. By utilizing the UCNPs-labeling, they observed the migration of mMSCs from lung to liver in a nude mouse model with the whole body imaging by a modified Maestro in vivo imaging system using a 980 nm optical fiber-coupled laser as the excitation source [63].
While unique, UCNPs have the following disadvantages as contrast agents for fluorescence imaging. First of all, the upconversion efficiencies have been relatively low (usually less than 1%) [64]. The excitation thresholds are quite high, and the investigated phosphors (generally fluorides) often presented poor chemical stability [65]. Secondly, the potential long-term toxicity of Ln3+ doped UCNPs is another significant concern [64]. A thorough and systematic investigation is highly needed to reveal their biocompatibility and biostability.
Stem cell tracking with nanoparticle-aided photoacoustic imaging
Photoacoustic (PA) imaging, also called optoacoustic imaging is a new biomedical imaging modality based on photoacoustic effect, in which the absorbed energy from the light is transformed into kinetic energy of the sample through energy exchange processes. It is a hybrid modality, combining the high-contrast and spectroscopic-based specificity of optical imaging with the high spatial resolution of ultrasound imaging. In essence, a PA image is an ultrasound image in which contrast depends on the optical absorption of samples. Thus biological tissues with optical properties such as hemoglobin could be visualized with PA imaging.
As stem cells usually don’t have obvious optical properties, in PA imaging, they are usually labeled with biocompatible materials with optical properties such as Au NPs or Au nanorods (NRs). In a recent study, hMSCs were pre-labeled with 20 nm Au NPs before their incorporation into PEGylated fibrin gel [66]. Then fibrin gel was injected intramuscularly in the lateral gastrocnemius of an anesthetized Lewis rat. The contrast brought by Au NPs allowed the researchers to visualize the in vivo differentiation and neovasularization of hMSCs using PA imaging. Au NRs, another attractive probe for PA imaging, have plasmon resonance absorption and scattering in the NIR region [67]. Jokerst et al used silica coated Au NRs (SiGNRs) to label and image hMSCs [68]. They found that the silica coating could dramatically increase the cellular uptake of SiGNRs (5-fold) without any change to the viability and function of hMSCs. Figure 4 showed the process of intramuscular implantation of SiGNR-labeled hMSCs to the hindlimb muscles of the mouse. PA imaging provides relative high spatial resolution (340 nm) and temporal resolution (0.2 s), which allows the real time monitoring of stem cell after transplantation.
Figure 4.
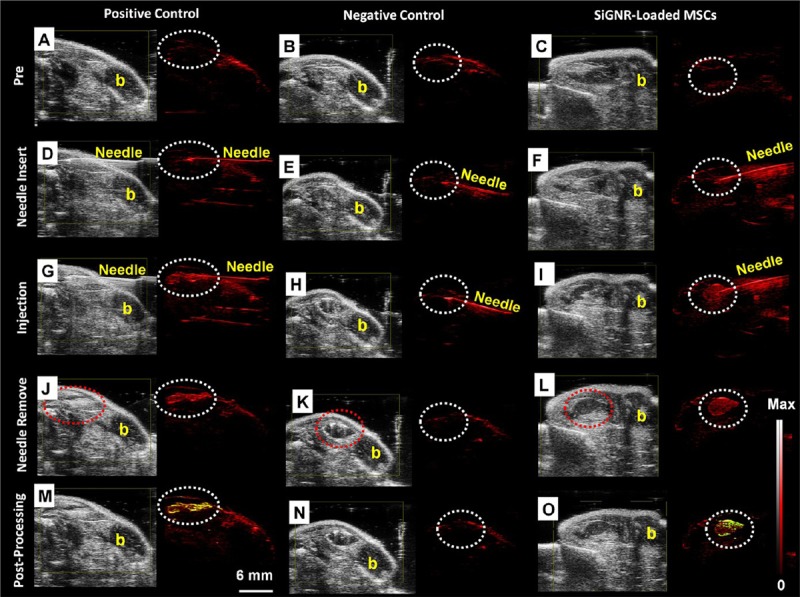
The backscatter (B-mode) (gray color) and PA (red color) images of the intramuscular injection of a positive control (0.7 nM SiGNRs; left), negative control (0 nM SiGNRs (no cells); middle), and 800 000 SiGNR-labeled MSCs (right) all in 50% matrigel/PBS into hind limb muscle of an athymic mouse. Imaging sequence is as follows: A-C. preinjection; D-F. needle insertion and position; G-I. postinjection; J-L. needle removal and final imaging, and M-O. contrast enhancement to illustrate increased signal. Pixels (signals) increased relative to preinjection image were coded yellow. Note significant signal increased in M and O at injection site relative to A and C (dashed circles highlight injection site). Also, note low signal in negative control (N). Scale bar in M and intensity scale in L and O applies to all images. The “b” in all panels indicated bone and the red dashed circle in J, K, L indicated that the injection bolus could also be seen with B-mode ultrasound. Reprinted with permission from [68]. Copyright © 2012 American Chemical Society.
Stem cell tracking with nanoparticle-aided Raman or surface enhanced Raman spectroscopy
The last technology we would like to discuss is Raman or surface enhanced Raman spectroscopy (SERS) based imaging. Raman scattering is the inelastic scattering of photons by molecular bonds that utilize the fact that every chemical bond in a molecule has a characteristic vibrational energy. This “molecular fingerprint” allows the nondestructive and label-free imaging of biological molecules such as DNA and protein in cells and tissue. In comparison to infrared (IR) and nuclear magnetic resonance (NMR) spectroscopy, Raman spectroscopy is not affected by the presence of water. However, Raman scattering from the natural molecules produces a very weak signal which is usually approximately 12-14 orders of magnitude weaker than fluorescence [69]. SERS could be used to further enhance the signal. Although Raman spectroscopy and SERS have been widely used from imaging to diagnoses [70-77], it is relatively a new direction to track stem cells with those techniques.
Stem cell tracking with nanoparticle-aided Raman imaging
Spontaneous Raman spectroscopy has been wildly used for monitoring the differentiation of human embryonic stem cells (hESC) [78-80], adult stem cells [81], and neural stem cells [80] due to the spectral characteristics of different types of cells. Notingher et al observed a clear reduction in DNA (786 cm-1 Raman peaks) and RNA (813 cm-1 Raman peaks) during living murine embryonic stem (ES) cell differentiation (over 16 days of differentiation) [78]. Chan et al. demonstrated specific Raman spectra could distinguish undifferentiated hESC from hESC-derived cardiomyocytes and human fetal left ventricle cardiomyocytes [79]. These results lay the foundation for the development of single cell Raman spectroscopy as a systematic method for sorting cardiomyocytes derived from reprogrammed, embryonic or adult stem cells for future cell-based heart therapies [79]. Spectral variations assigned to glycogen have also been reported for hESCs maintained under normal growth condition in vitro [82,83]. In addition, Raman spectroscopy has been employed to monitor osteoblast differentiation and in vitro mineralization capacity of MSC and osteoprogentior cells [84,85]. Cultured normal and abnormal stem cells including normal hESC, karyotypically abnormal hESC, normal and transformed hMSC can be identified by Raman spectroscopy. The changes of intensity peaks from phenylalanine (1005 cm-1), cytochrome C (1128 cm-1), protein, DNA/RNA, lipid in normal and abnormal stem cells show that Raman spectroscopy provides an alternative method allowing screening of cultured stem cells from abnormalities (abnormal and transformed stem cells) prior to cell transplantation [86].
As mentioned above, the fingerprint characteristics of Raman spectroscopy provides a way to distinguish stem cell types, differentiation processes, and abnormalism (chromosomal instability, development of cell lines, in vitro replicative senescence and etc). Statistic methods such as principal component analysis and linear discriminant analysis are usually required for the spectra analysis. However, due to the long acquisition time (normally several minutes for each Raman spectrum and hours for a single cell imaging), tracking the fate and the function of stem cells in vivo based on its intrinsic Raman information remain significant challenges [82]. Alternatively, if the Raman signal of certain probes within the cell is strong enough for fast imaging, tracking stem cell based on Raman signal could eventually be practical.
Unlike fluorescent imaging, Raman imaging does not suffer from photobleaching and autofluorescence background interference once NIR excitation is used. Among various NP-based contrast agents, single-walled carbon nanotubes (SWNTs) show an intense intrinsic Raman peak (G band at 1593 cm-1) produced by the strong electron photon coupling that causes efficient excitation of tangential vibration in the nanotubes quasi one-dimensional structure upon light exposure [87]. Strong and narrow signal of SWNTs not only enables fast mapping (with integration time of 0.1 s for each mapping point) but also provides easy differentiation from the tissue autofluoresence. So far SWNTs have been successfully used for specific tumor targeting in vivo within various tumor models [88-90] and whole-body, deep-tissue, small-animal imaging [91]. They are chemical and photo stable, and can be used for long-term tracking and imaging in biological systems [92]. Wang et al. found the protamine and PEG functionalized SWNTs (naturally accompany with Fe/Co metallic nanoparticles come from synthesis methods) were allowed to track hMSCs without interfering their proliferation and differentiation. Triple model Raman/MRI/PAT imaging of SWNT-labeled hMSCs was further demonstrated in living mice (Figure 5) [93]. Moreover, SWNTs with different isotope compositions (C12-SWNTs and C13-SWNTs) display well-shifted Raman G-band peaks (C12-SWNTs at around 1590 cm-1 and C13-SWNTs at around 1528 cm-1) [94]. This character enables them to be potentially used for multiplex tracking of different types of stem cells which are injected simultaneously.
Figure 5.
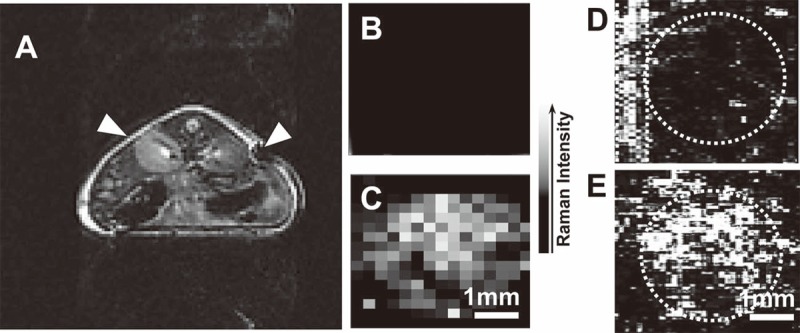
In vivo triple-modal imaging of SWNT-labeled hMSCs: unlabeled and SWNT-labeled hMSCs (10 nM of SWNTs during labeling) were subcutaneously injected into the back of a nude mouse before imaging. A. In vivo T2-weighted MR image. Arrows pointed to the sites where unlabeled (left) and SWNT-labeled (right) hMSCs were injected; B, C. In vivo Raman images of unlabeled (B) and SWNT-labeled (C) hMSCs; D, E. In vivo PAT images of unlabeled (D) and SWNT-labeled (E) hMSCs. The circles highlighted the locations were hMSCs were injected. PA signals in the rectangle highlighted in D were from a blood vessel crossing this area. Reprinted with permission from [93]. Copyright © 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Stem cell tracking with nanoparticle-aided surface enhanced Raman spectroscopy
Besides Raman spectroscopy, imaging with SERS has also gained significant interest over the last several years [95]. SERS is a phenomenon that the Raman scattering from a molecule is enhanced by many orders of magnitude due to its proximity to a metal surface (usually gold and silver) with surface plasmon efficiently coupling the energy of the incoming laser light [96]. Normally, two different methodologies, direct detection (with metallic NPs as SERS substrates for label-free detection of the analyte) and indirect detection (biotargeted research with SERS labels) are used for cell and biomedical research.
Similar to Raman spectroscopy, direct detection of SERS provides vibration information of stem cell while enhancing the signal sensitivity. By labeling mESC with Au NPs, Sathuluri and colleagues demonstrated that specific SERS fingerprint information of undifferentiated single cells, embryoid bodies and terminally differentiated cardiomyocytes has unique SERS fingerprints based upon nucleic acids, mitochondria, proteins, and cardiomyocytes adsorbed onto metal NPs. This information reflects the molecular changes accompanying with metabolic activities in each stage of differentiation [97].
By labeling cells with Raman probes (molecules with significant Raman activity) and biospecific antibodies, an indirect method to visualize the distribution of certain proteins within living cell can be engineered. The expressed proteins, including CD34, Sca-1, and SP-C in bronchioalveolar stem cells (BASCs) in the murine lung were quantitatively compared based on multifunctional NPs, namely fluorescent-surface-enhanced Raman spectroscopic dots (F-SERS dots). Conjugated with BASC-specific markers, with the dual signal detection model, F-SERS dots provided multiple target analysis and clarified the effective stem cell specific markers and therapy strategies for disease [98]. In addition to multiplex targeting, magnetic NP-based surface enhanced Raman spectroscopic dots (M-SERS Dots) could be used as a sorting system effectively isolate BASCs [99].
Perspectives and conclusion
With the growing demands of regenerative therapy, and the current lack of a sensitive and yet efficient method of tracking transplanted cells, there is an urgent need for the development of novel imaging technologies which allow the real-time and non-invasive monitoring of transplanted stem cells for their survival, biodistribution, migration, and differentiation. In comparison to other imaging modalities like MRI, PET, and CT, optical imaging is cost-effective, time-efficient, and are thus more accessible for the majority of researchers during the pre-clinical studies with small animals. In this mini-review, we discussed three modalities of NP-aided optical imaging that have been widely used for stem cell imaging and tracking.
However, we should be aware that opportunity and risk always co-exist. Several concerns about using NPs for stem cell tracking should be addressed before they are routinely used in clinical treatments. Specifically, those challenges include contrast agents transfer, signal dilution, interference from tissue autofluorescence, and inability to gather information on cellular functions [14,100].
In contrast agents (i.e. NPs) transfer, NPs leaked out from labeled cells due to normal cell processes (e.g. cell death and exocytosis) and were able to re-enter into adjacent cells over time. The possibility of NPs leakage, a common challenge in cell tracking studies, leads to possible uptake of NPs by surrounding cells and thus introduces false positive results [101]. For example, stem cells were labeled with QDs in order to noninvasively track their distribution after being seeded on scaffolds and transplanted into a nude rat that was critically sized femoral defects. As excepted, clear fluorescence was observed at implantation sites; however, signals were also observed at sites treated with acellular QD-free scaffolds after surgery for 7-10 days [102]. Sarah et al demonstrated the transfer of QDs from labeled hMSCs to adjacent unlabeled cells in an in vitro co-culture system. Their leakage was further examined in vivo by transplantation of QDs-labeled hMSCs to an animal model of spinal cord injury, which indicated the deposition of QDs into the host cells after 1 week was probably due to their active excretion from labeled cells or the release from the dead cells [36]. These studies show that the leakage of NPs from labeled cells must be tested to avoid false positive results. Meanwhile, valid conclusion of cell localization cannot be drawn only from in vivo imaging data. It is also expected that modifications (e.g. cationic lipids [32]) of NPs with some specific ligands which eliminate their leakage over a long period of time could provide significant potential in long term in vivo imaging.
Signal dilution, another issue for long-term in vivo stem cell tracking, is the signal loss resulted from cell division and exocytosis. For example, Sarah et al found that the fluorescence of QDs labeled hMSCs fell progressively with cell divisions. After four subculturing passage, the fluorescence intensity decreased by more than 80% [36]. One approach is to genetically engineer the cell to produce NPs [103,104], which would be similar to the expression of green fluorescence protein. However, it is still unclear whether genetic engineering of cells will have any long-term consequence on cell phenotype and function. Another approach is to enhance the signal of NPs which will still detectable after multiple cell divisions [105]. Single NP optical imaging technique holds the promise of in vivo stem cell tracking even when only one NP is left in one stem cell after cell divisions. The real time tracking of the transport and random walk of single NP (gold and silver) in zebrafish embryos have been achieved by Xu’s group [106,107].
Optical imaging modalities except PA imaging usually suffer from the background interference of autofluorescence of tissues [108]. NPs with tunable emission wavelength of light either generated from inherent properties (QDs) or encapsulated fluorescence probe (C-Dots) can avoid the autofluorescence background, which can also be reduced by UCNPs for fluorescence imaging and SWNTs for Raman imaging using NIR excitation light.
Although the location of stem cells post transplantation could be tracked in vivo with good temporal and spatial resolution, the functions (e.g. differentiation of stem cells) of the transplanted cells are also important. In the clinical treatments, if the viability and function of transplanted stem cells can be obtained simultaneously, it will help the physician to make a decision without the need for multiple biopsies. One potential approach is to design NP-aided sensors, which can respond to the secreted chemicals or expressed chemokines/cytokines during cell differentiation, the local pH changes during cell apoptosis and death. Another approach is using Raman and SERS imaging modalities to simultaneously track stem cells in vivo and record fingerprint spectrum of certain chemicals indicated different cellular conditions and functions.
Acknowledgements
This work was supported in part by Nanyang Technological University Start-Up Grant and Singapore MOE AcRF Tier 1 research fund (RG 64/12) to XCJ. JKYC received salary support from the Ministry of Health’s National Medical Research Council in Singapore (NMRC/CSA/042/2012).
Conflict of interest
The authors have declared that no competing interests exist.
References
- 1.Weissman IL. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 2.Weissman IL. Translating stem and progenitor cell biology to the clinic: Barriers and opportunities. Science. 2000;287:1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 3.Singec I, Jandial R, Crain A, Nikkhah G, Snyder EY. The leading edge of stem cell therapeutics. Annu Rev Med. 2007;58:313–328. doi: 10.1146/annurev.med.58.070605.115252. [DOI] [PubMed] [Google Scholar]
- 4.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 5.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 6.Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: Nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 8.Caplan AI, Bruder SP. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol Med. 2001;7:259–264. doi: 10.1016/s1471-4914(01)02016-0. [DOI] [PubMed] [Google Scholar]
- 9.Prochymal - First stem cell drug approved. 2012. http://www.medicalnewstoday.com/articles/ 245704.php.
- 10.2013. www.clinicaltrials.gov.
- 11.Halban PA, German MS, Kahn SE, Weir GC. Current Status of Islet Cell Replacement and Regeneration Therapy. J Clin Endocr Metab. 2010;95:1034–1043. doi: 10.1210/jc.2009-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen PK, Nag D, Wu JC. Methods to assess stem cell lineage, fate and function. Adv Drug Deliv Rev. 2010;62:1175–1186. doi: 10.1016/j.addr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagani FD, DerSimonian H, Zawadzka A, Wetzel K, Edge ASB, Jacoby DB, Dinsmore JH, Wright S, Aretz TH, Eisen HJ, Aaronson KD. Autologous skeletal myoblasts transplanted to ischemia-damaged myocardium in humans - Histological analysis of cell survival and differentiation. J Am Coll Cardiol. 2003;41:879–888. doi: 10.1016/s0735-1097(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 14.Xu CJ, Mu LY, Roes I, Miranda-Nieves D, Nahrendorf M, Ankrum JA, Zhao WA, Karp JM. Nanoparticle-based monitoring of cell therapy. Nanotechnology. 2011;22:494001. doi: 10.1088/0957-4484/22/49/494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, Otto C. Nonresonant Raman imaging of protein distribution in single human cells. Biopolymers. 2003;72:1–9. doi: 10.1002/bip.10246. [DOI] [PubMed] [Google Scholar]
- 16.Kneipp J, Kneipp H, Wittig B, Kneipp K. Novel optical nanosensors for probing and imaging live cells. Nanomedicine. 2010;6:214–226. doi: 10.1016/j.nano.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto JG, Farkas D. Biomedical optical imaging. USA: Oxford University Press; 2009. [Google Scholar]
- 18.Solanki A, Kim JD, Lee KB. Nanotechnology for regenerative medicine: nanomaterials for stem cell imaging. Nanomedicine. 2008;3:567–578. doi: 10.2217/17435889.3.4.567. [DOI] [PubMed] [Google Scholar]
- 19.Villa C, Erratico S, Razini P, Fiori F, Rustichelli F, Torrente Y, Belicchi M. Stem Cell Tracking by Nanotechnologies. Int J Mol Sci. 2010;11:1070–1081. doi: 10.3390/ijms11031070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deb KD, Griffith M, De Muinck E, Rafat M. Nanotechnology in stem cells research: advances and applications. Front Biosci. 2012;17:1747–1760. doi: 10.2741/4016. [DOI] [PubMed] [Google Scholar]
- 21.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharrna P, Brown S, Walter G, Santra S, Moudgil B. Nanoparticles for bioimaging. Adv Colloid Interface Sci. 2006;123:471–485. doi: 10.1016/j.cis.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Piao Y, Hyeon T. Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy. Chem Soc Rev. 2009;38:372–390. doi: 10.1039/b709883a. [DOI] [PubMed] [Google Scholar]
- 24.Hahn MA, Singh AK, Sharma P, Brown SC, Moudgil BM. Nanoparticles as contrast agents for in-vivo bioimaging: current status and future perspectives. Anal Bioanal Chem. 2011;399:3–27. doi: 10.1007/s00216-010-4207-5. [DOI] [PubMed] [Google Scholar]
- 25.Soo Lee D. In Vivo Neuronal Cell Differentiation Imaging From Transplanted Stem Cells. Curr Med Imaging Rev. 2012;8:278–283. [Google Scholar]
- 26.Buschke DG, Squirrell JM, Fong JJ, Eliceiri KW, Ogle BM. Cell death, non-invasively assessed by intrinsic fluorescence intensity of NADH, is a predictive indicator of functional differentiation of embryonic stem cells. Biol Cell. 2012;104:352–364. doi: 10.1111/boc.201100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao WA, Schafer S, Choi J, Yamanaka YJ, Lombardi ML, Bose S, Carlson AL, Phillips JA, Teo WS, Droujinine IA, Cui CH, Jain RK, Lammerding J, Love JC, Lin CP, Sarkar D, Karnik R, Karp JM. Cell-surface sensors for real-time probing of cellular environments. Nat Nanotechnol. 2011;6:524–531. doi: 10.1038/nnano.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996;271:933–937. [Google Scholar]
- 29.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 30.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 31.Chen FQ, Gerion D. Fluorescent CdSe/ZnS nanocrystal-peptide conjugates for long-term, nontoxic imaging and nuclear targeting in living cells. Nano Lett. 2004;4:1827–1832. [Google Scholar]
- 32.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat Med. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 33.Rosen AB, Kelly DJ, Schuldt AJT, Lu J, Potapova IA, Doronin SV, Robichaud KJ, Robinson RB, Rosen MR, Brink PR, Gaudette GR, Cohen IS. Finding fluorescent needles in the cardiac haystack: Tracking human mesenchymal stem cells labeled with quantum dots for quantitative in vivo three-dimensional fluorescence analysis. Stem Cells. 2007;25:2128–2138. doi: 10.1634/stemcells.2006-0722. [DOI] [PubMed] [Google Scholar]
- 34.Lei Y, Tang H, Yao L, Yu R, Feng M, Zou B. Applications of mesenchymal stem cells labeled with Tat peptide conjugated quantum dots to cell tracking in mouse body. Bioconjug Chem. 2008;19:421–427. doi: 10.1021/bc0700685. [DOI] [PubMed] [Google Scholar]
- 35.Chang JC, Hsu SH, Su HL. The regulation of the gap junction of human mesenchymal stem cells through the internalization of quantum dots. Biomaterials. 2009;30:1937–1946. doi: 10.1016/j.biomaterials.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 36.Ranjbarvaziri S, Kiani S, Akhlaghi A, Vosough A, Baharvand H, Aghdami N. Quantum dot labeling using positive charged peptides in human hematopoetic and mesenchymal stem cells. Biomaterials. 2011;32:5195–5205. doi: 10.1016/j.biomaterials.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Yukawa H, Kagami Y, Watanabe M, Oishi K, Miyamoto Y, Okamoto Y, Tokeshi M, Kaji N, Noguchi H, Ono K, Sawada M, Baba Y, Hamajima N, Hayashi S. Quantum dots labeling using octa-arginine peptides for imaging of adipose tissue-derived stem cells. Biomaterials. 2010;31:4094–4103. doi: 10.1016/j.biomaterials.2010.01.134. [DOI] [PubMed] [Google Scholar]
- 38.Yukawa H, Watanabe M, Kaji N, Okamoto Y, Tokeshi M, Miyamoto Y, Noguchi H, Baba Y, Hayashi S. Monitoring transplanted adipose tissue-derived stem cells combined with heparin in the liver by fluorescence imaging using quantum dots. Biomaterials. 2012;33:2177–2186. doi: 10.1016/j.biomaterials.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Lu S, Xu X, Zhao WX, Wu WW, Yuan H, Shen HB, Zhou CH, Li S, Ma L. Targeting of embryonic stem cells by peptide-conjugated quantum dots. PloS One. 2010;5:e12705. doi: 10.1371/journal.pone.0012075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derfus AM, Chan WCW, Bhatia SN. Probing the cytotoxicity of semiconductor quantum dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirchner C, Liedl T, Kudera S, Pellegrino T, Javier AM, Gaub HE, Stolzle S, Fertig N, Parak WJ. Cytotoxicity of colloidal CdSe and CdSe/ZnS nanoparticles. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 42.Lai CW, Wang YH, Chen YC, Hsieh CC, Uttam BP, Hsiao JK, Hsu CC, Chou PT. Homogenous, far-reaching tuning and highly emissive QD-silica core-shell nanocomposite synthesized via a delay photoactive procedure; their applications in two-photon imaging of human mesenchymal stem cells. J Mater Chem. 2009;19:8314–8319. [Google Scholar]
- 43.Chen N, He Y, Su YY, Li XM, Huang Q, Wang HF, Zhang XZ, Tai RZ, Fan CH. The cytotoxicity of cadmium-based quantum dots. Biomaterials. 2012;33:1238–1244. doi: 10.1016/j.biomaterials.2011.10.070. [DOI] [PubMed] [Google Scholar]
- 44.Wang F, Tan WB, Zhang Y, Fan XP, Wang MQ. Luminescent nanomaterials for biological labelling. Nanotechnology. 2006;17:R1–R13. [Google Scholar]
- 45.Burns A, Ow H, Wiesner U. Fluorescent core-shell silica nanoparticles: towards “Lab on a Particle” architectures for nanobiotechnology. Chem Soc Rev. 2006;35:1028–1042. doi: 10.1039/b600562b. [DOI] [PubMed] [Google Scholar]
- 46.van Blaaderen A, Vrij A. Synthesis and characterization of monodisperse colldidal organosilica spheres. J Colloid Interface Sci. 1993;156:1–18. [Google Scholar]
- 47.Yamauchi H, Ishikawa T, Kondo S. Surface characterization of ultramicro spherical-particles of silica prepared by w/o microemulsion method. Colloids Surf. 1989;37:71–80. [Google Scholar]
- 48.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Bright and stable core-shell fluorescent silica nanoparticles. Nano Lett. 2005;5:113–117. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 49.Burns AA, Vider J, Ow H, Herz E, Penate-Medina O, Baumgart M, Larson SM, Wiesner U, Bradbury M. Fluorescent silica nanoparticles with efficient urinary excretion for nanomedicine. Nano Lett. 2009;9:442–448. doi: 10.1021/nl803405h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alberto G, Miletto I, Viscardi G, Caputo G, Latterini L, Coluccia S, Martra G. Hybrid cyanine-silica nanoparticles: Homogeneous photoemission behavior of entrapped fluorophores and consequent high brightness enhancement. J Phys Chem C. 2009;113:21048–21053. [Google Scholar]
- 51.Accomasso L, Rocchietti EC, Raimondo S, Catalano F, Alberto G, Giannitti A, Minieri V, Turinetto V, Orlando L, Saviozzi S, Caputo G, Geuna S, Martra G, Giachino C. Fluorescent silica nanoparticles improve optical imaging of stem cells allowing direct discrimination between live and early-stage apoptotic cells. Small. 2012;8:3192–3200. doi: 10.1002/smll.201200882. [DOI] [PubMed] [Google Scholar]
- 52.Brijmohan SB, Swier S, Weiss RA, Shaw MT. Synthesis and characterization of cross-linked sulfonated polystyrene nanoparticles. Ind Eng Chem Res. 2005;44:8039–8045. [Google Scholar]
- 53.Jiang XE, Musyanovych A, Rocker C, Landfester K, Mailander V, Nienhaus GU. Specific effects of surface carboxyl groups on anionic polystyrene particles in their interactions with mesenchymal stem cells. Nanoscale. 2011;3:2028–2035. doi: 10.1039/c0nr00944j. [DOI] [PubMed] [Google Scholar]
- 54.Farrer RA, Butterfield FL, Chen VW, Fourkas JT. Highly efficient multiphoton-absorption-induced luminescence from gold nanoparticles. Nano Lett. 2005;5:1139–1142. doi: 10.1021/nl050687r. [DOI] [PubMed] [Google Scholar]
- 55.Thakor AS, Jokerst J, Zavaleta C, Massoud TF, Gambhir SS. Gold nanoparticles: A revival in precious metal administration to patients. Nano Lett. 2011;11:4029–4036. doi: 10.1021/nl202559p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagesha D, Laevsky GS, Lampton P, Banyal R, Warner C, DiMarzio C, Sridhar S. In vitro imaging of embryonic stem cells using multiphoton luminescence of gold nanoparticles. Int J Nanomedicine. 2007;2:813–819. [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F, Liu XG. Recent advances in the chemistry of lanthanide-doped upconversion nanocrystals. Chem Soc Rev. 2009;38:976–989. doi: 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]
- 58.Wang F, Banerjee D, Liu YS, Chen XY, Liu XG. Upconversion nanoparticles in biological labeling, imaging, and therapy. Analyst. 2010;135:1839–1854. doi: 10.1039/c0an00144a. [DOI] [PubMed] [Google Scholar]
- 59.Haase M, Schafer H. Upconverting Nanoparticles. Angew Chem Int Ed Engl. 2011;50:5808–5829. doi: 10.1002/anie.201005159. [DOI] [PubMed] [Google Scholar]
- 60.Liu Q, Sun Y, Yang TS, Feng W, Li CG, Li FY. Sub-10 nm hexagonal lanthanide-doped NaLuF4 upconversion nanocrystals for sensitive bioimaging in vivo. J Am Chem Soc. 2011;133:17122–17125. doi: 10.1021/ja207078s. [DOI] [PubMed] [Google Scholar]
- 61.Nyk M, Kumar R, Ohulchanskyy TY, Bergey EJ, Prasad PN. High contrast in vitro and in vivo photoluminescence bioimaging using near infrared to near infrared up-conversion in TM3+ and Yb3+ doped fluoride nanophosphors. Nano Lett. 2008;8:3834–3838. doi: 10.1021/nl802223f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Idris NM, Li ZQ, Ye L, Sim EK, Mahendran R, Ho PCL, Zhang Y. Tracking transplanted cells in live animal using upconversion fluorescent nanoparticles. Biomaterials. 2009;30:5104–5113. doi: 10.1016/j.biomaterials.2009.05.062. [DOI] [PubMed] [Google Scholar]
- 63.Wang C, Cheng L, Xu H, Liu Z. Towards whole-body imaging at the single cell level using ultra-sensitive stem cell labeling with oligoarginine modified upconversion nanoparticles. Biomaterials. 2012;33:4872–4881. doi: 10.1016/j.biomaterials.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 64.Cheng L, Wang C, Liu Z. Upconversion nanoparticles and their composite nanostructures for biomedical imaging and cancer therapy. Nanoscale. 2013;5:23–37. doi: 10.1039/c2nr32311g. [DOI] [PubMed] [Google Scholar]
- 65.Wang M, Abbineni G, Clevenger A, Mao C, Xu S. Upconversion nanoparticles: synthesis, surface modification and biological applications. Nanomedicine. 2011;7:710–729. doi: 10.1016/j.nano.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nam SY, Ricles LM, Suggs LJ, Emelianov SY. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PloS One. 2012;7:e37267. doi: 10.1371/journal.pone.0037267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tong L, Wei QS, Wei A, Cheng JX. Gold nanorods as contrast agents for biological imaging: Optical properties, surface conjugation and photothermal effects. Photochem Photobiol. 2009;85:21–32. doi: 10.1111/j.1751-1097.2008.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jokerst JV, Thangaraj M, Kempen PJ, Sinclair R, Gambhir SS. Photoacoustic imaging of mesenchymal stem cells in living mice via silica-coated gold nanorods. ACS Nano. 2012;6:5920–5930. doi: 10.1021/nn302042y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Ru EC, Schroeter LC, Etchegoin PG. Direct measurement of resonance Raman spectra and cross sections by a polarization difference technique. Anal Chem. 2012;84:5074–5079. doi: 10.1021/ac300763q. [DOI] [PubMed] [Google Scholar]
- 70.Wachsmann-Hogiu S, Weeks T, Huser T. Chemical analysis in vivo and in vitro by Raman spectroscopy-from single cells to humans. Curr Opin Biotechnol. 2009;20:63–73. doi: 10.1016/j.copbio.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kneipp K, Kneipp H, Kneipp J. Surface-enhanced Raman scattering in local optical fields of silver and gold nanoaggregatess - From single-molecule Raman spectroscopy to ultrasensitive probing in live cells. Acc Chem Res. 2006;39:443–450. doi: 10.1021/ar050107x. [DOI] [PubMed] [Google Scholar]
- 72.Chan J, Fore S, Wachsman-Hogiu S, Huser T. Raman spectroscopy and microscopy of individual cells and cellular components. Laser Photon Rev. 2008;2:325–349. [Google Scholar]
- 73.Li M, Xu J, Romero-Gonzalez M, Banwart SA, Huang WE. Single cell Raman spectroscopy for cell sorting and imaging. Curr Opin Biotechnol. 2012;23:56–63. doi: 10.1016/j.copbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 74.Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy can detect and monitor cancer at cellular level: Analysis of resistant and sensitive subtypes of testicular cancer cell lines. Appl Spectrosc Rev. 2012;47:571–581. [Google Scholar]
- 75.Vitol EA, Orynbayeva Z, Friedman G, Gogotsi Y. Nanoprobes for intracellular and single cell surface-enhanced Raman spectroscopy (SERS) J Raman Spectrosc. 2012;43:817–827. [Google Scholar]
- 76.Huh YS, Chung AJ, Erickson D. Surface enhanced Raman spectroscopy and its application to molecular and cellular analysis. Microfluid and Nanofluid. 2009;6:285–297. [Google Scholar]
- 77.Qian XM, Nie SM. Single-molecule and single-nanoparticle SERS: from fundamental mechanisms to biomedical applications. Chem Soc Rev. 2008;37:912–920. doi: 10.1039/b708839f. [DOI] [PubMed] [Google Scholar]
- 78.Notingher I, Bisson I, Bishop AE, Randle WL, Polak JM, Hench LL. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 79.Chan JW, Lieu DK, Huser T, Li RA. Label-free separation of human embryonic stem cells and their cardiac derivatives using Raman spectroscopy. Anal Chem. 2009;81:1324–1331. doi: 10.1021/ac801665m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan JW, Lieu DK. Label-free biochemical characterization of stem cells using vibrational spectroscopy. J Biophotonics. 2009;2:656–668. doi: 10.1002/jbio.200910041. [DOI] [PubMed] [Google Scholar]
- 81.Downes A, Mouras R, Elfick A. Optical spectroscopy for noninvasive monitoring of stem cell differentiation. J Biomed Biotechnol. 2010;2010:101864. doi: 10.1155/2010/101864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stewart S, Priore RJ, Nelson MP, Treado PJ. Raman imaging. Annu Rev Anal Chem. 2012;5:337–360. doi: 10.1146/annurev-anchem-062011-143152. [DOI] [PubMed] [Google Scholar]
- 83.Konorov SO, Schulze HG, Caron NJ, Piret JM, Blades MW, Turner RFB. Raman microspectroscopic evidence that dry-fixing preserves the temporal pattern of non-specific differentiation in live human embryonic stem cells. J Raman Spectrosc. 2011;42:576–579. [Google Scholar]
- 84.McManus LL, Burke GA, McCafferty MM, O’Hare P, Modreanu M, Boyd AR, Meenan BJ. Raman spectroscopic monitoring of the osteogenic differentiation of human mesenchymal stem cells. Analyst. 2011;136:2471–2481. doi: 10.1039/c1an15167c. [DOI] [PubMed] [Google Scholar]
- 85.Gentleman E, Swain RJ, Evans ND, Boonrungsiman S, Jell G, Ball MD, Shean TA, Oyen ML, Porter A, Stevens MM. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- 86.Harkness L, Novikov SM, Beermann J, Bozhevolnyi SI, Kassem M. Identification of abnormal stem cells using Raman spectroscopy. Stem Cells Dev. 2012;21:2152–2159. doi: 10.1089/scd.2011.0600. [DOI] [PubMed] [Google Scholar]
- 87.Jorio A, Saito R, Dresselhaus G, Dresselhaus MS. Determination of nanotubes properties by Raman spectroscopy. Philos Transact A Math Phys Eng Sci. 2004;362:2311–2336. doi: 10.1098/rsta.2004.1443. [DOI] [PubMed] [Google Scholar]
- 88.Liu Z, Cai WB, He LN, Nakayama N, Chen K, Sun XM, Chen XY, Dai HJ. In vivo biodistribution and highly efficient tumour targeting of carbon nanotubes in mice. Nat Nanotechnol. 2007;2:47–52. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 89.Bhirde AA, Patel V, Gavard J, Zhang GF, Sousa AA, Masedunskas A, Leapman RD, Weigert R, Gutkind JS, Rusling JF. Targeted killing of cancer cells in vivo and in vitro with EGF-directed carbon nanotube-based drug delivery. ACS Nano. 2009;3:307–316. doi: 10.1021/nn800551s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McDevitt MR, Chattopadhyay D, Kappel BJ, Jaggi JS, Schiffman SR, Antczak C, Njardarson JT, Brentjens R, Scheinberg DA. Tumor targeting with antibody-functionalized, radiolabeled carbon nanotubes. J Nucl Med. 2007;48:1180–1189. doi: 10.2967/jnumed.106.039131. [DOI] [PubMed] [Google Scholar]
- 91.Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc Natl Acad Sci U S A. 2008;105:5844–5849. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Z, Davis C, Cai WB, He L, Chen XY, Dai HJ. Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy. Proc Natl Acad Sci U S A. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang C, Ma XX, Ye SQ, Cheng L, Yang K, Guo L, Li CH, Li YG, Liu Z. Protamine functionalized single-walled carbon nanotubes for stem cell labeling and in vivo Raman/magnetic resonance/photoacoustic triple-modal imaging. Adv Funct Mater. 2012;22:2363–2375. [Google Scholar]
- 94.Liu ZA, Li XL, Tabakman SM, Jiang KL, Fan SS, Dai HJ. Multiplexed multicolor Raman imaging of live cells with isotopically modified single walled carbon nanotubes. J Am Chem Soc. 2008;130:13540–13541. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schlucker S. SERS microscopy: Nanoparticle probes and biomedical applications. ChemPhysChem. 2009;10:1344–1354. doi: 10.1002/cphc.200900119. [DOI] [PubMed] [Google Scholar]
- 96.Moskovits M. Surface-enhanced spectroscopy. Rev Mod Phys. 1985;57:783–826. [Google Scholar]
- 97.Sathuluri RR, Yoshikawa H, Shimizu E, Saito M, Tamiya E. Gold nanoparticle-based surface-enhanced Raman scattering for noninvasive molecular probing of embryonic stem cell differentiation. PloS One. 2011;6:e22802. doi: 10.1371/journal.pone.0022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Woo MA, Lee SM, Kim G, Baek J, Noh MS, Kim JE, Park SJ, Minai-Tehrani A, Park SC, Seo YT, Kim YK, Lee YS, Jeong DH, Cho MH. Multiplex immunoassay using fluorescent-surface enhanced Raman spectroscopic dots for the detection of bronchioalveolar stem cells in murine lung. Anal Chem. 2009;81:1008–1015. doi: 10.1021/ac802037x. [DOI] [PubMed] [Google Scholar]
- 99.Noh MS, Jun BH, Kim S, Kang H, Woo MA, Minai-Tehrani A, Kim JE, Kim J, Park J, Lim HT, Park SC, Hyeon T, Kim YK, Jeong DH, Lee YS, Cho MH. Magnetic surface-enhanced Raman spectroscopic (M-SERS) dots for the identification of bronchioalveolar stem cells in normal and lung cancer mice. Biomaterials. 2009;30:3915–3925. doi: 10.1016/j.biomaterials.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 100.Janowski M, Bulte JW, Walczak P. Personalized nanomedicine advancements for stem cell tracking. Adv Drug Deliv Rev. 2012;64:1488–1507. doi: 10.1016/j.addr.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pi QM, Zhang WJ, Zhou GD, Liu W, Cao YL. Degradation or excretion of quantum dots in mouse embryonic stem cells. BMC Biotechnol. 2010;10:36. doi: 10.1186/1472-6750-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dupont KM, Sharma K, Stevens HY, Boerckel JD, Garcia AJ, Guldberg RE. Human stem cell delivery for treatment of large segmental bone defects. Proc Natl Acad Sci U S A. 2010;107:3305–3310. doi: 10.1073/pnas.0905444107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 104.Zurkiya O, Chan AW, Hu XP. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59:1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu CJ, Miranda-Nieves D, Ankrum JA, Matthiesen ME, Phillips JA, Roes I, Wojtkiewicz GR, Juneja V, Kultima JR, Zhao WA, Vemula PK, Lin CP, Nahrendorf M, Karp JM. Tracking mesenchymal stem cells with iron oxide nanoparticle loaded poly(lactide-co-glycolide) microparticles. Nano Lett. 2012;12:4131–4139. doi: 10.1021/nl301658q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee KJ, Nallathamby PD, Browning LM, Osgood CJ, Xu XH. In vivo imaging of transport and biocompatibility of single silver nanoparticles in early development of zebrafish embryos. ACS Nano. 2007;1:133–143. doi: 10.1021/nn700048y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Browning LM, Lee KJ, Huang T, Nallathamby PD, Lowman JE, Xu XH. Random walk of single gold nanoparticles in zebrafish embryos leading to stochastic toxic effects on embryonic developments. Nanoscale. 2009;1:138–152. doi: 10.1039/b9nr00053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]


