Abstract
Multiwalled carbon nanotubes (MWCNT) are one of the most commonly produced nanomaterials, and pulmonary exposure during production, use, and disposal is a concern for the developing nanotechnology field. The airway epithelium is the first line of defense against inhaled particles. In a mouse model, MWCNT were reported to reach the alveolar space of the lung after in vivo exposure, penetrate the epithelial lining, and result in inflammation and progressive fibrosis. This study sought to determine the cellular and gene expression changes in small airway epithelial cells (SAEC) after in vitro exposure to MWCNT in an effort to elucidate potential toxicity mechanisms and signaling pathways. A direct interaction between SAEC and MWCNT was confirmed by both internalization of MWCNT and interaction at the cell periphery. Following exposure, SAEC showed time-dependent increases in reactive oxygen species production, total protein phosphotyrosine and phosphothreonine levels, and migratory behavior. Analysis of gene and protein expression suggested altered regulation of multiple biomarkers of lung damage, carcinogenesis, and tumor progression, as well as genes involved in related signaling pathways. These results demonstrate that MWCNT exposure resulted in the activation of SAEC. Gene expression data derived from MWCNT exposure provide information that may be used to elucidate the underlying mode of action of MWCNT in the small airway and suggest potential prognostic gene signatures for risk assessment.
Key Words: multiwalled carbon nanotubes, airway epithelial damage, signaling pathways, molecular mechanisms.
Nanotechnology, the manipulation of matter having at least one dimension less than 100nm, is a heavily invested technology for use within occupational and commercial fields. Novel properties of nanomaterials allow for the exploitation of melting points, electrical conductivity, magnetic properties, and chemical reactivity for the development of new technologies (http://nano.gov). Concurrent with the advancement of nanotechnology is the need for nanotoxicology, the evaluation of the interaction of nanomaterials with biological systems and their resultant responses and mechanisms (Castranova, 2011). As nanotechnology becomes increasingly integrated into products used for medicinal and commercial purposes, the risk of both occupational and commercial exposure to nanomaterials escalates.
One of the most predominant nanomaterials in use today are multiwalled carbon nanotubes (MWCNT), concentric cylindrical tubes of carbon with a diameter less than 100nm (Iijima, 1991). MWCNT exhibit unique physicochemical properties and are of great potential to the engineering and medical industries due to their small size, great strength, electronic conductivity, and strong capillary forces (Ajayan, 1999; Yakobson and Smalley, 1997). The small size and light weight of MWCNT make them easily aerosolized during production, use, and disposal. Upon in vivo aspiration exposure in a mouse model, MWCNT passed through the conducting airways of the lung and deposited into the alveolar regions (Mercer et al., 2010). An increase in lactate dehydrogenase, albumin level, and the number of polymorphonuclear leukocytes in the bronchoalveolar lavage fluid of mice after MWCNT exposure was indicative of an acute inflammatory response in the lung (Porter et al., 2010). Additionally, dose-dependent and persistent granulomatous lesions of progressive interstitial fibrosis were reported (Mercer et al., 2011; Porter et al., 2010).
The inflammatory and subsequent fibrotic response to MWCNT exposure was believed to correlate with their size, shape, and biopersistence. As particle size decreases, relative surface area increases, thus increasing the number of potential oxidant reactive sites (Nel et al., 2006). MWCNT have a high surface area per mass and were biopersistent, with their presence noted in the lung long after initial exposure (Mercer et al., 2010). The fibrous shape and bioactivity of MWCNT are similar to those of asbestos, which has been well characterized with regard to lung disease, and it is hypothesized that MWCNT have similar deleterious effects (Donaldson et al., 2010; Pacurari et al., 2010). Recently, gene signatures associated with these MWCNT-induced inflammatory and fibrotic responses in vivo were noted to involve major signaling pathways for lung injury and carcinogenesis (Pacurari et al., 2011). Pacurari et al. (2011) identified, from a set of biomarkers detecting lung damage, carcinogenesis, tumor progression, and related major signaling pathways genes, a 7-gene inflammatory signature and a 11-gene fibrotic signature, which could separate MWCNT-exposed mouse lungs from control animals over time. Additional gene signatures identified from MWCNT-exposed mouse lungs accurately predicted lung cancer risk and survival in sets of human lung cancer samples (Guo et al., 2012), suggesting the ability to develop a gene signature for early detection of MWCNT exposure and prognosis for MWCNT-induced pathology.
As MWCNT had the ability to bypass the conducting airways, deposit in the alveolar regions, and induce inflammation and fibrosis, this study sought to determine the cellular effects of MWCNT on small airway epithelial cells (SAEC). In addition to the determination of broad cellular effects, a comparison of gene expression in SAEC in vitro studies with previously determined in vivo expression was conducted to find gene expression results concordant in both settings. By using this concordant data, this study proposes that the systematic analysis of MWCNT-induced cellular signaling and gene expression in SAEC could be used to dissect the molecular mechanisms behind MWCNT-induced toxicity in an in vitro manner that would be relevant to the mechanism upon in vivo exposure.
MATERIALS AND METHODS
MWCNT.
MWCNT used in this study were a gift from Mitsui-&-Company (MWCNT-7, lot no. 05072001K28). Characterization of the MWCNT was previously described (Porter et al., 2010). Briefly, the bulk MWCNT exhibited a distinctive crystalline structure with the number of walls ranging from 20 to 50 walls. Overall, MWCNT trace metal contamination was 0.78%, including sodium (0.41%) and iron (0.32%) with no other metals present above 0.02%. Quantitative analysis of TEM micrographs revealed the median length of the MWCNT sample was 3.86 μm (GSD 1.94), and the count mean width was 49±13.4 (SD) nm. The zeta potential of the MWCNT in dispersion media (DM) was determined to be −11 mV.
MWCNT preparation.
For cell culture studies, MWCNT were prepared in DM (Porter et al., 2008). Transmission electron microscopy (TEM) micrographs of MWCNT dispersed in DM demonstrated that DM promotes significant dispersion of MWCNT. Briefly, DM consisted of PBS, pH 7.4, Ca/Mg free supplemented with 5.5mM D-glucose, 0.6mg/ml serum albumin, and 0.01mg/ml 1,2 dipalmitoyl-sn-glycero-3-phosphocholine (DPPC). DPPC was prepared fresh as a 1mg/ml stock solution in absolute ethanol. MWCNT were prepared in DM followed by indirect sonication at 4°C for 5min (Hielscher ultrasonic processor, UIS259L) at amplitutide 100% and cycle 1. Following indirect sonication, the suspension was directly sonicated for 5min at 5W output and 10% duty cycle (Branson Sonifier 450). The stock solution (0.5mg/ml) of MWCNT was kept at 4°C and used within 2–3 weeks. Prior to cell culture experiments, the MWCNT stock solution was directly sonicated for 1min at the setting indicated above.
MWCNT were used at a concentration of 1.2 μg/ml in all experiments except where indicated. Based upon in vivo alveolar surface area, occupationally observed MWCNT airborne concentrations, MWCNT mass median aerodynamic diameter, and minute ventilation, a 1.2 μg/ml concentration of MWCNT was extrapolated to correspond to previously identified in vivo exposures of MWCNT, which induced transient and chronic cellular signaling (Han et al., 2008; Mercer et al., 2010; Porter et al., 2010).
Cell culture.
SAEC were a kind gift from Dr Tom K. Hei (Columbia University, New York, NY) (Piao et al., 2005). SAEC were cultured in serum-free complete SABM medium supplemented with a SAGM SingleQuot kit of growth factors, cytokines, and supplements (Lonza Walkersville, Inc., Walkersville, MD). SAEC were maintained in an incubator at 37°C with 5% CO2.
Western blot analysis.
Whole-cell protein extraction was conducted using RIPA buffer (50mM tris, 150mM NaCl, 10% sodium dodecyl sulfate, 1% Triton X-100, 1% sodium deoxycholate [pH 7.4]) supplemented with 10 µg/ml protease inhibitor cocktail (Sigma, Aldrich, MO) and phosphatase inhibitors. Protein concentrations were determined using a Bio-Rad protein assay (Bio-Rad, Hercules, CA). Protein lysates were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride membrane. The membrane was blocked with 5% bovine serum albumin (Sigma)/PBS/0.1% Tween 20 at room temperature for 1h. The membranes were immunoblotted with primary antibodies against phosphotyrosine and phosphothreonine (Qiagen, CA) in blocking buffer at 4°C overnight at the concentration suggested by the manufacturer. Immunodetection was performed using horseradish peroxidase–labeled secondary antibodies and developed using a chemiluminescent detection system. The signal was captured using X-ray film (Kodak). Experiments were repeated at least thrice independently. Data reported are from one representative experiment. β-Actin was used as a loading control.
Reactive oxygen species measurement by confocal microscopy.
Reactive oxygen species (ROS) measurements by confocal microscopy were performed according to methods previously described (Qian et al., 2005). Briefly, cells were cultured on cover slips and grown until confluent, serum starved overnight, and exposed to 1.2 µg/ml MWCNT for varying time periods. During the exposure period, dihydroethidium (DHE) was added at a final concentration of 5µM for the last 30min of exposure. After incubation, cells were washed twice with PBS (pH 7.4), fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100/PBS, washed thrice with PBS, and mounted on slides with ProLong Gold anti-fade (Invitrogen). A Zeiss LSM 510 microscope was used to obtain images. DHE staining was determined by fluorescence at 546nm. Scale bars were generated and inserted by LSM Zen 2009 Light Edition software.
Cell migration assay.
Cell migration assays were performed according to manufacturer’s protocol (BD Biosciences, Franklin Lakes, NJ) with a slight modification. The top chamber of the transwell was loaded with 0.3ml of 1×106 cells/ml in serum-free media and MWCNT (1.2 µg/ml), and the bottom chamber was loaded with 0.5ml of 0.1% SAEC growth medium, followed by incubation at 37°C in 5% CO2 for 18h. Cell migration assay results were analyzed according to manufacturer’s protocol. Migrated cells were stained with 0.1% crystal violet followed by dye elution in 10% acetic acid. A microplate reader was used to measure the optical density (OD) of the eluted solutions to determine the migration values. The mean values were obtained from five individual experiments and were subjected to a t-test assuming unequal variance. Significance was determined by a p value less than 0.05.
Quantitative real-time PCR.
RNA was isolated from SAEC using Qiagen’s RNAprotect Cell Reagent according to the manufacturer’s protocol (Qiagen). RNA concentration was determined using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA (1 μg) was converted into complementary DNA (cDNA) using a High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Life Technologies, Carlsbad, CA). Thermal cycling conditions were as follows: 25°C for 10min, 2 cycles of 37°C for 60min, and 85°C for 5 s, followed by a programmed hold at 4°C.
Individual quantitative real-time PCR reactions were performed on a 7500 Real-Time PCR system from Applied Biosystems. Each treatment group consisted of three biological replicates. qRT-PCR analysis for each sample was performed in triplicate, and the Ct values obtained were normalized to the 18S housekeeping gene. Gene expression assay numbers are listed in Supplementary table 1. Thermal cycling conditions were as follows: 50°C for 2min, 95°C for 10min, followed by 40 cycles of 95°C for 15 s and 60°C for 10min. Additional PCR reactions using low-density arrays (LDA) were performed on a 7900HT Fast Real-Time PCR System from Applied Biosystems. Each treatment group consisted of two biological replicates and all samples were run in triplicate. Thermal cycling conditions were as follows: 50°C for 2min and 94.5°C for 10min, followed by 40 cycles of 97°C for 30 sand 59.7°C for 1min.
Fold change was determined using the 2–ΔΔCt method with 18S as an internal control. A two-sample t-test using SAS 9.2 was performed to determine statistical significance of p < 0.05.
TEM.
SAEC interaction with and uptake of MWCNT was analyzed by TEM. SAEC were grown to confluence and exposed to MWCNT (1.2 μg/ml) for 15min, 30min, 1h, 2h, 6h, and 22h. Cells were washed thrice with ice-cold Ca2+-/Mg2+-free PBS, scraped from their wells, and harvested by centrifugation at 109 × g for 5min. Cells were fixed in Karnovksy’s fixative (2.5% glutaraldehyde and 3% paraformaldehyde in 0.1M sodium cacodylate, pH 7.4), washed thrice in 0.1M sodium cacodylate, and postfixed in 1% osmium tetraoxide followed by washing with 0.1M sodium cacodylate and distilled water. The cells were dehydrated by sequential washings in 25, 50, and 100% ethanol and then embedded in LX-112 (Ladd, Williston, VT). The ultrathin sections were stained with uranyl acetate and lead citrate and examined with TEM (JEOL 1220, Tokyo).
Ingenuity Pathway Analysis.
Gene expression data were analyzed through the use of Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, www.ingenuity.com). A network/My Pathway is a graphical representation of the molecular relationships between molecules. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Only human molecules were represented. All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Knowledge Base. Nodes are displayed using various shapes that represent the functional class of the gene product. A data set containing gene identifiers and corresponding expression values was uploaded into the application. Each identifier was mapped to its corresponding object in the Ingenuity Knowledge Base. A fold change cutoff of 1.5-fold was set to identify molecules whose expression was significantly differentially regulated. These molecules, called Network Eligible molecules, were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base. Networks of Network Eligible molecules were then algorithmically generated based on their connectivity.
The Functional Analysis of a network identified the biological functions and/or diseases that were most significant to the molecules in the network. The network molecules associated with biological functions and/or diseases in the Ingenuity Knowledge Base were considered for the analysis. Right‐tailed Fisher’s exact test was used to calculate a p value determining the probability that each biological function and/or disease assigned to that network is due to chance alone.
ELISA.
Three biological replicates of SAEC were plated at 60,000 cells per well of a 24-well dish and grown at 37°C for 48h. SAEC were serum starved overnight and exposed to 1.2 μg/ml MWCNT for 24h. Conditioned media from each biological replicate were collected and assayed in triplicate for vascular endothelial growth factor A (VEGFA) and C-C motif chemokine 2 (CCL2) protein expression levels using DuoSet ELISA Development Systems from R&D Systems according to manufacturer’s protocol. Statistical analysis was done using a two-sample t-test assuming unequal variances.
RESULTS
MWCNT Engage SAEC After In Vitro Exposure
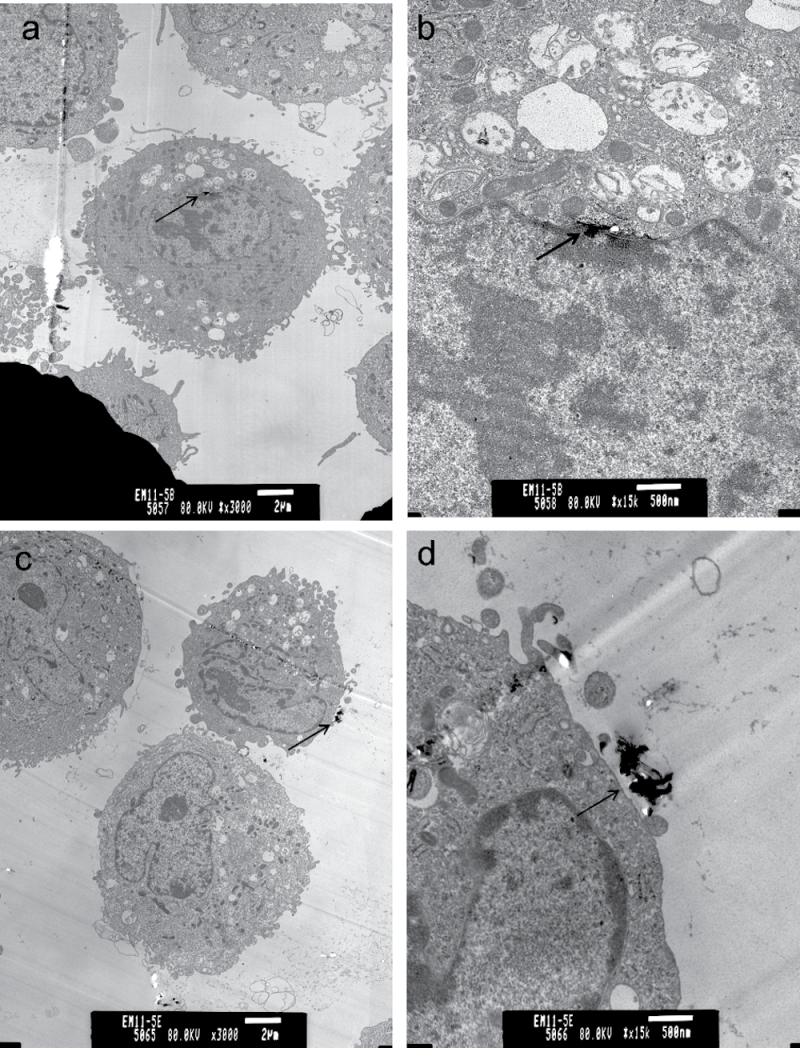
MWCNT were shown to penetrate through the alveolar epithelial cells into the interstitium of the alveolar septa after in vivo exposure (Mercer et al., 2010). To determine whether MWCNT associate with SAEC in vitro, SAEC were grown to a confluent monolayer, exposed to 1.2 μg/ml MWCNT for various time periods, washed thoroughly, and prepared for TEM. Analysis of ultrathin sections of cell preparations suggest that MWCNT enter SAEC after exposure (Figs. 1a and b). Additionally, MWCNT associated with SAEC through the cell periphery even after thorough washing, indicating a direct interaction (Figs. 1c and d).
Fig. 1.
Interaction and uptake of MWCNT by SAEC after exposure. SAEC were exposed to MWCNT for various time periods and prepared for TEM. Representative images show SAEC both internalizing MWCNT after exposure (a, b) and MWCNT interacting with the cell surface (c, d) after thorough washing.
MWCNT Exposure Stimulates ROS Production
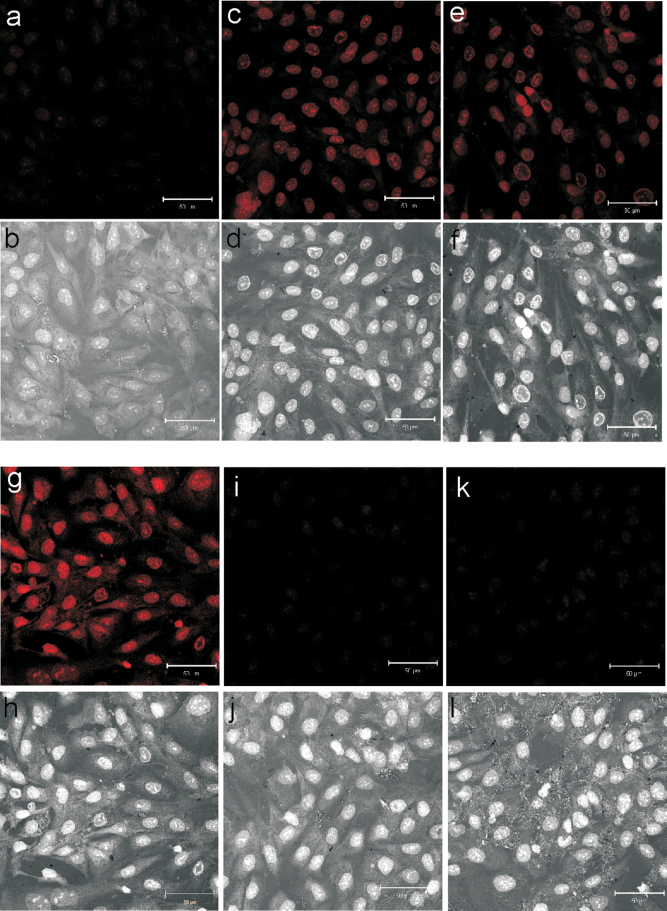
MWCNT induce the production of ROS in a variety of cell types (He et al., 2011; Pacurari et al., 2012; Ye et al., 2009). The potential for MWCNT exposure to produce ROS in SAEC was determined by DHE staining followed by confocal microscopy. SAEC were grown to a confluent monolayer and exposed to 1.2 μg/ml MWCNT for 1, 6, and 24h. DHE was added at a 5μM concentration during the last 30min of exposure. Upon oxidation, DHE exhibited a bright red fluorescence with nuclear localization. Under uniform confocal microscopy parameters, SAEC exhibited an increase in the production of ROS upon increasing exposure time to MWCNT (Figs. 2a–h). To confirm the specificity of ROS production, SAEC were pretreated for 1h with 2000U/ml of an ROS scavenger, catalase, before a 6- and 24-h MWCNT exposure. Catalase reduced the amount of DHE fluorescence produced by MWCNT-exposed SAEC (Figs. 2i–l).
Fig. 2.
MWCNT exposure induces the production of ROS. SAEC were grown on coverslips, serum starved, and treated with MWCNT 1.2 µg/ml for 0h (DM control) (a, b), 1h (c, d), 6h (e, f), and 24h (g, h). Additionally, SAEC were pretreated with 2000U/ml catalase before MWCNT exposure for 1h (i, j) and 6h (k, l). ROS production was determined by DHE incubation during the last 30min of exposure. Following exposure, cells were fixed, permeabilized, and analyzed by confocal microscopy under uniform parameters. Fluorescent images are presented in panels (a, c, e, g, i, and k) to show fluorescent ROS images. Bright field images are presented in panels (b, d, f, h, j, and l) to show that equal numbers of cells were analyzed to determine ROS production. This figure can be viewed in color online.
MWCNT Exposure Induces Cellular Protein Phosphorylation
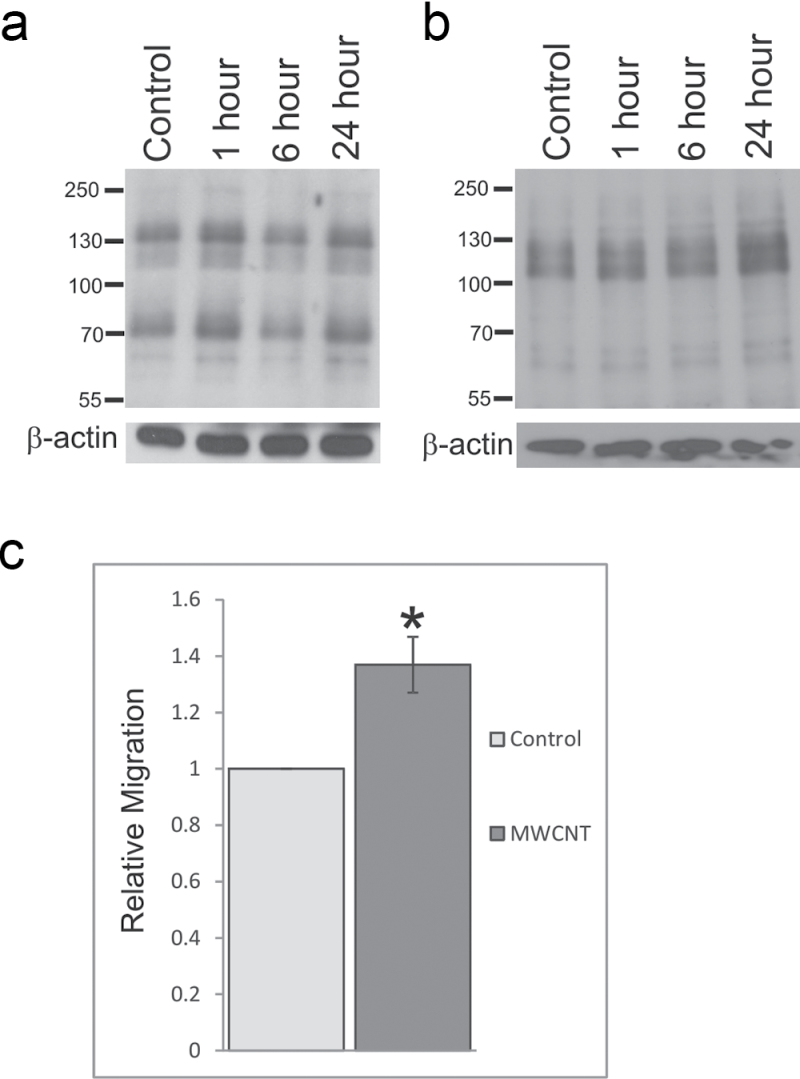
To determine whether exposure to MWCNT induced protein phosphorylation changes indicative of altered cellular signaling, SAEC were exposed to 1.2 μg/ml MWCNT for 1, 6, and 24h. SAEC were lysed in RIPA buffer, and 20 μg of total cellular lysates were resolved by SDS-PAGE and probed for changes in total phosphotyrosine and phosphothreonine levels. MWCNT exposure caused a biphasic increase in overall tyrosine phosphorylation at 1 and 24h (Fig. 3a) and an overall increase in threonine phosphorylation (Fig. 3b) with increasing time of exposure.
Fig. 3.
MWCNT induces epithelial cell activation. SAEC were serum starved, exposed to DM control or 1.2 µg/ml MWCNT for 1, 6, or 24h, and lysed for analysis by SDS-PAGE. Whole-cell lysates were resolved on an 8% polyacrylamide gel and probed for (a) phosphotyrosine or (b) phosphothreonine. β-Actin was used as a loading control. (c) SAEC migration after MWCNT exposure was determined using a modified Boyden chamber. Migrated cells were stained with 0.1% crystal violet, eluted with acetic acid, and signal intensity read on a microplate reader. Data are presented as mean ± SEM of five assays. * Statistically significant at p ≤ 0.05.
MWCNT Exposure Increases Migratory Potential
As MWCNT were previously indicated to affect cytoskeletal remodeling and migration in other cell types (Pacurari et al., 2012), SAEC were seeded into a modified Boyden chamber, exposed to 1.2 μg/ml MWCNT for 18h, and allowed to cross a porous membrane to determine the ability of MWCNT to induce migratory behavior in SAEC. Migrated cells were stained with 0.1% crystal violet and destained in acetic acid, and the OD of the eluted solution was determined by spectrophotometry. SAEC exposed to 1.2 μg/ml MWCNT had significantly (p < 0.05) increased migratory ability compared with control cells as determined by an overall increase in crystal violet staining (Fig. 3c) and cell visualization (data not shown). The invasive potential of SAEC after MWCNT exposure was also studied by invasion through a Matrigel plug in a modified Boyden chamber followed by crystal violet staining, cell visualization, and spectrophotometry. Although an increase in the overall invasive potential of SAEC exposed to MWCNT was noted, these levels were not altered significantly over control (data not shown).
MWCNT Exposure Alters Gene Expression of Prognostic Biomarkers of Lung Inflammation, Fibrosis, and Potential Carcinogenesis
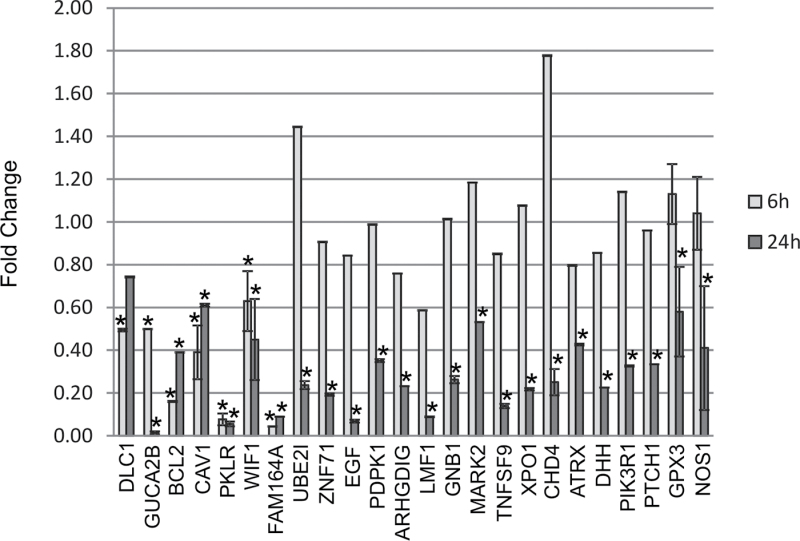
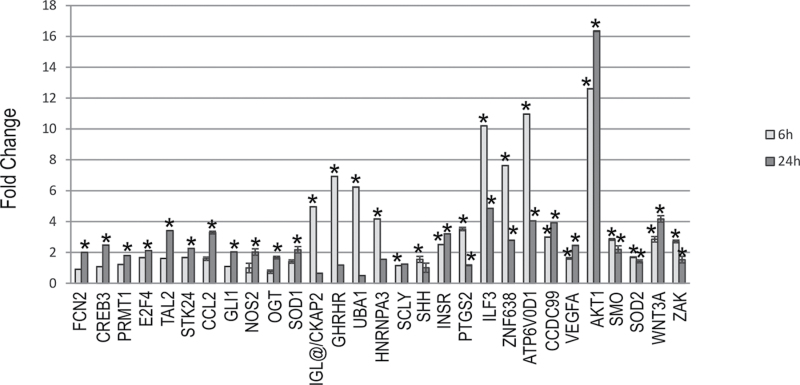
Previous work by our laboratory determined a set of genes that were identified to be potential prognostic biomarkers of lung damage and were able to accurately and specifically predict disease progression in mouse and human samples (Supplementary table 1) (Guo et al., 2006, 2008, 2012; Wan et al., 2010). To determine whether these genes were altered in response to SAEC exposure to MWCNT, differential gene expression was analyzed by quantitative PCR using both TaqMan custom LDA plates and individual assays after 6- and 24-h exposure to MWCNT. Twenty-four genes, dlc1, guca2b, bcl2, cav1, pklr, fam164a, ube2i, znf71, egf, pdpk1, arhgdig, lmf1, gnb1, mark2, tnfsf9, xpo1, chd4, gpx3, nos1, wif1, atrx, dhh, pik3r1, and ptch1, were significantly downregulated (greater than 1.5-fold) at either 6 or 24h or at both 6 and 24h after MWCNT exposure (Fig. 4). Twenty-nine genes, fcn2, creb3, prmt1, e2f4, tal2, stk24, ccl2, gli1, nos2, shh, wnt3a, zak, sod1, ilg@/ckap2, ghrhr, uba1, hnrnpa3, scly, insr, ptgs2, ilf3, znf638, atp6v0d1, ccdc99, vegfa, akt1, sod2, ogt, and smo, were significantly upregulated (greater than 1.5-fold) at either 6 or 24h or at both 6 and 24h after MWCNT exposure (Fig. 5). Nineteen genes, mapk1/erk2, cfhr2, atp5a1, atp8a2, fut7, arhgap19, mt3, vac14, ahnak, akap13, msx2, rer1, tuba1a, sod3, mapk3/erk1, nsfl, nos3, ihh, and smpd1 showed no significant change at any time point after exposure (data not shown). Control gene 18S values were not affected by MWCNT exposure (Supplementary fig. 1).
Fig. 4.
MWCNT exposure results in downregulation of gene expression. SAEC were exposed to DM control or 1.2 μg/ml MWCNT for 6 and 24h. Total RNA was collected, cDNA created, and qRT-PCR performed to analyze gene expression changes after MWCNT exposure. Twenty-four genes showed significant downregulation at one or both of the 6- and 24-h time points.
Fig. 5.
MWCNT exposure results in upregulation of gene expression. SAEC were exposed to DM control or 1.2 μg/ml MWCNT for 6 and 24h. Total RNA was collected, cDNA created, and qRT-PCR performed to analyze gene expression changes after MWCNT exposure. Twenty-nine genes showed significant upregulation at one or both of the 6- and 24-h time points.
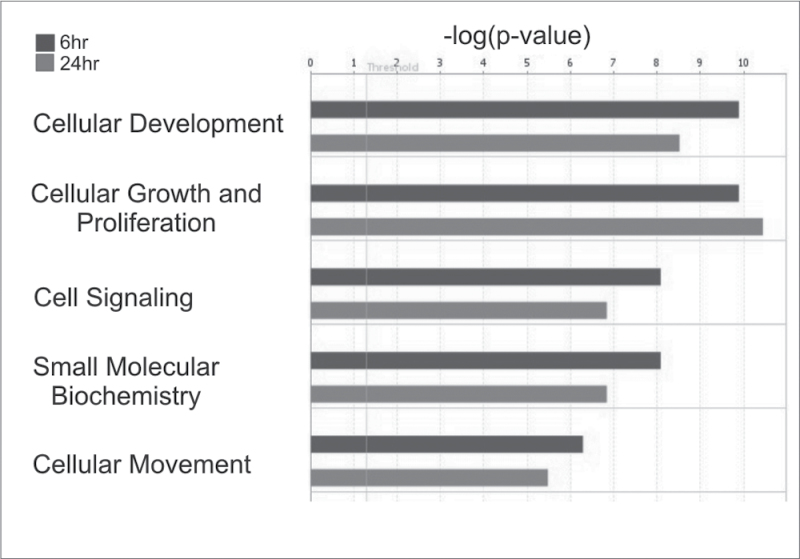
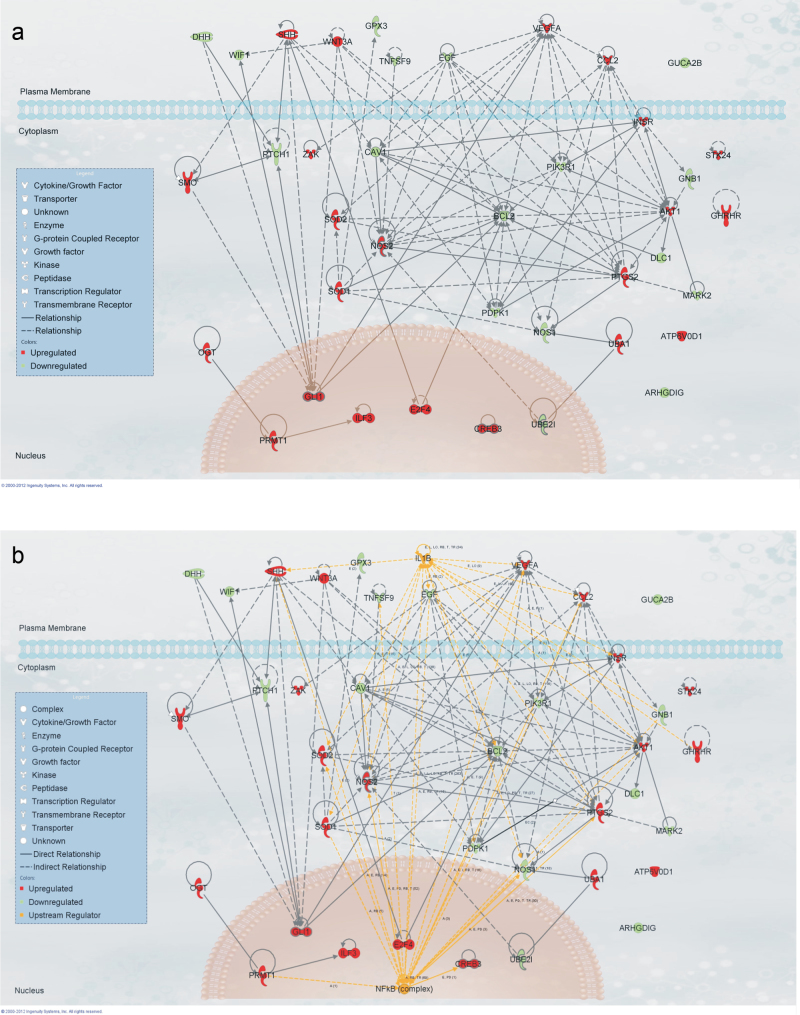
A Functional Analysis of both up- and downregulated genes with a significant fold change of 1.5-fold through IPA revealed significantly overrepresented biological functions in the gene set, which were most significant to the molecules in the network. The overall top five predicted biological functions at both 6 and 24h (cellular development, cellular growth and proliferation, cell signaling, small molecule biochemistry, and cellular movement) are shown in Figure 6. To establish the interaction of the genes in these functions so as to determine potential mechanistic pathways for future study, all genes in the five significant functions were added to a pathway in IPA and connected using the Build-Connect tool (Fig. 7a). An Upstream Regulator Analysis employing these significant genes predicted the active involvement of two potential upstream regulators, the NFκB complex and IL1β (Fig. 7b). These potential upstream regulators of the significantly altered genes may be used for future elucidation of the mechanistic pathways behind MWCNT exposure. Comparing mRNA expression in SAEC after MWCNT exposure and gene expression in mouse lungs following MWCNT aspiration analyzed by both microarray or RT-PCR (Pacurari et al., 2011) (Table 1, Supplementary table 2), a set of the biomarker genes was concordant between in vivo and in vitro expression. The study of these concordant genes could be used to determine the mechanism behind MWCNT-induced toxicity in an in vitro manner that would be relevant to in vivo exposure.
Fig. 6.
Altered gene expression indicates the activation of multiple biological functions. The top five significantly altered biological functions based upon gene expression direction at both 6 and 24h were determined by IPA.
Fig. 7.
IPA analysis of significantly activated functions. (a) All genes in the predicted activated functions were connected by direct and indirect interactions using the Build-Connect tool. (b) Significant upstream regulators of the predicted activated functions were determined by IPA and connected to the gene set by both direct and indirect interactions.
Table 1.
Comparison of MWCNT-Induced In Vivo and In Vitro Gene Expression Changes
| Gene symbol | Gene name | Function | Expression in vivo (RT-PCR) | Expression in vivo (Microarray) | Expression in SAEC |
|---|---|---|---|---|---|
| Bcl2 | Apoptosis-regulator Bcl2 | Suppresses apoptosis | Decrease | No significant change | Decrease |
| Cav1 | Caveolin-1 | Scaffolding protein | Decrease | Decrease | Decrease |
| Ccdc99 | Coiled-coil domain containing 99 | Mitotic spindle orientation | Increase | Increase | Increase |
| Ccl2 | C-C motif chemokine 2 | Monoctye and basophil chemoattraction | No significant change | Increase | Increase |
| Cdh4 | Chromodomain helicase DNA binding protein-4 | Transcription regulation | No significant change | Decrease | Decrease |
| Ckap2 | Cytoskeletal-associated protein 2 | Microtubule stabilization | No significant change | Increase | Increase |
| Dhh | Desert hedgehog protein | Cell-cell signaling, development | Decrease | No significant change | Decrease |
| Gli1 | Glioma-associated oncogene-1 | DNA binding, protein binding | No significant change | Increase | Increase |
| Gpx3 | Glutathione peroxidase 3 | Oxidative damage protection | Decrease | No significant change | Decrease |
| Nos1 | Nitric oxide synthase, brain | Nitric oxide signaling | No significant change | Decrease | Decrease |
| Nos2 | Nitric oxide synthase, inducible | Nitric oxide signaling | Increase | No significant change | Increase |
| Pik3r1 | Phosphatidyl inositol 3-kinase regulatory subunit alpha | Protein adapter | Decrease | No significant change | Decrease |
| Prmt1 | Protein arginine methyltransferase -1 | Transferase histone methyltransferase | No significant change | Increase | Increase |
| Ptch1 | Protein patched homolog 1 | Receptor, tumor suppressor | Decrease | No significant change | Decrease |
| Shh | Sonic hedgehog protein | Transcription | Increase | Increase | Increase |
| Sod2 | Super oxide dismutase [Mn], mitochondrial | Radical scavenger | No significant change | Increase | Increase |
| Tal2 | T-cell acute lymphocytic leukemia-2 | DNA binding, protein binding | No significant change | Increase | Increase |
| Wif1 | Wnt inhibitory factor 1 | Inhibits Wnts | Decrease | Decrease/Increasea | Decrease |
| Xpo1 | Exportin-1 | Protein transport | No significant change | Decrease | Decrease |
Notes. Gene symbol, gene name, function, and expression direction of genes, which correlated between SAEC in vitro exposure and in vivo microarray analysis and RT-PCR. Gene symbol, name, and function were derived from UniProtKB (www.uniprot.org). In vivo expression was determined by microarray analysis and RT-PCR as described in Pacurari et al. (2011).
a wif1 expression decreased on day 1 dose 40 and day 7 dose 40. wif1 expression increased on day 56 dose 80.
MWCNT Alters Protein Expression of Inflammatory Markers
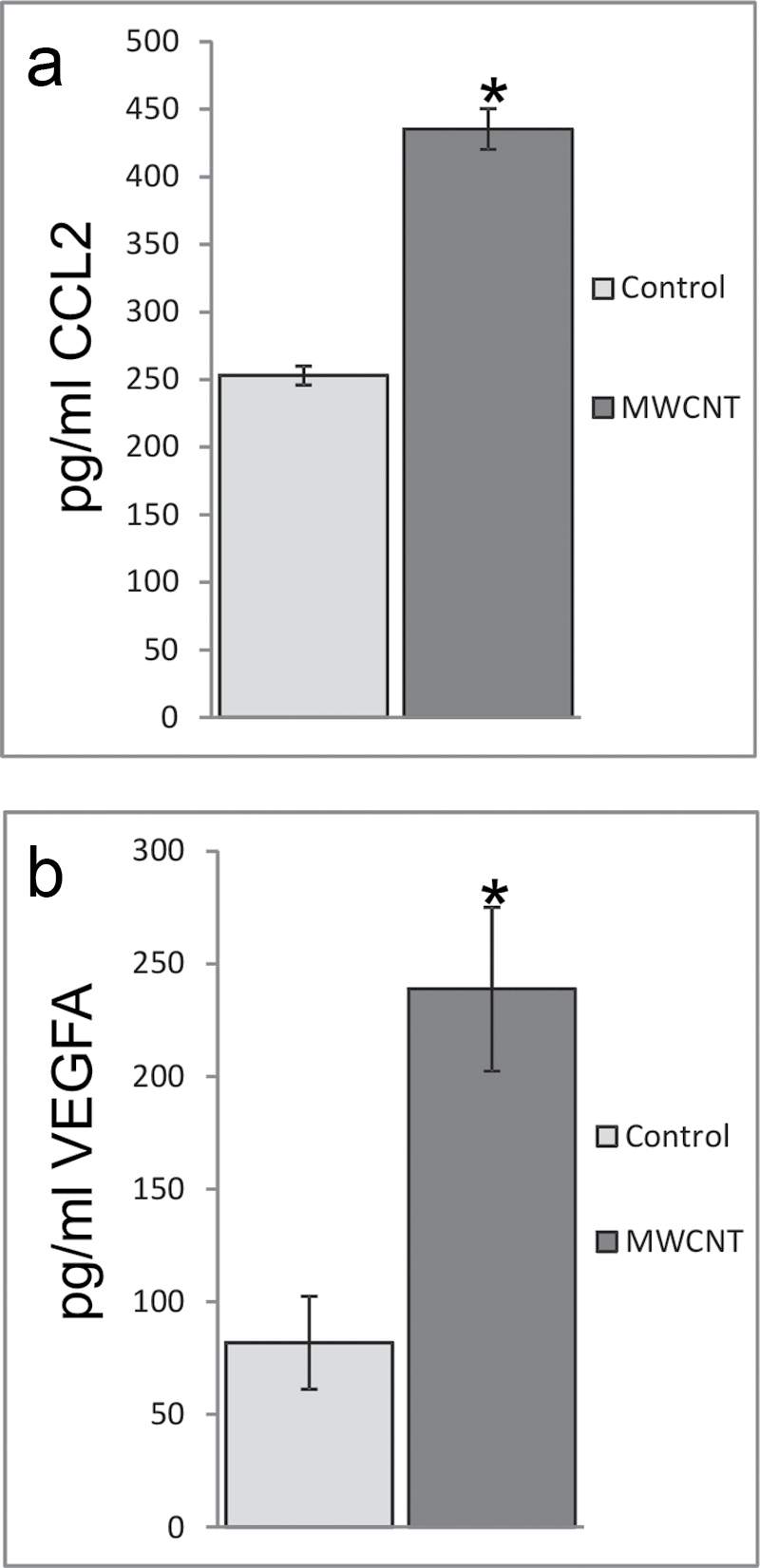
To further examine the predicted changes in mRNA expression and their association to protein expression, two genes, ccl2 and vegfa, were analyzed for their concurrent protein levels after MWCNT exposure. Conditioned media were collected from SAEC after exposure to 1.2 μg/ml MWCNT for 24h and analyzed by ELISA. CCL2 protein levels increased significantly from 253 pg/ml in control to 435 pg/ml after 24h of exposure (Fig. 8a), which was concordant with a significant increase in mRNA expression at 24h (Fig. 5). VEGFA protein levels increased significantly from 82 pg/ml in control to 239 pg/ml after 24-h exposure to MWCNT (Fig. 8b), which was also concordant with a significant increase in mRNA expression at 24h (Fig. 5).
Fig. 8.
MWCNT exposure induces protein expression of VEGFA and CCL2. ELISA analysis of cell supernatants after MWCNT exposure for 24h indicated an increase in (a) CCL2 protein expression (control: 253±7 pg/ml; MWCNT: 435±15 pg/ml) and (b) VEGFA protein expression (control: 82±21 pg/ml; MWCNT: 239±36 pg/ml). *p < 0.05.
DISCUSSION
MWCNT have many unique physicochemical properties that make them desirable for both occupational and commercial applications. Due to their potential to easily aerosolize, respiratory exposure to MWCNT remains a predominant hazard to the growing field, and the resulting cellular effects in the lung must be determined.
Composed of a variety of cells with diverse functions, the airway epithelium acts as a first line of defense to the environment to maintain normal airway function and structural integrity. The conducting airways consist of ciliated epithelial cells and mucous secreting cells, which sweep inhaled particles and invading pathogens from the lung, whereas the alveolar region is involved in gas exchange. Upon injury, the airway epithelium releases inflammatory mediators to trigger an immune response and result in clearance of the offending material (Tam et al., 2011).
Initial injury of the airway epithelium results in a rapid response coordinated between the epithelium itself, the immune system, and the underlying extracellular matrix (ECM) networks and endothelium. Immune cells extravasate through the vascular endothelium and engage the injurious particle, after which the basement membrane is re-established and acts as a scaffold onto which dedifferentiated epithelial cells spread and redifferentiate to rebuild an intact epithelial barrier (Strieter, 2008). On the occasion of repetitive injury, a state of chronic inflammation can develop in which there is a loss of basement membrane structure, loss of airway epithelial and vascular endothelial architecture, and proliferation of fibroblasts with aberrant ECM deposition through which pulmonary fibrosis may occur (Strieter, 2008). This loss of airway architecture coupled with increased ECM production results in thickening of the airways and difficulty breathing as gas exchange between the lung and vasculature is compromised.
As the epithelial lining is the first line of defense after inhalation exposure to MWCNT, and MWCNT have been shown previously to penetrate alveolar epithelial cells and enter the interstitium of alveolar septa in vivo (Mercer et al., 2010), this study sought to determine whether MWCNT had the ability to activate SAEC, resulting in changes to cellular signaling and gene expression. It was noted that SAEC had the ability to both internalize MWCNT and associate with MWCNT through the cell membrane in vitro. In vivo, MWCNT exposure resulted in an initial inflammatory response that progressed to a fibrotic state as witnessed by a gradual thickening of the alveolar connective tissue and hyperplasia of alveolar epithelial cells (Mercer et al., 2011; Porter et al., 2010). The association between SAEC and MWCNT in vitro suggests that MWCNT may elicit a similar cellular response to that seen in vivo, and this interaction may initiate toxicological responses following MWCNT exposure, which are relevant to in vivo exposure.
ROS are well established in their role in the immune response and play a key role in cellular signaling. A delicate balance must be maintained between beneficial levels of ROS production and oxidative stress, which may result in potential disease states and cell death (Ma, 2010). Excessive ROS production may elicit unwanted cellular signaling and DNA damage, which may in turn increase the amount of ROS produced and therefore enter the cell into a cycle of excess ROS production. The increased production of ROS in SAEC with increasing time of MWCNT exposure suggests a potential for protein and DNA damage and oxidative stress and altered cellular signaling (Lipinski, 2011; Ma, 2010). MWCNT, by virtue of their nanoscale size, have a large surface area to mass ratio, which allows for many surface reactant sites that may interact with cells to generate ROS. ROS production was suggested to be a considerable contributing factor to inflammation and disease progression in multiple in vitro and in vivo studies. Indeed, ROS production has been proposed to be a major source of nanoparticle-induced toxicity (Gwinn and Vallyathan, 2006; Nel et al., 2006). The interaction of iron with hydrogen peroxide, the Fenton reaction, has the ability to produce hydroxyl radicals. MWCNT, by nature of their production, contain metal contaminants, and it was suggested that ROS production was due to metal contaminants intrinsic to the material (Pulskamp et al., 2007). The MWCNT used in the current study contained only a trace amount of iron (0.32%), and these MWCNT produced no detectable •OH in the presence of H2O2 in an acellular Fenton reaction ESR experiment (data not shown). Iron contamination in the MWCNT preparations, therefore, was not a significant source of ROS. ROS production could consequently be attributed to the cellular association with and subsequent response to MWCNT and may play a role in both potential oxidative stress of the cell and cellular signaling.
Integral to cellular signaling is the ability of a cell to respond to a cue and produce a signal in response that is highly regulated in specificity, reversibility, and temporality. Cellular protein phosphorylation is one example of a precisely regulated protein-protein binding mechanism. The phosphorylation of tyrosine, followed by its selective binding to preferred domain sequences, allows for the assembly and control of complex signaling cascades (Pawson, 2004). Phosphorylation and dephosphorylation of tyrosine residues have been well documented as integral to dynamic cellular signaling (Hunter, 2009). Additionally, phosphorylation of serine and threonine residues is also necessary for the formation of complex signaling cascades and specific interactions between proteins (Yaffe and Elia, 2001). The ability of MWCNT to increase the overall phosphorylation state of phosphotyrosine and phosphothreonine suggests alteration of cellular signaling pathways and activation of SAEC in response to MWCNT exposure.
One of the main functions of type II alveolar cells is conservation of the lung epithelial lining. Whether by normal turnover or proliferation and transdifferentiation after epithelial damage, type II alveolar cells are responsible for restoration of the lung epithelium after injury for efficient gas exchange. Motility of type II cells is necessary for maintenance of the lung epithelial lining (Mason, 2006). The ability of SAEC to respond to MWCNT exposure with an increase in cellular migration was an additional indication of an increase in cellular signaling and a concerted response to epithelial injury.
In addition to the normal wound healing process, inflammation and the subsequent progression to fibrosis were common occurrences after pulmonary exposure to fibrous material, and MWCNT in a mouse model induced these disease states (Porter et al., 2010). Recent work suggests a correlation between gene expression in MWCNT-exposed mouse lungs and previously identified prognostic biomarkers of lung damage, carcinogenesis, and tumor progression. Identified by genome-wide expression profiling of non–small cell lung cancer tumor tissues, these biomarkers were prognostic of patient survival, indicating an association with tumor progression and the risk for developing metastasis and recurrence in lung cancer tumors (Guo et al., 2006, 2008; Wan et al., 2010). These genes also showed differential expression in lung cancer tumor versus normal tissues, suggesting their potential involvement in carcinogenesis. For example, ccdc99 was overexpressed in human lung tumors versus normal lung tisssues and in MWCNT-treated in vivo animal lung tissues (Pacurari et al., 2011) and in vitro SAEC cells (Table 1). As the results of MWCNT-induced phosphorylation and kinase activation had implications for carcinogenesis and signaling pathway activation, it was important to examine MWCNT-induced expression changes in these lung cancer biomarkers and genes in the related signaling pathways. Both up- and downregulation of these prognostic genes were noted in SAEC after exposure to MWCNT. Table 2 details the signaling pathways highlighted in this article, ROS generation, cell motility, and protein phosphorylation, and those disease states affected by MWCNT exposure, inflammation, and fibrosis, with regard to those prognostic biomarkers with significant alteration following MWCNT exposure. The use of IPA to determine significant network functions, connected pathways, and upstream regulators for the combination of up- and downregulated genes provided a comprehensive picture of the interaction of the genes with each other and their link to signaling pathways. These signaling pathways could be extrapolated from IPA and subsequently tested in vitro to determine the mode of action of MWCNT exposure and the downstream cellular effects. These significant genes were concordant with gene expression in mouse lungs after MWCNT exposure. Therefore, exposure of SAEC to MWCNT may help to identify cellular toxicity mechanisms of MWCNT exposure and potential gene signatures for risk assessment.
Table 2.
Function and Disease Overlay function of IPA Was Used to Determine Those Prognostic Genes With a Significant Up- or Downregulation, Which Have a Role in ROS Production, Cellular Migration, Phosphorylation, Fibrosis, Inflammation, and Adenocarcinoma
| IPA function and disease | Genes |
|---|---|
| Free radical production—Production of ROS | il1β, insr, cav1, sod2, egf, sod1, nos2, bcl2, nos1, pik3r1, akt1 |
| Cellular movement—Migration of cells | shh, wnt3a, smo, tnfsf9, egf, cav1, sod2, sod1, nos2, gli1, pdpk1, bcl2, creb3, ube2i, ptgs2, pik3r1, dlc1, mark2, stk24, akt1, insr, ccl2, vegfa, il1β, nos1 |
| Posttranslational modification—Phosphorylation | stk24, insr, mark2, akt1, pik3r1, bcl2, pdpk1, il1β, egf, vegfa, zak, ccl2 |
| Organismal injury and abnormalities—Fibrosis | ccl2, egf, dhh, cav1, sod2, nos2, nos1, ptgs2, vegfa, il1β |
| Inflammatory response—Inflammation | ccl2, sod2, sod1, cav1 |
| Cancer—Adenocarcinoma | vegfa, gpx3, shh, smo, ptch1, zak, cav1, nos2, gli1, bcl2, ptgs2, dlc1, akt1, pik3r1 |
In summary, MWCNT exposure in vitro induced changes to cellular signaling and gene expression in SAEC. ROS production, as well as alterations in the phosphorylation status of tyrosine and threonine and an increase in migratory ability, suggests activation of SAEC signaling pathways following MWCNT exposure. Gene and protein expression analyses following MWCNT exposure indicate integral cellular changes. The use of IPA to analyze the altered gene expression provided a comprehensive evaluation of significantly perturbed signaling pathways and potential disease mechanisms for future evaluation. Assessment of the gene expression changes in these prognostic biomarkers in cell culture suggests the ability to determine the underlying basis of toxicity due to MWCNT exposure in vitro and provides a basis for the identification of potential prognostic gene signatures for MWCNT risk assessment.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences at the National Institutes of Health (RO1 ES021764 to N.L.G.).
Supplementary Material
REFERENCES
- Ajayan P. M. (1999). Nanotubes from carbon. Chem. Rev. 99, 1787–1800 [DOI] [PubMed] [Google Scholar]
- Castranova V. (2011). Overview of current toxicological knowledge of engineered nanoparticles. J. Occup. Environ. Med. 53 (6 Suppl), S14–S17 [DOI] [PubMed] [Google Scholar]
- Donaldson K., Murphy F. A., Duffin R., Poland C. A. (2010). Asbestos, carbon nanotubes and the pleural mesothelium: A review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma. Part. Fibre Toxicol. 7, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ma Y., Ward R., Castranova V., Shi X., Qian Y. (2006). Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin. Cancer Res. 12(11 Pt 1), 3344–3354 [DOI] [PubMed] [Google Scholar]
- Guo N. L., Wan Y. W., Denvir J., Porter D. W., Pacurari M., Wolfarth M. G., Castranova V., Qian Y. (2012). Multiwalled carbon nanotube-induced gene signatures in the mouse lung: Potential predictive value for human lung cancer risk and prognosis. J. Toxicol. Environ. Health Part A. 75, 1129–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N. L., Wan Y. W., Tosun K., Lin H., Msiska Z., Flynn D. C., Remick S. C., Vallyathan V., Dowlati A., Shi X., et al. (2008). Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin. Cancer Res. 14, 8213–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn M. R., Vallyathan V. (2006). Nanoparticles: Health effects–pros and cons. Environ. Health Perspect. 114, 1818–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. H., Lee E. J., Lee J. H., So K. P., Lee Y. H., Bae G. N., Lee S. B., Ji J. H., Cho M. H., Yu I. J. (2008). Monitoring multiwalled carbon nanotube exposure in carbon nanotube research facility. Inhal. Toxicol. 20, 741–749 [DOI] [PubMed] [Google Scholar]
- He X., Young S. H., Schwegler-Berry D., Chisholm W. P., Fernback J. E., Ma Q. (2011). Multiwalled carbon nanotubes induce a fibrogenic response by stimulating reactive oxygen species production, activating NF-κB signaling, and promoting fibroblast-to-myofibroblast transformation. Chem. Res. Toxicol. 24, 2237–2248 [DOI] [PubMed] [Google Scholar]
- Hunter T. (2009). Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima S. (1991). Helical microtubules of graphitic carbon. Nature. 354, 56–58 [Google Scholar]
- Lipinski B. (2011). Hydroxyl radical and its scavengers in health and disease. Oxid. Med. Cell. Longev. 2011, 809696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. (2010). Transcriptional responses to oxidative stress: Pathological and toxicological implications. Pharmacol. Ther. 125, 376–393 [DOI] [PubMed] [Google Scholar]
- Mason R. J. (2006). Biology of alveolar type II cells. Respirology. 11(Suppl), S12–S15 [DOI] [PubMed] [Google Scholar]
- Mason R. P., Hanna P. M., Burkitt M. J., Kadiiska M. B. (1994). Detection of oxygen-derived radicals in biological systems using electron spin resonance. Environ. Health Perspect. 102(Suppl. 10), 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Hubbs A. F., Scabilloni J. F., Wang L., Battelli L. A., Friend S., Castranova V., Porter D. W. (2011). Pulmonary fibrotic response to aspiration of multi-walled carbon nanotubes. Part. Fibre Toxicol. 8, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R. R., Hubbs A. F., Scabilloni J. F., Wang L., Battelli L. A., Schwegler-Berry D., Castranova V., Porter D. W. (2010). Distribution and persistence of pleural penetrations by multi-walled carbon nanotubes. Part. Fibre Toxicol. 7, 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nel A., Xia T., Mädler L., Li N. (2006). Toxic potential of materials at the nanolevel. Science. 311, 622–627 [DOI] [PubMed] [Google Scholar]
- Pacurari M., Castranova V., Vallyathan V. (2010). Single- and multi-wall carbon nanotubes versus asbestos: Are the carbon nanotubes a new health risk to humans?. J. Toxicol. Environ. Health Part A. 73, 378–395 [DOI] [PubMed] [Google Scholar]
- Pacurari M., Qian Y., Fu W., Schwegler-Berry D., Ding M., Castranova V., Guo N. L. (2012). Cell permeability, migration, and reactive oxygen species induced by multiwalled carbon nanotubes in human microvascular endothelial cells. J. Toxicol. Environ. Health Part A. 75, 129–147 [DOI] [PubMed] [Google Scholar]
- Pacurari M., Qian Y., Porter D. W., Wolfarth M., Wan Y., Luo D., Ding M., Castranova V., Guo N. L. (2011). Multi-walled carbon nanotube-induced gene expression in the mouse lung: Association with lung pathology. Toxicol. Appl. Pharmacol. 255, 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. (2004). Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 116, 191–203 [DOI] [PubMed] [Google Scholar]
- Piao C. Q., Liu L., Zhao Y. L., Balajee A. S., Suzuki M., Hei T. K. (2005). Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis. 26, 725–731 [DOI] [PubMed] [Google Scholar]
- Porter D. W., Hubbs A. F., Mercer R. R., Wu N., Wolfarth M. G., Sriram K., Leonard S., Battelli L., Schwegler-Berry D., Friend S., et al. (2010). Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology. 269, 136–147 [DOI] [PubMed] [Google Scholar]
- Porter D., Sriram K., Wolfarth M., Jefferson A., Schwegler-Berry D., Andrew M., Castranova V. (2008). A biocompatible medium for nanoparticle dispersion. Nanotoxicology. 2, 144–154 [Google Scholar]
- Pulskamp K., Diabaté S., Krug H. F. (2007). Carbon nanotubes show no sign of acute toxicity but induce intracellular reactive oxygen species in dependence on contaminants. Toxicol. Lett. 168, 58–74 [DOI] [PubMed] [Google Scholar]
- Qian Y., Liu K. J., Chen Y., Flynn D. C., Castranova V., Shi X. (2005). Cdc42 regulates arsenic-induced NADPH oxidase activation and cell migration through actin filament reorganization. J. Biol. Chem. 280, 3875–3884 [DOI] [PubMed] [Google Scholar]
- Strieter R. M. (2008). What differentiates normal lung repair and fibrosis? Inflammation, resolution of repair, and fibrosis. Proc. Am. Thorac. Soc. 5, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A., Wadsworth S., Dorscheid D., Man S. F., Sin D. D. (2011). The airway epithelium: More than just a structural barrier. Ther. Adv. Respir. Dis. 5, 255–273 [DOI] [PubMed] [Google Scholar]
- Wan Y. W., Sabbagh E., Raese R., Qian Y., Luo D., Denvir J., Vallyathan V., Castranova V., Guo N. L. (2010). Hybrid models identified a 12-gene signature for lung cancer prognosis and chemoresponse prediction. PLoS ONE. 5, e12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. B., Elia A. E. (2001). Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13, 131–138 [DOI] [PubMed] [Google Scholar]
- Yakobson B. I., Smalley R. E. (1997). Fullerene nanotubes: C-1000000 and beyond. Am. Scientist. 85, 324–337 [Google Scholar]
- Ye S. F., Wu Y. H., Hou Z. Q., Zhang Q. Q. (2009). ROS and NF-kappaB are involved in upregulation of IL-8 in A549 cells exposed to multi-walled carbon nanotubes. Biochem. Biophys. Res. Commun. 379, 643–648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.