Abstract
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)–elicited time- and dose-dependent differential gene expression was compared in human, mouse, and rat primary hepatocytes. Comprehensive time course (10 nM TCDD or dimethyl sulfoxide vehicle control for 1, 2, 4, 8, 12, 24, and 48h) studies identified 495, 2305, and 711 differentially expressed orthologous genes in human, mouse, and rat hepatocytes, respectively. However, only 16 orthologs were differentially expressed across all three species, with the majority of orthologs exhibiting species-specific expression (399 human, 2097 mouse, and 533 rat), consistent with species-specific expression reported in other in vitro and in vivo comparative studies. TCDD also elicited the dose-dependent induction of 397 human, 100 mouse, and 443 rat genes at 12h and 615 human, 426 mouse, and 314 rat genes at 24h. Comparable EC50 values were obtained for AhR battery genes including Cyp1a1 (0.1 nM human, 0.05 nM mouse, 0.08 nM rat at 24h) and Tiparp (0.97 nM human, 0.63 nM mouse, 0.14 nM rat at 12h). Overrepresented functions and pathways included amino acid metabolism in humans, immune response in mice, and energy homeostasis in rats. Differentially expressed genes functionally associated with lipid transport, processing, and metabolism were overrepresented in all three species but exhibited species-specific expression consistent with the induction of hepatic steatosis in mice but not in rats following a single oral gavage of TCDD. Furthermore, human primary hepatocytes showed lipid accumulation following 48h of treatment with TCDD, suggesting that AhR-mediated steatosis in mice more closely resembles human hepatic fat accumulation compared with that in rats. Collectively, these results suggest that species-specific gene expression profiles mediate the species-specific effects of TCDD despite the conservation of the AhR and its signaling mechanism.
Key Words: dioxin, microarray, primary hepatocyte, interspecies.
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) elicits a broad spectrum of species-specific effects including endocrine disruption, hepatotoxicity, and immunotoxicity that are mostly mediated by the aryl hydrocarbon receptor (AhR) (Denison and Heath-Pagliuso, 1998; Gonzalez and Fernandez-Salguero, 1998; Hankinson, 1995; Poland and Knutson, 1982; Vorderstrasse et al., 2001). Briefly, ligand binding causes a conformational change that results in the dissociation of the chaperone protein complex and translocation of the AhR to the nucleus where it heterodimerizes with the AhR nuclear translocator (ARNT). The AhR:ARNT heterodimer binds to dioxin response elements (DREs) in the regulatory regions of target genes and affects their transcriptional response (Denison et al., 2002; Hankinson, 1995). Recent studies also suggest that noncanonical mechanisms independent of DREs serve an underappreciated role in AhR-mediated effects (Beischlag et al., 2008; Denison et al., 2011; Dere et al., 2011a, c; Huang and Elferink, 2012; Patel et al., 2009; Tanos et al., 2012a, b).
With the exception of polymorphic differences in selected strains (Okey et al., 2005), AhR structure, TCDD binding affinity, and complex stability are comparable across species (Denison et al., 1986; Okey et al., 1989; Olson et al., 1980; Poland et al., 1976). For example, human and mouse AhRs exhibit 87% amino acid similarity, with the rat AhR having comparable similarity. Despite its evolutionarily conserved mechanism of action, structure, and function, the AhR mediates a broad spectrum of species-specific responses (Boverhof et al., 2006; Flaveny et al., 2010; Schmidt and Bradfield, 1996) that not only confound extrapolations in support of human risk assessment but also challenge the assumption of a common mechanism of toxicity for TCDD and related compounds beyond ligand binding.
Significant differences in TCDD sensitivity exist between species, with LD50s ranging from 1 μg/kg for guinea pigs to > 5000 μg/kg for hamsters, whereas mice (~200 μg/kg) and rats (~30 μg/kg) exhibit LD50s in the mid range (McConnell, 1985). Hepatic responses including species-specific histopathology and metabolite, gene expression, and serum biochemistry changes have been observed in response to TCDD (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Forgacs et al., 2012; Silkworth et al., 2005). Comparative in vitro and in vivo toxicogenomic studies report TCDD-elicited lipid accumulation consistent with AhR-mediated hepatic steatosis in mice but not in rats (Boverhof et al., 2005; Forgacs et al., 2012). Complementary differential gene expression profiles suggest that TCDD-elicited hepatic steatosis in mice is due to the downregulation of de novo fatty acid (FA) biosynthesis with the induction of hepatic lipid uptake and metabolism of saturated FAs (SFAs) to mono-unsaturated FAs (MUFAs) and poly-unsaturated FAs (PUFAs) (Angrish et al., 2012b; Forgacs et al., 2012). Although a single oral gavage of TCDD also elicited an increase in relative liver weight in rats, histopathology and gene expression studies are consistent with hepatocyte hypertrophy with only modest hepatic lipid accumulation (Boverhof et al., 2006; Forgacs et al., 2012). Unfortunately, comparative studies examining the relevance of these differences in rodent hepatic responses to human models are lacking.
Computational genome-wide DRE analyses in the human, mouse, and rat genomes also indicate species-specific DRE distribution that may further contribute to species-specific AhR-mediated gene expression and metabolite profiles (Dere et al., 2011a; Sun et al., 2004). For example, < 15% of TCDD-responsive orthologous genes in C57BL/6 mouse liver are also differentially expressed in Sprague Dawley rats (Boverhof et al., 2006). Likewise, comparisons between human HepG2, mouse Hepa1c1c7, and rat H4IIE hepatoma cells report < 8% of differentially expressed orthologs are conserved, with examples of divergent regulation (e.g., ortholog induced in one species, repressed in another) despite the conserved induction of “AhR battery” genes (e.g., Cyp1a1, Cyp1a2, and Tiparp) (Dere et al., 2011b; Dere et al., 2006; Nebert et al., 2000). However, the significance of divergent gene expression between hepatoma cell lines and their relevance to human toxicity are debatable due to possible mutation differences, genetic instability, and clonal selection under differing culture conditions.
In order to further investigate the human relevance of TCDD-elicited hepatic steatosis and metabolic disruption in rodents, systematic time- and dose-dependent whole-genome transcriptomic comparisons were conducted in human, mouse, and rat primary hepatocytes. Overall, TCDD elicited species-specific hepatic gene expression profiles, consistent with reported in vitro and in vivo studies. Conserved overrepresented functions and pathways were also identified including lipid transport, processing, and metabolism. However, further examination revealed species-specific differences within the orthologs comprising lipid transport, processing, and metabolism gene expression that contribute to TCDD-elicited hepatic steatosis in mice but not in rats. Moreover, comparisons to human primary hepatocytes suggest that TCDD-induced hepatic FA accumulation in mice may be more similar to human responses compared with rats.
Materials and Methods
Primary cell culture and treatment.
Cancer-free human, mouse, and rat primary hepatocytes preplated in six-well plates with collagen type I substratum and Matrigel overlay were obtained from CellzDirect (Invitrogen, Carlsbad, CA). Human primary hepatocytes were isolated from cancer-free portions of a hepatic resection as determined by the vendor. All donors were Caucasian, postmenopausal, alcohol-free, nonsmoking females with body mass indexes < 30, experiencing cancer at the time of hepatic resection. Three independent biological replicates were used for all human time course and dose-response studies. Rodent primary hepatocytes were pooled from four 12-week-old CD-1 male mice and two 9-week-old male Sprague Dawley rats, respectively, to obtain three technical replicates (Table 1). The vendor reported rodent primary hepatocyte viabilities as 95%, whereas human primary hepatocyte viability was > 84% for all donor isolations. Black et al. reported no significant cytotoxicity to 100 nM TCDD in rat and human primary hepatocytes from the same vendor after 24h. Upon receipt, cells were incubated overnight in fresh DMEM/F-12 media (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Thermo Scientific HyClone, Logan, UT), 100U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), and 50 μg/ml gentamicin (Invitrogen) under standard conditions (5% CO2, 37°C). Primary hepatocytes from all species were cultured with the same media and culture conditions. For dose-response studies, hepatocytes were treated with dimethyl sulfoxide (DMSO) vehicle (Sigma, St Louis, MO), 0.001, 0.01, 0.1, 1, 10, or 100 nM TCDD (provided by S. Safe, Texas A&M University, College Station, TX) for 12 and 24h. Time course studies were conducted for 1, 2, 4, 8, 12, 24, and 48h, and hepatocytes were treated with DMSO vehicle or 10 nM TCDD.
Table 1 .
Primary Hepatocyte Donor Information
| Species | Sex | Age | Viability |
|---|---|---|---|
| Humana | Female | 54–64 years | > 84% |
| Mouseb | Male | 12 weeks | 95% |
| Ratc | Male | 9 weeks | 95% |
Notes. aCancer-free primary hepatocytes were isolated from three separate Caucasian postmenopausal human donors. Each donor represents one independent biological replicate.
bFour CD-1 mouse livers were pooled. All studies were conducted with the same preparation, with three technical replicates.
cTwo Sprague Dawley rat livers were pooled. All studies were conducted with the same preparation, with three technical replicates.
RNA isolation.
Hepatocytes were harvested by homogenization in RLT lysis buffer (Qiagen, Valencia, CA) using a Polytron PT2100 homogenizer (Kinematica, Bohemia, NY). Total RNA was isolated with RNeasy Mini Kits (Qiagen) according to the manufacturer’s protocol and stored at −80°C. RNA was quantified spectrophotometrically (NanoDrop; Thermo Scientific, Wilmington, DE), and purity was assessed by A260/A280 ratio.
Microarray experimental design.
Total RNA (500ng) isolated from TCDD- and DMSO vehicle–treated primary hepatocytes were individually hybridized to 4×44K Agilent oligonucleotide microarrays (human, G4112F; mouse, G4122F; rat, G4131F; Agilent Technologies, Santa Clara, CA). One-color labeling (Cy3) and hybridization of three replicates for all doses and time points were carried out according to the manufacturer’s protocol (Agilent manual: G4140-90040 v5.7). Independent reactions were performed per array whereby Cy3 dye was incorporated into TCDD- and vehicle-treated samples, respectively. One-color labeling produces comparable data with no significant variation compared with two-color (Cy3 and Cy5) labeling studies (Patterson et al., 2006). The 4×44K microarrays have four arrays per slide allowing four samples to be evaluated per slide. For the dose-response studies, each slide was hybridized with three TCDD-treated samples and one vehicle control, whereas time course microarrays were hybridized with two TCDD-treated samples and two time-matched vehicle control samples. Microarrays were scanned at 532nm with a GenePix 400B scanner (Molecular Devices, Union City, CA) using GenePix Pro 6.0 to extract feature and background intensities. All images and data were managed in TIMS dbZach data management system (Burgoon and Zacharewski, 2007).
Microarray analysis.
Each treatment and time point (n = 3) for the 12- and 24-h dose-response studies and the time course study for human, mouse, and rat, respectively, resulted in nine separate microarray experiments that were analyzed independently. All microarray data passed the quality assurance protocol (Burgoon et al., 2005) and were normalized in SAS v9.1 (SAS Institute) using a semiparametric approach (Eckel et al., 2005) adapted for one-color microarray analysis. Statistical values, herein referred to as P1(t) values, were obtained by evaluation of posterior probabilities on a per gene and per dose or time point basis (Eckel et al., 2004) to determine model-based t values in SAS v9.1. An empirical Bayes method was then used to calculate P1(t) in R (R, Institute for Statistics and Mathematics, WU Wien). Data were ranked and prioritized based on a |fold change| > 1.5 and P1(t) > 0.90 to identify differentially expressed genes in mice and rats. Because human dose-response and time course datasets exhibited greater variability, the filtering criteria were relaxed to |fold change| > 1.4 and statistical cutoff of P1(t) > 0.80. All filtering, prioritization, and comparisons of datasets were performed in a MySQL v5.1.40 database. The complete microarray datasets are available as supplementary tables, time course for human (Supplementary table S1), mouse (Supplementary table S2), and rat (Supplementary table S3), 12-h dose-response for human (Supplementary table S4), mouse (Supplementary table S5), and rat (Supplementary table S6), and 24-h dose-response for human (Supplementary table S7), mouse (Supplementary table S8), and rat (Supplementary table S9).
Comparative analyses and functional annotation.
Agilent 4×44K oligonucleotide arrays are annotated using Entrez Gene ID, RefSeq, Ensembl and/or GenBank accession numbers, and gene symbols as defined by NCBI using human genome build hg19, mouse genome build mm9, and rat genome build rn4. Only genes with known human, mouse, and rat orthologs were included in interspecies comparisons. HomoloGene identifiers (HIDs; www.ncbi.nlm.nih.gov/HomoloGene), which compare protein and DNA sequences between species to calculate alignment and distance metric scores for orthologous gene groups, were used to identify orthologous human, mouse, and rat genes. Each ortholog group (i.e., genes arising from a common ancestor) is assigned a unique HID. HomoloGene contains 18,981, 21,766, and 19,229 unique HIDs for human, mouse, and rat, respectively, of which 17,258 human, 17,578 mouse, and 15,013 rat HIDs are represented on the corresponding species microarray. Profile similarities between differentially expressed orthologs in time course studies were evaluated by examining fold change (direction of regulation and magnitude relative to vehicle control) and significance (based on the statistical P1(t) values) using a correlation plot on a per ortholog basis.
Overrepresented functions and enriched pathways within differentially expressed datasets were investigated using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, Redwood City, CA; www.ingenuity.com). Differentially expressed genes from time course studies were uploaded, and unions were created to include all time points in one analysis. Overrepresented functional annotation categories, identified as Bio Functions by the Ingenuity Knowledge Base, and canonical biochemical pathways were considered significant using a p value cutoff (p > 0.05).
Quantitative real-time PCR.
Assays were performed as previously described (Dere et al., 2011b). Briefly, 2 µg of total RNA was reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) using an anchored oligo-dT primer, producing cDNA that was used as template for quantitative real-time PCR (QRTPCR). Reactions consisting of 1 µl of cDNA, 0.15 µM forward and reverse gene-specific primers (Supplementary table S10), 3 mM MgCl2, 1.0 mM dNTPs, 0.025 IU AmpliTaq Gold, and 1× SYBR Green PCR Buffer (Applied Biosystems) were amplified using an Applied Biosystems PRISM 7500 Sequence Detection System. Transcript copy numbers were calculated from a standard curve and normalized to the geometric mean of three housekeeping genes to control for differences in RNA loading, quality, and cDNA synthesis (Vandesompele et al., 2002). For graphing purposes, relative expression levels are scaled such that the expression level of the vehicle control group is equal to 1. QRTPCRs from time course and dose-response data were analyzed using ANOVA followed by Tukey’s or Dunnett’s post hoc test, respectively (SAS v9.1, SAS Institute). Differences between TCDD treatment compared with vehicle control samples were considered significant when p < 0.05.
Dose-response modeling.
Microarray dose-response data meeting the filtering criteria (|fold change| > 1.5 and P1(t) > 0.90 for mouse and rat, |fold change| > 1.4 and P1(t) > 0.80 for human) were evaluated on a per gene basis by particle swarm optimization to determine the best-fit model among five classes (sigmoidal, exponential, linear, quadratic, or Gaussian) using the ToxResponse modeler (Burgoon and Zacharewski, 2008). EC50 values were calculated for genes exhibiting a sigmoidal dose-response and reported if within the experimental dose range (0.001 − 100 nM).
Gas chromatography-mass spectrometry of FA methyl esters.
FA extraction, derivatization to methyl esters, and analysis by gas chromatography-mass spectrometry (GC-MS) were performed as previously described (Angrish et al., 2011; Forgacs et al., 2012). Briefly, human hepatocytes were treated with 10 nM TCDD for 48h and collected in PBS. Cells were pelleted by centrifugation, and supernatant was discarded. Pellets were weighed, homogenized in 40% methanol, and acidified with concentrated hydrochloric acid. Lipids were extracted with chloroform:methanol (2:1) containing 1 mM 2,6-di-tert-butyl-4-methylphenol. The organic phase was removed, and protein and aqueous phases were re-extracted with chloroform. Organic phases were pooled and solvents evaporated under nitrogen. Samples were resuspended in 3 N nonaqueous methanolic hydrochloric acid and held at 60°C overnight and then cooled to room temperature followed by addition of 0.9% (wt/vol) sodium chloride and hexane. The organic phase was separated by centrifugation, collected, dried under nitrogen, and resuspended in hexane. Samples were analyzed with an Agilent 6890N GC with a 30mDB23 column interfaced to an Agilent 5973 MS. 19:1n9 free FA and 19:0 triglyceride were added as extraction efficiency controls with 17:1n1 FA methyl ester added as a loading control (Nu-chek, Elysian, MN). GC-MS data files were converted to Waters MassLynx format and analyzed with MassLynx software. Data are reported as nmol/mg wet cell pellet based upon peak areas from total ion chromatograms obtained from a linear calculation based on a calibration curve and normalized to wet cell pellet weight.
Results
Microarray Analysis
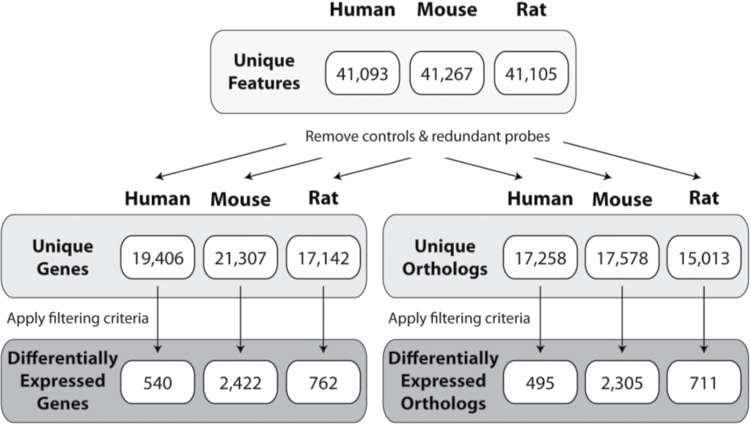
Each microarray species platform contained ~41,000 features representing 19,406 human, 21,307 mouse, and 17,142 rat unique genes (Fig. 1). Human datasets exhibited greater variability, likely due to biological variation between individual donors, and were filtered using |fold change| > 1.4 and P1(t) > 0.80 compared with |fold change| > 1.5 and P1(t) > 0.90 criteria for rat and mouse datasets. Analysis of the time course microarray data identified the TCDD-elicited differential expression of 540 human, 2422 mouse, and 762 rat genes over the 48-h period. Mapped orthologs were then examined using HIDs in order to compare the differential expression of equivalent genes across human, mouse, and rat primary hepatocytes. The HomoloGene database contains 18,981 human, 21,766 mouse, and 19,229 rat unique HIDs, of which 17,258, 17,578, and 15,013 HIDs are represented on the respective microarray platforms. In the time course study, 10 nM TCDD elicited the differential expression of 495 human, 2305 mouse, and 711 rat orthologs.
Fig. 1.
Summary of temporal TCDD-elicited differential gene and ortholog expression in human, mouse, and rat primary hepatocytes. Primary hepatocytes were treated with 10 nM TCDD for 1, 2, 4, 8, 12, 24, and 48h. Differential expression was assessed using 4×44K Agilent oligonucleotide microarrays. Human, mouse, and rat orthologs were identified using HomoloGene. Human hepatocyte responses exhibited greater variability and therefore were filtered using more relaxed criteria (|fold change| > 1.4 and P1(t) > 0.80) compared with the criteria used for rat and mouse datasets (|fold change| > 1.5 and P1(t) > 0.90).
Dose-Dependent Differential Gene Expression and EC50 Determination
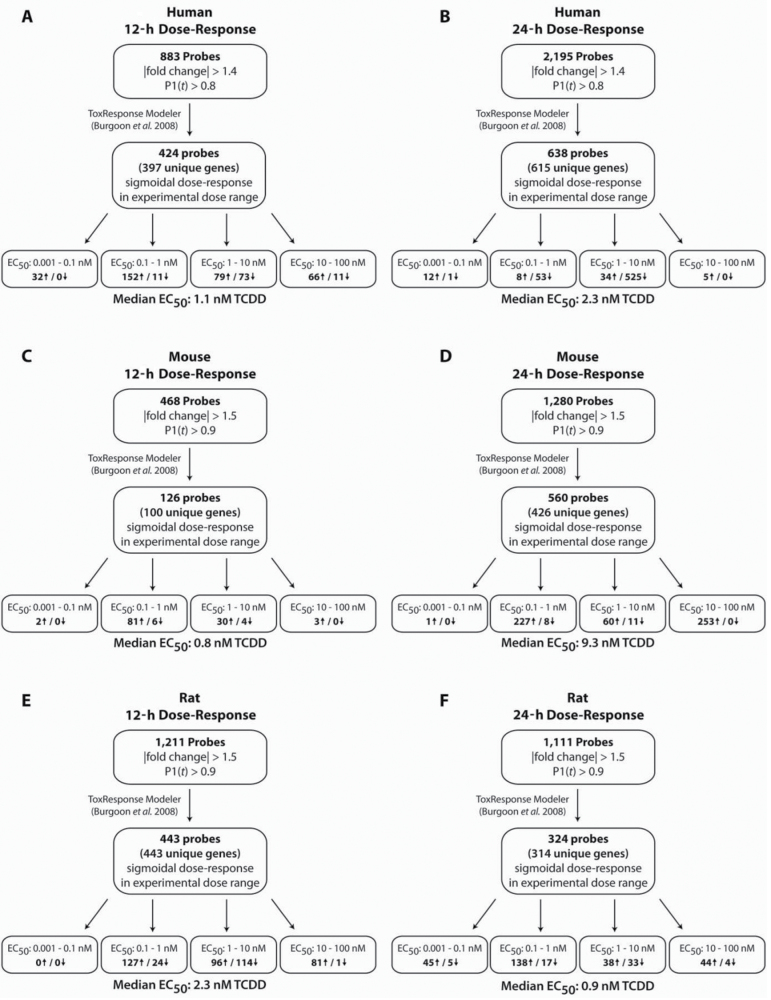
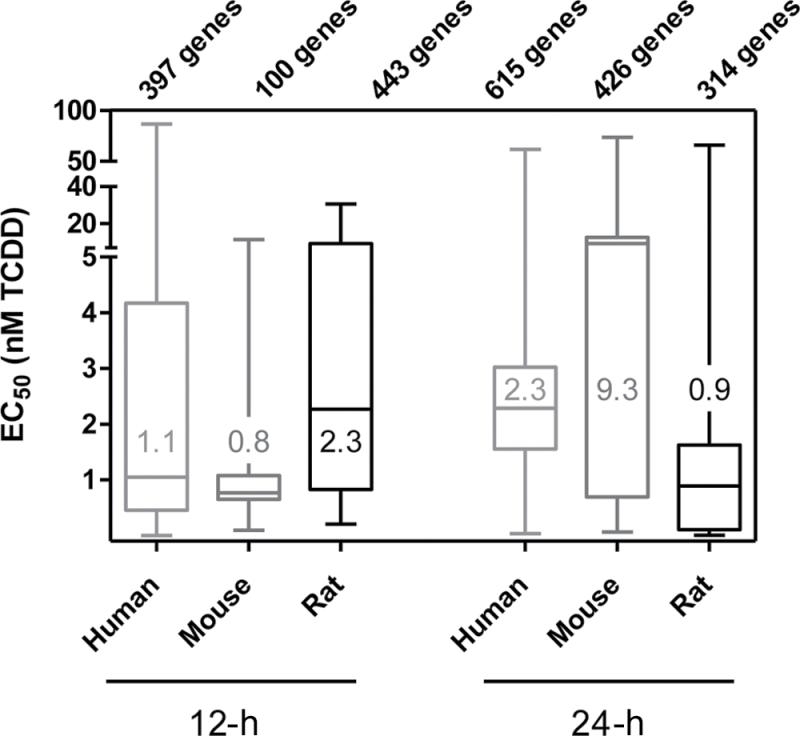
Dose-response studies were conducted at 12 and 24h, with EC50 values calculated for all differentially expressed features (human: |fold change| > 1.4 and P1(t) > 0.80; mouse and rat: |fold change| > 1.5 and P1(t) > 0.90) exhibiting a sigmoidal dose-response profile as identified by the ToxResponse modeler (Fig. 2)(Burgoon and Zacharewski, 2008). In total, 397 and 615 human, 100 and 426 mouse, and 443 and 314 rat genes exhibited a sigmoidal dose-response profile at 12 and 24h, respectively. Box and whisker plots of the EC50 distributions for these genes (Fig. 3) indicate that human, mouse, and rat primary hepatocytes have similar overall median EC50s at 12h (mouse: 0.8 nM, human: 1.1 nM, rat: 2.3 nM) and 24h (mouse: 9.3 nM, human: 2.3 nM, rat: 0.9 nM) that ranged over five orders of magnitude.
Fig. 2.
Automated dose-response modeling and EC50 determination. ToxResponse modeler calculated EC50 values for each probe that exhibited a sigmoidal dose response at 12 and 24h for human (|fold change| > 1.4 and P1(t) > 0.80) (A, B), mouse (|fold change| > 1.5 and P1(t) > 0.90) (C, D), and rat (|fold change| > 1.5 and P1(t) > 0.90) (E, F). Total number of sigmoidal dose-responsive genes, EC50 range, and direction of regulation are provided.
Fig. 3.
Differential gene expression EC50 distribution. Sigmoidal dose-response curves and EC50 values elicited by TCDD at 12 and 24h were analyzed using ToxResponse modeler, the number of sigmoidal dose-responsive genes (from Figure 2) are provided above each box and whisker. The 25th and 75th percentiles are denoted by the length of the boxes, and the whiskers demonstrate the EC50 range. The median EC50 values are provided within the graph and are denoted by the horizontal line within the boxes.
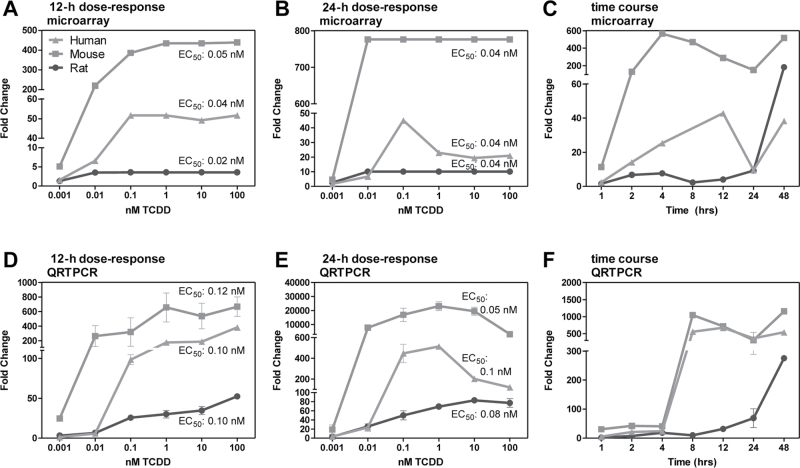
In general, EC50 values for AhR battery genes were comparable across species. For example, Cyp1a1 EC50s at 24h were 0.04 nM in all species based on microarray data (Fig. 4). QRTPCR-based Cyp1a1 EC50s at 24h were also similar across all species (0.1, 0.05, and 0.08 nM in human, mouse, and rat, respectively) and comparable to other reported human (0.14 − 0.37 nM) and rat (0.003 − 0.012 nM) primary hepatocyte EC50 values (Fig. 4; Table 2) (Budinsky et al., 2011; Silkworth et al., 2005). Differences in fold change between microarray and QRTPCR have been previously reported and attributed to data compression (Burgoon et al., 2005). Mouse hepatocytes showed the greatest Cyp1a1 efficacy (~400- to 800-fold), followed by human (~40- to 100-fold) and rat (~6- to 60-fold). Tiparp EC50 values of 0.97 nM for human, 0.63 nM for mouse, and 0.14 nM for rat at 12h exhibited the greatest variability among AhR battery genes but were still within an order of magnitude.
Fig. 4.
TCDD-elicited temporal and dose-dependent Cyp1a1 induction. Microarray (A–C) and QRTPCR (D–F) evaluation of TCDD-mediated induction of Cyp1a1 in human (light grey, triangles), mouse (medium grey, squares), and rat (dark grey, circles) primary hepatocytes. Comparable EC50s were calculated across human, mouse, and rat primary hepatocytes using microarray and QRTPCR assays although there were differences in efficacy. EC50s were calculated using the ToxResponse modeler (Burgoon and Zacharewski, 2008).
Table 2 .
Comparison of EC50 Values for TCDD-Elicited Induction of Cyp1a1 mRNA in Primary Hepatocytes
| Study | Human EC50 | Mouse EC50 | Rat EC50 |
|---|---|---|---|
| 24-h dose-response | 0.1 nM | 0.05 nM | 0.08 nM |
| Budinsky et al. (2010) | 0.37 nM | NA | 0.012 nM |
| Silkworth et al. (2005) | 0.14 nM | NA | 0.003 nM |
Notes. All EC50 values are from QRTPCR evaluation of mRNA expression in primary hepatocytes after 24h of treatment with TCDD in primary hepatocytes. NA, not available.
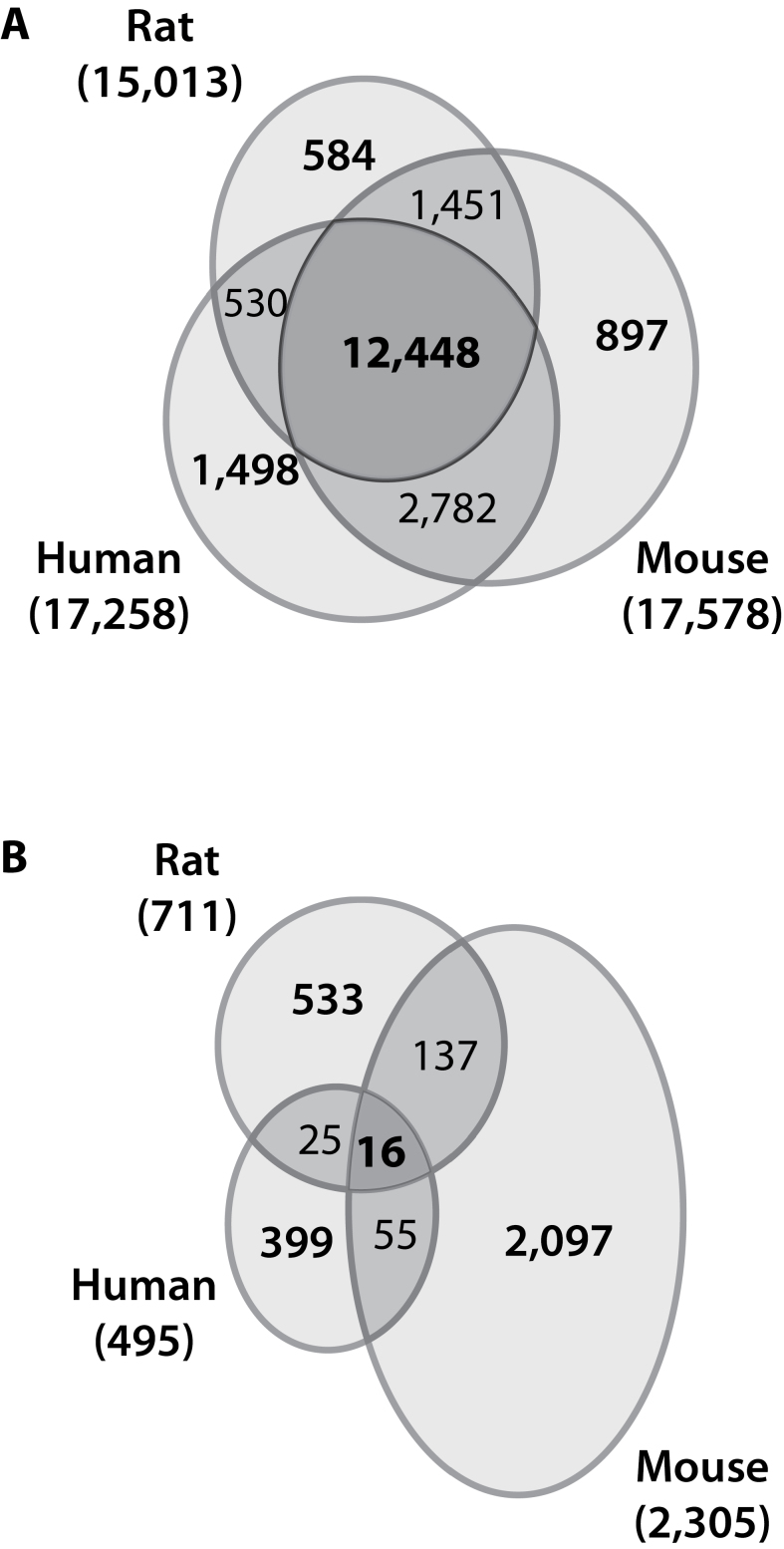
Interspecies Comparison of Temporal Gene Expression
A total of 12,448 unique HIDs are represented on the Agilent microarray platforms across all three species (Fig. 5A). However, only 16 orthologs were differentially expressed in response to TCDD in all three species (Fig. 5B; Table 3), representing < 3% of common orthologs. The majority of orthologs exhibited species-specific expression. More specifically, 81% (399/495) of human, 91% (2,097/2,305) of mouse, and 75% (533/711) of rat orthologs exhibited species-specific differential expression (Fig. 5B). Eleven of the 16 orthologs differentially expressed in all three species exhibited a comparable expression pattern (Table 3). This included the induction of prototypical “AhR battery” genes such as Cyp1a1, Cyp1a2, Tiparp, and Ugt1a6, as well as the induction of Ptgs2 (also known as Cox-2) involved in inflammation, the cell adhesion gene Lmo7, the tumor suppressor Bmf, and the oxidative stress response regulator Nfe2l2 (also known as Nrf2). Repression of the FA transporter Slc27a1, Nr1h4 (also known as the farnesoid X receptor) essential for hepatic bile acid and carbohydrate metabolism, and Cited2 involved in the regulation of liver development also exhibited comparable temporal expression. However, several orthologs exhibited divergent expression. For example, Igf1 and Kank1 involved in cell growth were repressed in rodents but induced in human, whereas the chemokine Cxcl10 and the transcription factor Litaf were repressed in human and mouse but induced in rat primary hepatocytes. The alcohol dehydrogenase Adh1c was repressed in human and rat but induced in the mouse. These examples suggest that divergent expression may also contribute to AhR-mediated species-specific effects and differing sensitivities to TCDD and related compounds.
Fig. 5.
Orthologs represented on human, mouse, and rat microarrays and their differential expression. (A) The total number of orthologs (identified as the number of unique HIDs) represented on each species Agilent microarray. (B) The total number of orthologs (identified as the number of unique HIDs) differentially expressed in response to 10 nM TCDD in the time course study.
Table 3 .
Conserved Temporal TCDD-Elicited Ortholog Differential Expression
| Gene symbol | HID | Human | Mouse | Rat |
|---|---|---|---|---|
| Adh1c | 73888 | ▼3.2 (48) | ▼7.7 (24) | ▼3.1 (48) |
| Bmf | 14130 | ▲4.5 (4) | ▲3.4 (2) | ▲3.0 (48) |
| Cited2 | 4433 | ▼1.5 (4) | ▼3.3 (12) | ▼3.6 (8) |
| Cxcl10 | 1203 | ▲2.5 (12) | ▲1.8 (48) | ▲2.9 (24) |
| Cyp1a1 | 68062 | ▼42.8 (12) | ▼564.7 (4) | ▼18.4 (48) |
| Cyp1a2 | 68082 | ▲72.5 (48) | ▲9.5 (4) | ▲50.0 (24) |
| Igf1 | 515 | ▼1.8 (48) | ▼2.8 (48) | ▼3.6 (8) |
| Kank1 | 17706 | ▲1.6 (4) | ▲2.3 (12) | ▲2.8 (8) |
| Litaf | 37974 | ▼1.7 (12) | ▼1.5 (48) | ▼1.5 (24) |
| Lmo7 | 83924 | ▲1.6 (4) | ▲3.7 (4) | ▲1.8 (4) |
| Nfe2l2 (Nrf2) | 2412 | ▼1.6 (4) | ▼ 4.3 (1) | ▼ 2.4 (2) |
| Nr1h4 (FXRa) | 3760 | ▲ 1.8 (12) | ▲1.9 (12) | ▲2.5 (24) |
| Ptgs2 (Cox2) | 31000 | ▼1.5 (12) | ▼ 1.6 (12) | ▼1.7 (8) |
| Slc27a2 (Fatp2) | 37830 | ▲1.6 (12) | ▲2.4 (12) | ▲4.4 (48) |
| Tiparp | 9167 | ▼6.6 (48) | ▼21.0 (8) | ▼8.5 (24) |
| Ugt1a6 | 85959 | ▲2.2 (48) | ▲2.5 (24) | ▲9.8 (24) |
Notes. Maximum fold changes are indicated; the time point (in hours) of the maximum fold change is indicated in brackets. Mouse and rat datasets were filtered using |fold change| > 1.5 and P1(t) > 0.90, and human microarray data were filtered using |fold change| > 1.4 and P1(t) > 0.80.
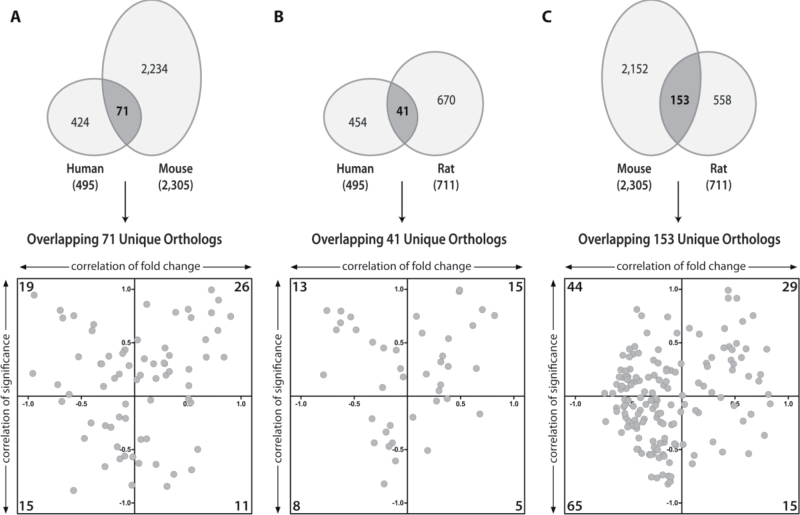
Pairwise comparisons did not identify better correlations between any two species (Fig. 6). For example, only 26 of 71 commonly expressed human and mouse orthologs exhibited a positive correlation (quadrant I), suggesting variable magnitude and/or direction of regulation. Collectively, these results provide compelling evidence that despite the conservation of the AhR and its signaling mechanism, species-specific gene expression profiles likely underlie the species-specific effects of TCDD and related compounds.
Fig. 6.
Pairwise correlation analysis of differentially expressed orthologs. Differentially expressed orthologs were evaluated for significance and fold change correlation between two species. The number of orthologs is provided in the corner of each quadrant. There was no correlation between differentially expressed orthologs between (A) human and mouse, (B) human and rat, or (C) mouse and rat primary hepatocytes indicating species-specific differential ortholog expression.
Functional Annotation and Pathway Enrichment
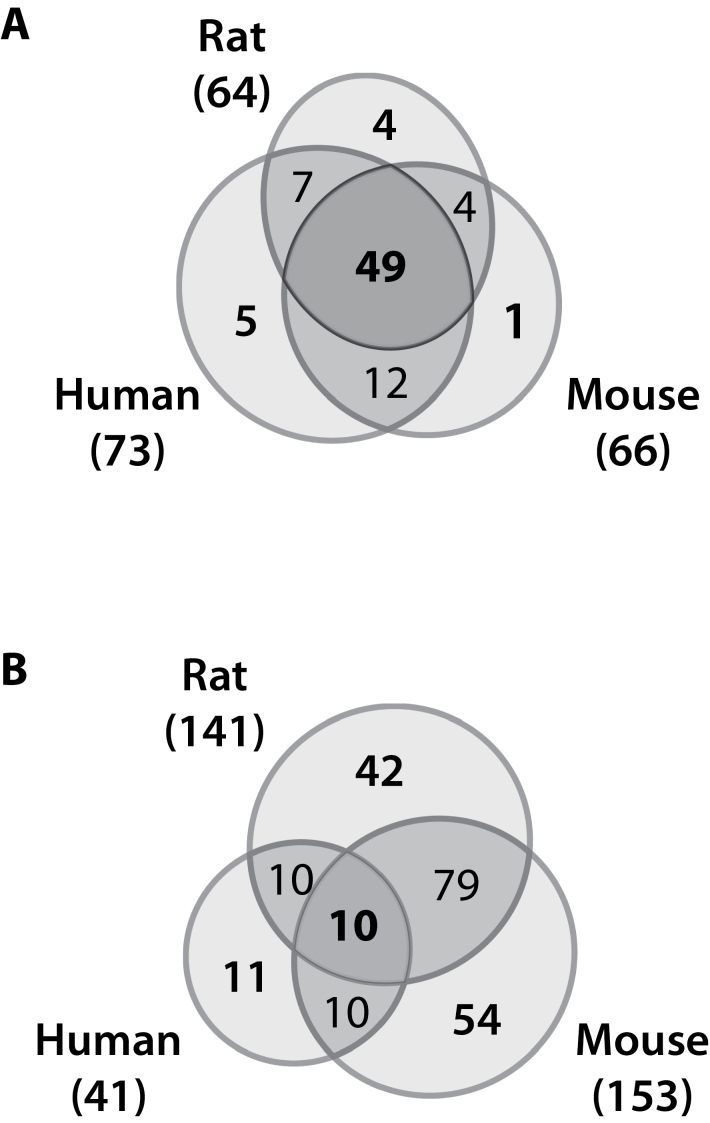
All differentially expressed genes from the time course studies were analyzed for overrepresentation within specific functional categories and canonical pathways. Differential gene expression associated with cholestasis, steatosis, and necrosis/cell death was overrepresented in all three species (data not shown). The number of significantly enriched Bio Functions (IPA; Ingenuity Knowledge Base) was comparable (66 human, 73 mouse, and 64 rat overrepresented functional categories) in the time course datasets (Fig. 7A). Overall, 49 categories were overrepresented in all three species including functions associated with carbohydrate metabolism, gene expression regulation, lipid homeostasis, metabolic disease, and protein synthesis. Species-specific overrepresented functions included hypersensitivity and immune response in human hepatocytes, RNA damage and repair in mouse primary hepatocytes, and energy production and protein folding in rat primary hepatocytes. Note that the number of genes included in these functional analyses varied between species, requiring further analysis to elucidate potential consequences on pathways to determine consistency with reported phenotypic effects.
Fig. 7.
Comparison of overrepresented functional annotation terms using time-dependent differential gene expression. (A) The total number of unique enriched functional categories identified in the time course across species. (B) Comparison of enriched canonical pathways. All terms and pathways included in the comparisons were significantly enriched (p value < 0.05).
TCDD-mediated differential gene expression mapped to 41 human, 153 mouse, and 142 rat canonical pathways, of which 11 were specific to human primary hepatocytes and < 50% were in common between mice and rats (Fig. 7B). Pathways in common to all three species included AhR and Nrf2 signaling, as well as lipid transport, processing, and metabolism. Mouse-specific differential gene expression mapped to immune response pathways and included the activation of CD40, IL-2, IL-3, IL-9, IL-10, and IL-17 signaling pathways, consistent with reports of immune cell infiltration (Boverhof et al., 2006). In the rat, energy homeostasis pathways such as oxidative phosphorylation, pyruvate metabolism, citrate cycle, and mitochondrial dysfunction predominated. Most of the genes within these rat-specific pathways were downregulated including electron transport chain constituents such as complex I NADH dehydrogenase subunit (Ndufa2) and complex V ATPase subunits (Atp6v02, Atp1a, Atp1c1, Atp5a1), as well as ATP citrate lyase (Acly) and pyruvate kinase (Pklr). Xenobiotic metabolism signaling (p value 3.34E−05) was one of the most significantly enriched canonical pathways in humans. This included CYP1A1 and CYP1A2 induction and the downregulation of several dehydrogenases (ADH1A, ADH1C, and ALDH1B1). Amino acid metabolism pathways, such as tyrosine and histidine metabolism, the catabolism of valine, leucine, and isoleucine, and the repression of serine dehydrogenase (SDS) and histidine ammonia-lysase (HAL) were also uniquely enriched in human primary hepatocytes.
Comparative Analysis of Lipid Transport, Processing, and Metabolism
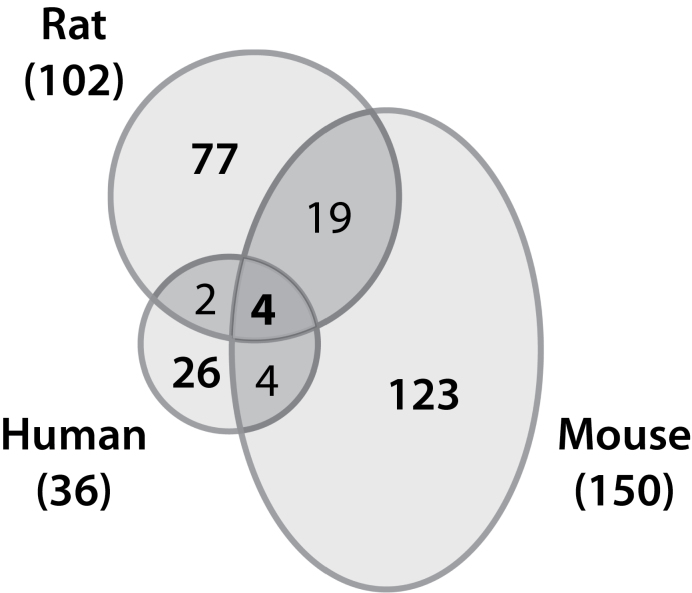
A single oral gavage of TCDD elicits hepatic steatosis in C57BL/6 mice but not in Sprague Dawley rats within 7 days (Boverhof et al., 2006; Forgacs et al., 2012). However, the human relevance of this response is unknown. Differential gene expression was functionally enriched for lipid transport, processing, and metabolism genes in human (p value 1.64E−07), mouse (p value 1.08E−08), and rat (p value 1.02E−07) primary hepatocytes. There are ~300 genes functionally associated with lipid transport, processing, and metabolism, according to DAVID (http://david.abcc.ncifcrf.gov/), represented on the human, mouse, and rat Agilent microarrays. In the time course study, 39 human, 176 mouse, and 133 rat lipid genes were differentially expressed, corresponding to 36, 150, and 102 orthologs, respectively, of which only 4 were differentially expressed in all three species (Fig. 8). The majority of these genes exhibited species-specific regulation with 26, 123, and 77 orthologs specific to human, mouse, and rat, respectively. Lipid transport, processing, and metabolism genes were also identified in dose-response datasets. For example, in the 24-h dose-response study, 13 human (median EC50 2.2 μM), 31 mouse (median EC50 1.3 μM), and 9 rat (median EC50 0.4 μM) genes were associated with lipid functions.
Fig. 8.
Comparison of time-dependent differentially expressed orthologs associated with lipid transport, processing, and metabolism. Functional annotation terms related to lipid metabolism were identified by IPA; terms were considered significantly enriched at p < 0.05.
In order to determine whether these species-specific lipid-related gene expression changes are consistent with reported phenotypes, the direction of regulation within overrepresented functions and pathways was further examined. Overrepresentation of functional annotation terms such as lipid hydroxylation, lipid quantity, and synthesis of lipid was observed in all species. Mouse-specific enrichment included lipid generation and metabolism genes such as phospholipases that cleave lipids to release free FA and are implicated in FA uptake, including the induction of Pla2g6 (fourfold) and Pla2g15 (twofold). In contrast, rat-specific functions were associated with intracellular lipid transport and localization such as the retinol binding protein 7 (Rbp7 formerly known as CRBP-III). Rbp7 is an intracellular lipid-binding protein that exhibited both temporal and dose-dependent induction up to threefold at 24h solely in the rat. Rat hepatocytes also exhibited phospholipid and choline metabolism differential gene expression (Acsl1, Acaca, Agpat6, Chka, Pck1, Pcyt1a, and Pemt). These species-specific categories are consistent with hepatic lipid accumulation in the mouse but not in the rat.
Human-specific functions in lipid transport, processing, and metabolism include the TCDD-mediated temporal repression of Fasn (biosynthesis), Acly (biosynthesis), Ppap2a (processing), and Elovl6 (metabolism) and dose-dependent induction of Acsl5 (processing, EC50 9.1 nM), Lipg (processing, EC50 1.0 nM), and Ffar3 (transport, EC50 12.3 nM) all supporting decreased lipid metabolism in human primary hepatocytes, consistent with TCDD-elicited hepatic steatosis due to increased FA uptake and decreased de novo FA biosynthesis (Angrish et al., 2012b). These species-specific lipid metabolism effects demonstrate that although the same functional categories and pathways may be affected by TCDD, differences in which genes are differentially expressed within a category and their direction of regulation can significantly influence the resulting phenotype.
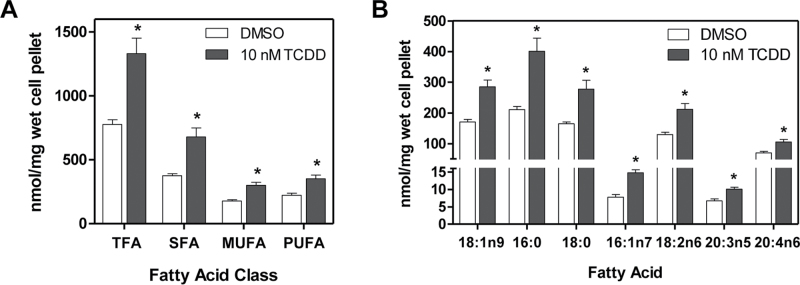
FA Accumulation in Human Primary Hepatocytes
The human relevance of differentially expressed genes associated with lipid transport, processing, and metabolism was examined by investigating the effect of TCDD on FA accumulation in human primary hepatocytes using GC-MS (Fig. 9). TCDD induced total FA with increases in SFAs, MUFAs, and PUFAs. More specifically, palmitate (16:0), stearate (18:0), linoleic (18:2n6), and oleic (18:1n9) acids increased ~twofold after 48h of 10 nM TCDD treatment. Interestingly, these were also the most abundant FA species detected by GC-MS in the FBS supplement used in the primary hepatocyte culture media (data not shown). This suggests that TCDD-elicited lipid accumulation in hepatocytes is due to increased FA uptake from FBS, consistent with our previous studies that reported the hepatic accumulation of radiolabeleld FA in mice gavaged with 14C-oleate (18:1n9), the dose-dependent hepatic accumulation of linoleic acid in diet studies, and the increase of unsaturated FAs in Scd1 knockout mice following TCDD treatment (Angrish et al., 2011 , 2012a , b). TCDD also induced moderate increases in palmitoleic (16:1n7) and arachidonic (20:4n6) acids. Collectively, these results demonstrate that TCDD-elicited lipid accumulation in human hepatocytes is similar to hepatic steatosis reported in mice but not in rats.
Fig. 9.
TCDD-elicited FA accumulation in human primary hepatocytes. Human primary hepatocytes were treated with 10 nM TCDD for 48h and analyzed for FA accumulation by GC-MS. (A) Total FA, SFA, MUFA, and PUFA levels. (B) TCDD-elicited accumulation of individual FA species. Data are reported as mean ± SEM of nmol FA methyl ester per milligram wet cell pellet weight and were analyzed by Dunnet’s t-test where asterisk (*) represents p < 0.05 for TCDD versus DMSO, n = 3.
Discussion
The current risk assessment paradigm assumes a common mechanism or mode of action, as well as similar effects between species. Although the conserved activation of AhR is generally accepted as the primary mode of action for TCDD-elicited effects across all species, toxicogenomic studies indicate that TCDD elicits species-specific AhR-mediated changes in differential gene expression and metabolite levels, consistent with the species-specific effects elicited by TCDD and related compounds (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Dere et al., 2011b; Flaveny et al., 2010; Forgacs et al., 2012). Previous cross-species hepatic gene expression studies examining TCDD focused on a limited number of genes (e.g., AhR gene battery: Cyp1a1, Cyp1a2, Ugt1a6, Tiparp, etc.) or used transformed cell lines that may not accurately reflect normal human responses. Consequently, comparative studies using primary hepatocytes provide a unique opportunity to distinguish causative mechanisms from adaptive responses, especially for species-specific responses. In this study, human, mouse, and rat primary hepatocytes were used to comprehensively evaluate genome-wide time- and concentration-dependent TCDD-elicited gene expression with a focus on the potential human relevance of TCDD-elicited hepatic steatosis in rodents.
It is important to note that all comparative studies may be confounded by study design limitations. For example, the accuracy and quality of the genome builds and their associated annotation varies between species, with the human and mouse genomes considered to be more complete compared with the rat. This difference is reflected in the total number of annotated genes represented on each respective microarray. Incomplete annotation also reduced the number of comparable orthologs across all three species, again with the rat limiting the number of mapped orthologs. Although the use of primary hepatocytes facilitate the inclusion of human samples in these comparative studies, the use of the same culture conditions may have inadvertently biased the results and confounded data interpretation. More specifically, the same incubation temperature (37°C) was used for all primary hepatocytes despite differences in normal in vivo body temperatures between humans and mice (~37°C) compared with rats (~39°C). Furthermore, media composition may also affect the availability and response to TCDD, which could differ between species. This study also included cancer-free primary hepatocytes isolated from postmenopausal female human donors compared with hepatocytes from male rodents. Although the human and rat donors were of comparable sexual maturity, mouse hepatocytes were from older donors. Despite these biases, the hallmark measure of TCDD sensitivity, the induction of Cyp1a1, and the induction of Tiparp were comparable among species, suggesting that study design differences likely did not confound the < 3% conserved expression of common orthologs across human, mouse, and rat primary hepatocytes.
Previous studies have also compared the effect of TCDD and related compounds on gene expression in human and rat primary hepatocytes but excluded mouse primary hepatocytes and reported similar species-specific expression (Black et al., 2012; Budinsky et al., 2011; Carlson et al., 2009; Silkworth et al., 2005). For example, using a comparable dosing regimen and microarray platform as used herein, only 12 genes were identified as differentially expressed in both human and rat primary hepatocytes following treatment with 0.01–100 nM TCDD for 24h (Black et al., 2012). Our results further demonstrate that TCDD elicited minimal overlap between human, mouse, and rat primary hepatocyte differential gene expression with only 16 orthologs (< 3%) in common between all three species. This mainly included the induction of “AhR battery” genes such as Cyp1a1, Cyp1a2, Tiparp, and Ugt1a6 with Cyp1a1 EC50s similar to values previously reported in other human and rat primary hepatocyte studies (Budinsky et al., 2011; Silkworth et al., 2005). These results are also consistent with other comparative in vivo and in vitro studies that report TCDD-elicited species-specific histopathology as well as species-specific gene expression and metabolomic effects (Black et al., 2012; Boutros et al., 2008; Boverhof et al., 2006; Carlson et al., 2009; Dere et al., 2011b; Forgacs et al., 2012). The consistency of these species-specific responses across different models and independent studies provides further evidence that the < 3% conserved expression of common orthologs across human, mouse, and rat primary hepatocytes cannot be attributed to study design differences.
In vivo studies have shown that TCDD elicits marked hepatic FA accumulation in mice but not in rats. A single oral gavage of TCDD induced histological and metabolite changes consistent with hepatic steatosis in mice, whereas minimal metabolite changes and no histological evidence of lipid accumulation are reported in rats (Boverhof et al., 2006; Forgacs et al., 2012). The differential expression of lipid transport, processing, and metabolism genes in mouse hepatocytes is consistent with the reported hepatic steatosis in mice. This included the induction of lipid uptake genes and the concomitant inhibition of de novo lipogenesis gene expression (Angrish et al., 2012b). Meanwhile, differential gene expression was associated with lipid localization and trafficking, and choline metabolism was overrepresented in rat primary hepatocytes, consistent with the in vivo accumulation of hepatic phosphocholine levels (Angrish et al., 2012b; Boutros et al., 2008; Boverhof et al., 2005 , 2006; Fletcher et al., 2005; Forgacs et al., 2012). TCDD-elicited gene expression in mice was also enriched with functions related to immune response pathways, consistent with immune cell infiltration and the emergence of steatohepatitis, which was not observed in rat liver following a single oral gavage of TCDD (Boverhof et al., 2005, 2006; Lu et al., 2011a).
Despite the conservation of the structure and function of the human, mouse, and rat AhR, TCDD-elicited species-specific differential gene expression leads to species-specific responses and sensitivities as illustrated with hepatic fat accumulation in mice but not in rats. In humans, dyslipidemia has been reported following exposure to TCDD and related compounds (Bertazzi et al., 2001; Calvert et al., 1996; Lee et al., 2006, 2007; Pelclová et al., 2002, 2009). The human relevance of hepatic fat accumulation was further evaluated by comparing overrepresented differential gene expression functions and pathways identified in primary hepatocytes. To determine whether these alterations are consistent with reported phenotypes, the direction of gene expression regulation within overrepresented functions or pathways was evaluated. Only three of the conserved differentially expressed orthologs (Nfe2l2, Nr1h4, and Cited2) have been implicated in hepatic steatosis (Kong et al., 2009; Lu et al., 2011a, b; Qu et al., 2007; Yang et al., 2010; Zhang et al., 2010). The differential expression of other lipid transport, processing, and metabolism genes in human primary hepatocytes is also consistent with hepatic fat accumulation. This included gene expression changes associated with lipid conversion/metabolism such as Ppapa2, Acly, Acsl5, and Elovl6 supporting triacylglycerol synthesis for FA storage, the induction of lipase (Lipg), and FA transport (Ffar3, Fabp5) expression in support of uptake and intracellular retention. In addition, the functional significance of these gene expression changes was phenotypically anchored to FA accumulation in human primary hepatocytes. More specifically, GC-MS analysis identified an increase in linoleic acid (18:2n6), an essential dietary FA that cannot be synthesized in human cells. TCDD-elicited accumulation of linoleic acid in human primary hepatocytes, consistent with the increased uptake of linoleic acid that is present at high levels in FBS. It is also consistent with recent studies reporting that FAs from the diet are the primary source of lipids in AhR-mediated hepatic steatosis (Angrish et al., 2012b). Moreover, functional analysis identified immune response gene enrichment in human primary hepatocytes, whereas canonical pathway analysis and histological examination identified immune response enrichment in mouse primary hepatocytes and mouse hepatic tissue (Boverhof et al., 2005). Collectively, these results suggest mice may be a better model than rats for AhR-mediated hepatic steatosis in humans.
The induction of hepatic steatosis is not unique to TCDD and related compounds. Many chemicals, drugs, and natural products also induce lipid accumulation in the liver. Furthermore, continuous TCDD exposure increases serum triglyceride levels in a variety of species including mice, guinea pigs, rabbits, and hamsters (Boverhof et al., 2006; Brewster and Matsumura, 1989) and hepatic fat accumulation in fish, chicken, mice, rats, and monkeys (Boverhof et al., 2006; Kanzawa et al., 2004; Korenaga et al., 2007; Lee et al., 2010; Volz et al., 2006; Walter et al., 2000). Consequently, TCDD and related compounds may use species-specific mechanisms to induce hepatic steatosis following a single bolus dose and after repeated doses or following chronic exposure. Nevertheless, the human relevance of AhR-mediated steatosis and its potential contributions in the development of metabolic diseases including nonalcoholic fatty liver disease, metabolic syndrome, hepatocellular carcinoma, and diabetes warrant further investigation.
Supplementary Data
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
Funding
National Institute of Environmental Health Sciences Superfund Research Program (NIEHS SRP P42ES04911).
Supplementary Material
Acknowledgments
The authors would like to thank Dr Lyle Burgoon and Dr Suntae Kim for their support and Dr Anna Kopec and Rance Nault for the critical review of this manuscript.
References
- Angrish M. M., Dominici C. Y., Zacharewski T. R. (2012a). TCDD-elicited effects on liver, serum, and adipose lipid composition in C57BL/6 mice. Toxicol. Sci. 131, 108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrish M. M., Jones A. D., Harkema J. R., Zacharewski T. R. (2011). Aryl hydrocarbon receptor-mediated induction of Stearoyl-CoA desaturase 1 alters hepatic fatty acid composition in TCDD-elicited steatosis. Toxicol. Sci. 124, 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrish M. M., Mets B. D., Jones A. D., Zacharewski T. R. (2012b). Dietary fat is a lipid source in 2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD)-elicited hepatic steatosis in C57BL/6 mice. Toxicol. Sci. 128, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertazzi P. A., Consonni D., Bachetti S., Rubagotti M., Baccarelli A., Zocchetti C., Pesatori A. C. (2001). Health effects of dioxin exposure: A 20-year mortality study. Am. J. Epidemiol. 153, 1031–1044 [DOI] [PubMed] [Google Scholar]
- Black M. B., Budinsky R. A., Dombkowski A., Cukovic D., LeCluyse E. L., Ferguson S. S., Thomas R. S., Rowlands J. C. (2012). Cross-species comparisons of transcriptomic alterations in human and rat primary hepatocytes exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 127, 199–215 [DOI] [PubMed] [Google Scholar]
- Boutros P. C., Yan R., Moffat I. D., Pohjanvirta R., Okey A. B. (2008). Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: Comparison of rat and mouse. BMC Genomics. 9, 419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Chittim B., Harkema J. R., Jump D. B., Zacharewski T. R. (2005). Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 85, 1048–1063 [DOI] [PubMed] [Google Scholar]
- Boverhof D. R., Burgoon L. D., Tashiro C., Sharratt B., Chittim B., Harkema J. R., Mendrick D. L., Zacharewski T. R. (2006). Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 94, 398–416 [DOI] [PubMed] [Google Scholar]
- Brewster D. W., Matsumura F. (1989). Differential effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on adipose tissue lipoprotein lipase activity in the guinea pig, rat, hamster, rabbit, and mink. Comp. Biochem. Physiol. C, Comp. Pharmacol. Toxicol. 93, 49–53 [DOI] [PubMed] [Google Scholar]
- Budinsky R. A., LeCluyse E. L., Ferguson S. S., Rowlands J. C., Simon T. (2011). Human and rat primary hepatocyte CYP1A1 and 1A2 induction with 2,3,7,8-tetrachlorodibenzo-p-dioxin, 2,3,7,8-tetrachlorodibenzofuran, and 2,3,4,7,8-pentachlorodibenzofuran. Toxicol. Sci. 118, 224–235 [DOI] [PubMed] [Google Scholar]
- Burgoon L. D., Eckel-Passow J. E., Gennings C., Boverhof D. R., Burt J. W., Fong C. J., Zacharewski T. R. (2005). Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 33, e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon L. D., Zacharewski T. R. (2007). dbZach toxicogenomic information management system. Pharmacogenomics. 8, 287–291 [DOI] [PubMed] [Google Scholar]
- Burgoon L. D., Zacharewski T. R. (2008). Automated quantitative dose-response modeling and point of departure determination for large toxicogenomic and high-throughput screening data sets. Toxicol. Sci. 104, 412–418 [DOI] [PubMed] [Google Scholar]
- Calvert G. M., Willie K. K., Sweeney M. H., Fingerhut M. A., Halperin W. E. (1996). Evaluation of serum lipid concentrations among U.S. workers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch. Environ. Health. 51, 100–107 [DOI] [PubMed] [Google Scholar]
- Carlson E. A., McCulloch C., Koganti A., Goodwin S. B., Sutter T. R., Silkworth J. B. (2009). Divergent transcriptomic responses to aryl hydrocarbon receptor agonists between rat and human primary hepatocytes. Toxicol. Sci. 112, 257–272 [DOI] [PubMed] [Google Scholar]
- Denison M. S., Heath-Pagliuso S. (1998). The Ah receptor: A regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. Environ. Contam. Toxicol. 61, 557–568 [DOI] [PubMed] [Google Scholar]
- Denison M. S., Pandini A., Nagy S. R., Baldwin E. P., Bonati L. (2002). Ligand binding and activation of the Ah receptor. Chem. Biol. Interact. 141, 3–24 [DOI] [PubMed] [Google Scholar]
- Denison M. S., Soshilov A. A., He G., DeGroot D. E., Zhao B. (2011). Exactly the same but different: Promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Vella L. M., Okey A. B. (1986). Structure and function of the Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Species difference in molecular properties of the receptors from mouse and rat hepatic cytosols. J. Biol. Chem. 261, 3987–3995 [PubMed] [Google Scholar]
- Dere E., Boverhof D. R., Burgoon L. D., Zacharewski T. R. (2006). In vivo-in vitro toxicogenomic comparison of TCDD-elicited gene expression in Hepa1c1c7 mouse hepatoma cells and C57BL/6 hepatic tissue. BMC Genomics. 7, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Forgacs A. L., Zacharewski T. R., Burgoon L. D. (2011a). Genome-wide computational analysis of dioxin response element location and distribution in the human, mouse, and rat genomes. Chem. Res. Toxicol. 24, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Lee A. W., Burgoon L. D., Zacharewski T. R. (2011b). Differences in TCDD-elicited gene expression profiles in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells. BMC Genomics. 12, 193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Lo R., Celius T., Matthews J., Zacharewski T. R. (2011c). Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genomics. 12, 365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel J. E., Gennings C., Chinchilli V. M., Burgoon L. D., Zacharewski T. R. (2004). Empirical bayes gene screening tool for time course or dose-response microarray data. J. Biopharm. Stat. 14, 647–670 [DOI] [PubMed] [Google Scholar]
- Eckel J. E., Gennings C., Therneau T. M., Burgoon L. D., Boverhof D. R., Zacharewski T. R. (2005). Normalization of two-channel microarray experiments: A semiparametric approach. Bioinformatics. 21, 1078–1083 [DOI] [PubMed] [Google Scholar]
- Flaveny C. A., Murray I. A., Perdew G. H. (2010). Differential gene regulation by the human and mouse aryl hydrocarbon receptor. Toxicol. Sci. 114, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher N., Wahlström D., Lundberg R., Nilsson C. B., Nilsson K. C., Stockling K., Hellmold H., Håkansson H. (2005). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: A microarray study. Toxicol. Appl. Pharmacol. 207, 1–24 [DOI] [PubMed] [Google Scholar]
- Forgacs A. L., Kent M. N., Makley M. K., Mets B., DelRaso N., Jahns G. L., Burgoon L. D., Zachareweski T. R., Reo N. V. (2012). Comparative metabolomic and genomic analyses of TCDD-elicited metabolic disruption in mouse and rat liver. Toxicol. Sci. 125, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. J., Fernandez-Salguero P. (1998). The aryl hydrocarbon receptor: Studies using the AHR-null mice. Drug Metab. Dispos. 26, 1194–1198 [PubMed] [Google Scholar]
- Hankinson O. (1995). The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340 [DOI] [PubMed] [Google Scholar]
- Huang G., Elferink C. J. (2012). A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Mol. Pharmacol. 81, 338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Kondo M., Okushima T., Yamaguchi M., Temmei Y., Honda M., Tsuchiya T. (2004). Biochemical and molecular biological analysis of different responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin in chick embryo heart and liver. Arch. Biochem. Biophys. 427, 58–67 [DOI] [PubMed] [Google Scholar]
- Kong B., Luyendyk J. P., Tawfik O., Guo G. L. (2009). Farnesoid X receptor deficiency induces nonalcoholic steatohepatitis in low-density lipoprotein receptor-knockout mice fed a high-fat diet. J. Pharmacol. Exp. Ther. 328, 116–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenaga T., Fukusato T., Ohta M., Asaoka K., Murata N., Arima A., Kubota S. (2007). Long-term effects of subcutaneously injected 2,3,7,8-tetrachlorodibenzo-p-dioxin on the liver of rhesus monkeys. Chemosphere. 67, S399–S404 [DOI] [PubMed] [Google Scholar]
- Lee C. C., Yao Y. J., Chen H. L., Guo Y. L., Su H. J. (2006). Fatty liver and hepatic function for residents with markedly high serum PCDD/Fs levels in Taiwan. J. Toxicol. Environ. Health Part A. 69, 367–380 [DOI] [PubMed] [Google Scholar]
- Lee D. H. Lee I. K. Porta M. Steffes M. Jacobs D. R.Jr. (2007). Relationship between serum concentrations of persistent organic pollutants and the prevalence of metabolic syndrome among non-diabetic adults: Results from the National Health and Nutrition Examination Survey 1999-2002. Diabetologia. 50, 1841–1851 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Wada T., Febbraio M., He J., Matsubara T., Lee M. J., Gonzalez F. J., Xie W. (2010). A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 139, 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Crawford R. B., Kaplan B. L., Kaminski N. E. (2011a). 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated disruption of the CD40 ligand-induced activation of primary human B cells. Toxicol. Appl. Pharmacol. 255, 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Cui W., Klaassen C. D. (2011b). Nrf2 protects against 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced oxidative injury and steatohepatitis. Toxicol. Appl. Pharmacol. 256, 122–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell E. E. (1985). Comparative toxicity of PCBs and related compounds in various species of animals. Environ. Health Perspect. 60, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W., Roe A. L., Dieter M. Z., Solis W. A., Yang Y., Dalton T. P. (2000). Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 59, 65–85 [DOI] [PubMed] [Google Scholar]
- Okey A. B., Franc M. A., Moffat I. D., Tijet N., Boutros P. C., Korkalainen M., Tuomisto J., Pohjanvirta R. (2005). Toxicological implications of polymorphisms in receptors for xenobiotic chemicals: The case of the aryl hydrocarbon receptor. Toxicol. Appl. Pharmacol. 207(2 Suppl ) 43–51 [DOI] [PubMed] [Google Scholar]
- Okey A. B., Vella L. M., Harper P. A. (1989). Detection and characterization of a low affinity form of cytosolic Ah receptor in livers of mice nonresponsive to induction of cytochrome P1-450 by 3-methylcholanthrene. Mol. Pharmacol. 35, 823–830 [PubMed] [Google Scholar]
- Olson J. R., Holscher M. A., Neal R. A. (1980). Toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in the golden Syrian hamster. Toxicol. Appl. Pharmacol. 55, 67–78 [DOI] [PubMed] [Google Scholar]
- Patel R. D., Murray I. A., Flaveny C. A., Kusnadi A., Perdew G. H. (2009). Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab. Invest. 89, 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson T. A., Lobenhofer E. K., Fulmer-Smentek S. B., Collins P. J., Chu T. M., Bao W., Fang H., Kawasaki E. S., Hager J., Tikhonova I. R., et al. (2006). Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 24, 1140–1150 [DOI] [PubMed] [Google Scholar]
- Pelclová D., Fenclová Z., Preiss J., Procházka B., Spácil J., Dubská Z., Okrouhlík B., Lukás E., Urban P. (2002). Lipid metabolism and neuropsychological follow-up study of workers exposed to 2,3,7,8- tetrachlordibenzo- p-dioxin. Int. Arch. Occup. Environ. Health. 75(Suppl ) S60–S66 [DOI] [PubMed] [Google Scholar]
- Pelclova D., Fenclova Z., Urban P., Ridzon P., Preiss J., Kupka K., Malik J., Dubska Z., Navratil T. (2009). Chronic health impairment due to 2,3,7,8-tetrachloro-dibenzo-p-dioxin exposure. Neuro Endocrinol. Lett. 30(Suppl. 1 ) 219–224 [PubMed] [Google Scholar]
- Poland A., Glover E., Kende A. S. (1976). Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J. Biol. Chem. 251, 4936–4946 [PubMed] [Google Scholar]
- Poland A., Knutson J. C. (1982). 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22, 517–554 [DOI] [PubMed] [Google Scholar]
- Qu X., Lam E., Doughman Y. Q., Chen Y., Chou Y. T., Lam M., Turakhia M., Dunwoodie S. L., Watanabe M., Xu B., et al. (2007). Cited2, a coactivator of HNF4alpha, is essential for liver development. EMBO J. 26, 4445–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. V., Bradfield C. A. (1996). Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12, 55–89 [DOI] [PubMed] [Google Scholar]
- Silkworth J. B., Koganti A., Illouz K., Possolo A., Zhao M., Hamilton S. B. (2005). Comparison of TCDD and PCB CYP1A induction sensitivities in fresh hepatocytes from human donors, sprague-dawley rats, and rhesus monkeys and HepG2 cells. Toxicol. Sci. 87, 508–519 [DOI] [PubMed] [Google Scholar]
- Sun Y. V., Boverhof D. R., Burgoon L. D., Fielden M. R., Zacharewski T. R. (2004). Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 32, 4512–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos R., Murray I. A., Smith P. B., Patterson A., Perdew G. H. (2012a). Role of the Ah receptor in homeostatic control of fatty acid synthesis in the liver. Toxicol. Sci. 129, 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos R., Patel R. D., Murray I. A., Smith P. B., Patterson A. D., Perdew G. H. (2012b). Aryl hydrocarbon receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology. 55, 1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz D. C., Hinton D. E., Law J. M., Kullman S. W. (2006). Dynamic gene expression changes precede dioxin-induced liver pathogenesis in medaka fish. Toxicol. Sci. 89, 524–534 [DOI] [PubMed] [Google Scholar]
- Vorderstrasse B. A., Steppan L. B., Silverstone A. E., Kerkvliet N. I. (2001). Aryl hydrocarbon receptor-deficient mice generate normal immune responses to model antigens and are resistant to TCDD-induced immune suppression. Toxicol. Appl. Pharmacol. 171, 157–164 [DOI] [PubMed] [Google Scholar]
- Walter G. L., Jones P. D., Giesy J. P. (2000). Pathologic alterations in adult rainbow trout, Oncorhynchus mykiss, exposed to dietary 2,3,7,8-tetrachlorodibenzo-p-dioxin. Aquat. Toxicol. 50, 287–299 [DOI] [PubMed] [Google Scholar]
- Yang Z. X., Shen W., Sun H. (2010). Effects of nuclear receptor FXR on the regulation of liver lipid metabolism in patients with non-alcoholic fatty liver disease. Hepatol. Int. 4, 741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. K., Yeager R. L., Tanaka Y., Klaassen C. D. (2010). Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol. Appl. Pharmacol. 245, 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.