Editor’s Highlight: Byproducts of constitutive metabolism may themselves be toxic, complicating the risk assessment of the same chemicals encountered from external sources. The application of stable labeled compounds offers insight into the source of chemicals producing biological effects and provides a basis to quantify the contribution of exogenous exposure to biological events. This report describes the concentration dependent contributions of exogenous [13C2]-acetaldehyde and endogenously produced acetaldehyde to adduct formation in human lymphoblastoid cells in vitro. — Jeffrey Fisher
Key Words: acetaldehyde, DNA adduct, micronucleus, biomarker of exposure, biomarker of effect, liquid chromatography–, mass spectrometry.
Abstract
The dose-response relationship for biomarkers of exposure (N2-ethylidene-dG adducts) and effect (cell survival and micronucleus formation) was determined across 4.5 orders of magnitude (50nM–2mM) using [13C2]-acetaldehyde exposures to human lymphoblastoid TK6 cells for 12h. There was a clear increase in exogenous N 2-ethylidene-dG formation at exposure concentrations ≥ 1µM, whereas the endogenous adducts remained nearly constant across all exposure concentrations, with an average of 3.0 adducts/107 dG. Exogenous adducts were lower than endogenous adducts at concentrations ≤ 10µM and were greater than endogenous adducts at concentrations ≥ 250µM. When the endogenous and exogenous adducts were summed together, statistically significant increases in total adduct formation over the endogenous background occurred at 50µM. Cell survival and micronucleus formation were monitored across the exposure range and statistically significant decreases in cell survival and increases in micronucleus formation occurred at ≥ 1000µM. This research supports the hypothesis that endogenously produced reactive species, including acetaldehyde, are always present and constitute the majority of the observed biological effects following very low exposures to exogenous acetaldehyde. These data can replace default assumptions of linear extrapolation to very low doses of exogenous acetaldehyde for risk prediction.
Acetaldehyde is a high-volume industrial chemical used in a diverse array of consumer and industrial applications. Humans are exposed to acetaldehyde through a wide variety of sources, including tobacco smoke, automotive emissions, and flavoring agents in foods and beverages (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, 1999; National Toxicology Program, 2011). It is used as the primary or intermediate chemical in the manufacture of a large number of industrial reagents including acetic acid, butylene glycol, crotonaldehyde, paraldehyde, and metaldehyde (National Toxicology Program, 2011). Acetaldehyde is listed as possible human carcinogen by the National Toxicology Program and by the International Agency for Research on Cancer (IARC Monographs on the Evaluation of Carciongenic Risks to Humans, 2010; National Toxicology Program, 2011). It is carcinogenic in animals, causing nasopharyngeal and laryngeal carcinomas in rats and hamsters following inhalation exposure (National Toxicology Program, 2011; Woutersen and Feron, 1987; Woutersen et al., 1984, 1986). Acetaldehyde is produced following metabolism of vinyl acetate and ethanol (Bogdanffy and Valentine, 2003; IARC Monographs on the Evaluation of Carciongenic Risks to Humans, 2010). In addition to its exogenous sources, acetaldehyde is endogenously produced as a byproduct of cellular respiration and metabolism (Hazen et al., 1998; O’Brien et al., 2005; Shin et al., 2009).
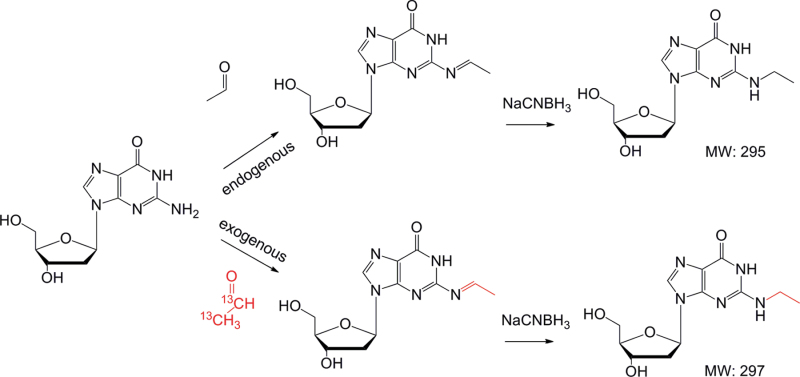
Acetaldehyde is a highly reactive 2-carbon aldehyde that can adduct to macromolecules including proteins and DNA (Balbo et al., 2008, 2012a, b; Brooks and Theruvathu, 2005; Chen et al., 2007; Fang and Vaca, 1995; Hecht et al., 2001a, b; Kuykendall and Bogdanffy, 1992a, b, 1994; Mabuchi et al., 2012; Setshedi et al., 2010; Takeshita et al., 1997). Acetaldehyde reacts with DNA at primarily the N2 position of deoxyguanosine (dG) and forms the N2-ethylidene-dG adduct, which can be reduced to N2-ethyl-dG following reaction with a strong reducing agent (Fig. 1) (Hecht et al., 2001a, b; Wang et al., 2000). Acetaldehyde can also react twice with dG to form the 1,N2-propano-dG adduct in the presence of amino acids (Garcia et al., 2011; Hecht et al., 2001b). The N2-ethylidene-dG adduct has been detected in a number of biological systems including human and rodent liver, lung, buccal cells, lymphocytes, and granulocytes, with the majority of work focusing on the formation of the adduct following ingestion of ethanol in humans or rodents (Chen et al., 2007; Fang and Vaca, 1997; Matsuda et al., 2007; Nagayoshi et al., 2009; Oyama et al., 2010; Vaca et al., 1998). Recent studies monitoring the formation of DNA adducts following [13C2]-acetaldehyde exposures in a human lung fibroblast cell line (IMR-90) have shown the formation of exogenous 1,N2-propano-dG along with the N2-ethylidene-dG adducts though no values for the N2-ethylidene-dG adduct were reported (Garcia et al., 2011). In addition to directly forming DNA adducts, acetaldehyde has been shown to increase reactive species from oxidative stress and lipid peroxidation, which may also form DNA lesions (Garcia et al., 2011; Millonig et al., 2011).
Fig. 1.
Reaction scheme for the formation of endogenous and exogenous N2-ethylidene-dG using [13C2]-acetaldehyde.
Biomarkers of effect have been measured in a number of in vitro systems exposed to acetaldehyde. Acetaldehyde has been shown to be genotoxic in a wide range of cell lines and in animal models with induction of sister-chromatid exchange, micronuclei, DNA adducts, DNA-protein crosslinks, DNA-DNA crosslinks, and chromosomal aberrations (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, 1999; Kuykendall and Bogdanffy, 1992a; Speit et al., 2008). Acetaldehyde is a weak mutagen, showing positive results in a number of mammalian systems (Grafström et al., 1994; He and Lambert, 1990). Site-directed mutagenesis studies have investigated both the N2-ethyl-dG and the 1,N2-propano-dG adducts, finding that the N2-ethyl-dG has a weak effect on blockage of replication, whereas the 1,N2-propano-dG is a potentially stronger mutagen (Choi and Guengerich, 2006; Stein et al., 2006; Terashima et al., 2001; Upton et al., 2006a, b; Yang et al., 2002).
Both endogenous and exogenous exposures to acetaldehyde are common, and there is a need to differentiate between both, especially at low exposures. This study is the first to use stable isotope exposures of [13C2]-acetaldehyde to differentiate between endogenous and exogenous formation of the N2-ethylidene-dG adduct and to study the effects of acetaldehyde on relative survival and micronucleus (MN) formation across exposure concentrations ranging over 4.5 orders of magnitude in human lymphoblastoid TK6 cells. To accomplish these goals, we developed a sensitive and selective liquid chromatography-tandem mass spectrometry (LC-MS/MS) method for the quantitation of the reduced DNA adduct (N2-ethyl-dG), and a flow cytometry method for assessing cell survival and MN formation to determine the effects of acetaldehyde exposures in cell culture. Data generated will enable the determination of the endogenous background of both biomarkers of exposure and effect and the contribution of exogenous acetaldehyde to those biomarkers in establishing possible threshold values.
MATERIALS AND METHODS
Chemicals and materials.
Acetonitrile, acetic acid, acetaldehyde, alkaline phosphatase, phosphodiesterase, dG, DNase I, ethyl methanesulfonate, high-performance liquid chromatography (HPLC)-grade water, methanol, MgCl2, N2-ethyl-dG, RPMI 1640 media, sodium phosphate, and sodium cyanoborohydride (NaCNBH3) were all purchased from Sigma (St Louis, MO). [15N5]-dG and [13C2]-acetaldehyde were ordered from Cambridge Isotope Laboratories (Andover, MA). Sodium pyruvate, penicillin, streptomycin, and 0.1% pluronics were purchased from Life Technologies Corporation (Carlsbad, CA).
Chemical hazards.
Acetaldehyde is a highly reactive compound and a possible human carcinogen and should be handled within a fume hood.
Preparation of internal standard.
The [15N5]-ethyl-dG internal standard was prepared using a modified version of previously published procedures (Kanaly et al., 2006; Sako et al., 1999). Briefly, a solution containing [15N5]-dG (5.2mM), NaCNBH3 (7.5mM), acetaldehyde (2.7M), and sodium phosphate buffer (60mM, pH 7.2) was incubated at 37°C for 15h. The reaction mixture was dried using a speedvac vacuum concentrator and redissolved in an aqueous solution of 10% methanol. The reaction mixture was injected onto an Agilent 1200 HPLC system, and N2-ethyl-dG was purified using a Thermo Scientific ODS-Hypersil column (4.6×250mm, 5 µm) with a flow rate of 1.5ml/min of mobile phases water (A) and methanol (B). [15N5]-N2-Ethyl-dG was separated using a gradient from 5% B to 30% B for the first 20min, followed by an increase to 70% B for 1min that was reduced to 30% B and held for 2min, followed by re-equilibration for 7min at 5% B. HPLC fractions were collected containing [15N5]-N2-ethyl-dG with a retention time of 16.5min and dried. The residue was redissolved and concentration determined by UV absorbance at 254nm with the extinction coefficient of 15,000/M cm (Wang et al., 2006).
Determination of adduct stability.
For the nucleoside stability of N2-ethylidene-dG, a solution of dG (5.8mM) and acetaldehyde (2.8M) was made to react in phosphate buffer (60mM, pH 7.2) for 1h at 37°C. An aliquot of the reaction mixture was injected onto the Agilent 1200 HPLC system with mobile phases of water with 0.1% acetic acid (A) and acetonitrile with 0.1% acetic acid (B). The flow rate was 1.0ml/min with a starting condition of 2% B, which was held for 5min, followed by a linear gradient of 4% B at 20min, 10% B at 30min, 12.5% B at 35min, followed by 2min at 80% B, and re-equilibration at the starting conditions for 7min. N2 -Ethylidene-dG eluted, with a retention time of 28.5min, which was collected using the fraction collector. Sodium phosphate buffer was added to the solution, and the aliquots were stored at −80°C until being used to determine the initial amount and the stability of N2-ethylidene-dG. For this experiment, aliquots were held at 15°C inside the HPLC and injections made every 45min. The experiment was run in triplicate. Aliquots were injected onto the HPLC system as mentioned previously, and peak areas were used to determine relative amounts of both dG and N2-ethylidene-dG.
For determination of N2-ethylidene-dG stability in DNA, calf thymus DNA (n = 3 experiments) was made to react with acetaldehyde (0.5M) in phosphate buffer for 1h at 37°C. DNA was concentrated using an Amicon 10kD MW filter, allowing the reaction solution to be removed. DNA was recovered by adding 300 µl of water to the filter, followed by mixing, and the DNA was cleaned by adding isopropyl alcohol. Precipitated DNA was allowed to dry, then dissolved in 650 µl of water, and aliquoted (~100 µg DNA per tube) into six tubes containing phosphate buffer (100mM, pH 7.2) for incubation at 37°C. Amount of N2-ethyl-dG adducts was quantitated at the following time points: 0, 3, 18, 24, 48, and 72h. At each time point, NaCNBH3 (50mM) was added to reduce N2-ethylidene-dG to N2-ethyl-dG. The solution was incubated for an additional 6h to enable the formation of more stable N2-ethyl-dG adduct. Following reduction, the samples were processed similarly to DNA samples from the cell culture experiments, and N2-ethyl-dG was quantitated by LC-MS/MS.
[13C2]-Acetaldehyde exposures in cell culture.
Human TK6 cells were originally obtained in 2008 from the American Type Culture Collection (Manassas, VA). Cells were routinely cultured in 75cm2 tissue culture flasks (Corning, NY) and maintained in RPMI 1640 medium with 10% heat-inactivated horse serum plus 0.1% Pluronic, sodium pyruvate, and antibiotics (penicillin at 20 units/ml and streptomycin 20 μg/ml) at 37±1°C, with 6±1% CO2 in air.
TK6 human lymphoblastoid cells (~2×107 cells/exposure, n = 6 per level) were exposed to [13C2]-acetaldehyde for 12h at the following concentrations: 0 (negative control), 0.05, 0.1, 1.0, 5.0, 10, 50, 250, 500, 1000, and 2000µM. Following completion of the exposure, the media were removed, and cells were washed and frozen at −80°C prior to DNA extraction. Cell survival and MN formation were determined using the identical exposure concentrations mentioned above in 12-well plates seeded at 8.0×105 cells/exposure concentration (n = 3).
Determination of cytotoxicity and MN frequency.
A flow cytometry–based cytotoxicity and MN assay developed by Litron Laboratories (Rochester, NY) was used to assess the cytotoxic and genotoxic effects of acetaldehyde in human TK6 cells (Bryce et al., 2007, 2008). This assay was chosen because it uses p53-competent human cells and has regulatory acceptance as a biomarker of chromosomal aberrations, and the flow-based MN assay has increased sensitivity compared with microscopy-based counting by scoring of 20,000 cells for MN assessment (OECD, 2010). The flow cytometry–based cytotoxicity and MN assay were performed using the In Vitro MicroFlow Kit and reagents (Litron Laboratories). Sample preparation, staining, and other methods were performed according to the In Vitro MicroFlow Instructional Manual. The data were collected using a Becton-Dickinson FACSCalibur 2-laser 4-color instrument (Becton Dickinson, San Jose, CA) as directed in the In Vitro MicroFlow Instructional Manual.
This flow cytometry–based method determines percent survival relative to unexposed controls and MN frequency in the same cell sample (Bryce et al., 2007, 2008). Relative cell survival was determined simultaneously on the same sample used for MN determination using an absolute counting technique with 6-μm latex counting beads as internal standards added during the cell preparation for flow cytometry. Relative survival was calculated using the ratio of counting beads to intact viable nuclei as a measure of the number of cells containing intact nuclei after exposures compared with that in the vehicle controls. The MN frequency was determined from 20,000 (± 2000) cells analyzed from each sample. Media and vehicle controls were run along with the positive controls and study samples.
ALDH2 genotyping of human TK6 cells.
DNA was isolated from human TK6 cells using Qiagen Blood & Cell Culture DNA Mini Kit (Qiagen, http://www.qiagen.com/). The genotype of the TK6 cell line for the aldehyde dehydrogenase 2 (ALDH2) gene was determined to be wild type using direct sequencing of a PCR amplification product of the region of ALDH2 (exon 12) containing the ALDH2*1/*2 SNP. The ALDH2*2 is known to affect ALDH2 enzyme activity toward ethanol (SNP reference: ALDH2 rs671 SNP = A; wild type = G; rs671: GAA to AAA at codon 487) (Yoshida et al., 1984). The following ALDH2-specific primers (5′→3′) were used: CCACACTCACAGTTTTCACT—forward primer and CAAATTACAGGGTCAACTG—reverse primer for the PCR reaction; the primer TATGATGTGTTTGGAGCC was used for sequencing the PCR product (Tanaka et al., 1996). The PCR conditions were as follows: 94°C for 3min, then 35 cycles of 94°C for 1min, 56°C for 1min, and 72°C for 1min (for the last cycle, 72°C was for 5min). Agarose gel electrophoresis demonstrated a PCR DNA band of the correct length, and this PCR product was submitted to Eton Biosciences (RTP, NC) for DNA sequence analysis. The DNA sequence from our TK6 cells matched the wild-type allele of ALDH2 (GAA = glutamine) at the rs671 SNP location.
DNA isolation.
DNA was isolated from cell pellets using the NucleoBond DNA Isolation Kit (Machery Nagel, Bethlehem, PA) per the manufacturer’s instructions. Briefly, cells were washed and lysed with the supplied buffers (G1 and G2), followed by a 1-h incubation at 50°C with proteinase K. Following incubation, the DNA was extracted using the anion exchange column and stored at −80°C prior to analysis.
DNA reduction and digestion.
DNA amounts from cells ranged from 21 to 159 µg. Lower amounts of DNA were used in the high exposures (> 500µM) due to the high amount of exogenous adducts formed. DNA was thawed and incubated with NaCNBH3 (50mM) and NaPO4 (100mM, pH 7.2) for 6h at 37°C to reduce N2-ethylidene-dG to N2-ethyl-dG. Following reduction, the DNA was frozen until digested (−80°C). Reduced DNA was thawed, and 200 µl of Tris/MgCl2 buffer (40/10mM final concentration, pH 7.2) along with 5fmol of the internal standard ([15N5]-N2-ethyl-dG) was added. DNA was digested with DNAse I (200 units), alkaline phosphatase (5 units), and phosphodiesterase (0.005 units) with a 10-min preincubation with DNase I for 1h at 37°C. Following digestion, hydrolyzed DNA was filtered with a Pall Nanosep 3kDa filter (Port Washington, NY) at 8000rpm for 30min.
HPLC.
Hydrolyzed DNA was injected onto an Agilent 1200 HPLC fraction collection system equipped with a diode-array detector (Santa Clara, CA). Analytes were separated by reverse-phase liquid chromatography using an Atlantis C18 T3 (150×4.6mm, 3 µm) column from Waters Corporation (Milford, MA). The mobile phases were water with 0.1% acetic acid (A) and acetonitrile with 0.1% acetic acid (B). The flow rate was 1.0ml/min with a starting condition of 2% B, which was held for 5min, followed by a linear gradient of 4% B at 20min, 10% B at 30min, 20% B at 40min, 80% B at 45min, which was held for 6min, followed by re-equilibration to the starting conditions for 12min. Deoxyguanosine and N2-ethyl-dG eluted, with retention times of 13.0 and 33.8min, respectively. The amount of dG in samples was quantitated by the UV peak area (λ = 254nm) at the corresponding retention time using a calibration curve ranging from 0.1 to 50 nmol dG on column.
LC-MS/MS.
LC-MS/MS analyses were performed on a TSQ-Quantum Ultra triple-stage quadrupole mass spectrometer (Thermo Scientific, San Jose, CA) operated in selected reaction monitoring (SRM) mode to detect and quantify N2-ethyl-dG. The mass spectrometer was interfaced with a nano-acquity ultra performance liquid chromatography system from Waters Corporation. A 0.18×20mm Symmetry C18 trap column (5 µm particle size) and a 0.1×100mm HSS T3 analytical column (1.8 µm particle size) from Waters were used. Mobile phases were comprised of water with 0.1% acetic acid (A) or acetonitrile with 0.1% acetic acid (B). Analytes were first retained on a trap column with a flow rate of 5 µl/min of 2% mobile phase B, followed by transfer to the analytical column with an initial starting condition of 2% B at 0.6 µl/min for 5min followed by a linear gradient to 30% B over 12.5min and to 80% B over 1.5min. The flow was then held at 80% B for 1min followed by re-equilibration for an additional 5min. The analytes were introduced to the MS using positive-mode electrospray ionization with a source voltage of 2200V and no additional gases. The ion transfer tube was held at 300ºC and skimmer offset set to zero. Scan speed was set at 75ms, scan width at 0.1 m/z, and a peak width at 0.7 m/z for Q1 and Q3. Argon was used as the collision gas and was set at 1.5 arbitrary units. The mass transitions used for detection and quantification are shown in Supplementary table SI-1. Linear calibration curves (R2 > 0.99) using the analyte to internal standard ratio were used to quantify the amount of N2-ethyl-dG in the samples (Supplementary fig. SI-1).
Statistical analysis and modeling.
The R statistical program was utilized for the statistical modeling (Version 2.11.0). Biomarkers of exposure and effect across groups were compared using Student’s t-tests (R function t-test). The Bonferroni-corrected p value threshold was used to account for multiple testing, with p < 0.05/(number of tests) used to declare significance.
RESULTS
Stability of N2-Ethylidene-dG as a Nucleoside and in DNA
The stability of the N2-ethylidene-dG adduct at the nucleoside level was assessed following the isolation of the unstable N2-ethylidene-dG from a reaction mixture of dG and acetaldehyde. The isolated adduct was injected onto the HPLC every 45min and the peak area used to determine the relative abundance of N2-ethylidene-dG and dG (Supplementary fig. SI-2a). The calculated half-life (t 1/2) of N2-ethylidene-dG at 15°C in phosphate buffer (pH 7.2) was 20.0min. N2-Ethylidene-dG was unstable, with the primary degradation product being dG as shown by the increase in dG area during the incubation. No N2-ethyl-dG was observed without the incorporation of the reduction step. In contrast to the instability of the N2-ethylidene-dG adduct at the nucleoside level, the adduct was more stable in calf thymus DNA incubated for up to 72h at 37°C with a calculated t 1/2 of 69.3h (Supplementary fig. SI-2b).
LC-MS/MS Validation
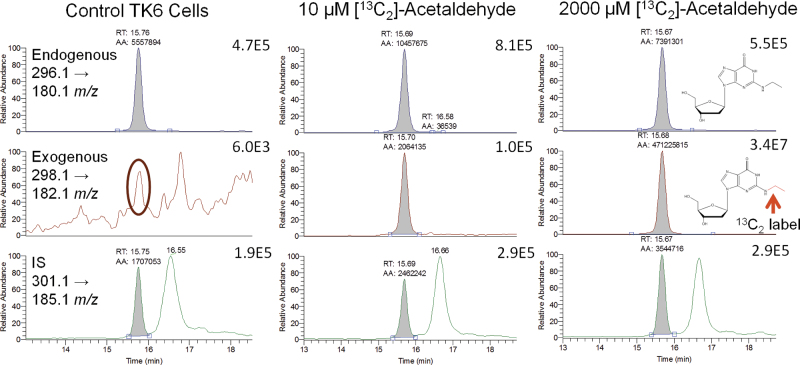
The method was validated, with accuracy, precision, recovery, and matrix effects assessed. The limits of detection (signal:noise > 3) and quantitation (signal:noise > 10) of N2-ethyl-dG were 20 and 40 amol on-column, respectively (Supplementary fig. SI-1). Accuracy and precision were determined at three concentrations (2, 10, and 30fmol) by adding N2-ethyl-dG to nonreduced control calf thymus DNA, which was processed similarly to study samples (Supplementary table SI-2). For determination of recovery and matrix effects, nonreduced calf thymus DNA was processed with both internal (5fmol) and analytical (30fmol) standards added postprocessing, along with analytical and internal standards in the absence of matrix. The recovery was 67%, and no matrix effects were observed with a % matrix effect of 100. Interassay (n = 6) precision and accuracy were also assessed using the 30fmol concentration with a % expected and % CV of 98 and 8.8, respectively. The possible artifact formation of N2-ethyl-dG was monitored with each run using reagent and fraction collection blanks. When artifact formation was detected, the amount was small (ranging from nondetectable to 400 amol) and was corrected in the determination of N2-ethyl-dG amounts. No artifactual formation of [13C2]-N2-ethyl-dG was observed, though with increasing amounts of N2-ethyl-dG, the natural isotopic distribution (0.7% of the 296.1 → 180.1 m/z peak area) resulted in an observable peak for the 298.1 → 182.1 m/z transition (Fig. 2). Accordingly, the area counts for [13C2]-N2-ethyl-dG chromatographic peaks were corrected by subtracting the expected isotopic peak area from the observed values to account for the isotopic contribution from endogenous N2-ethyl-dG.
Fig. 2.
Representative chromatograms. The 296.1 → 180.1 m/z transition was used to quantitate the amount of endogenous adducts, whereas the 298.1 → 182.1 m/z transition was used to quantitate the exogenous adducts. The transition for the internal standard was 301.1 → 185.1 m/z. The natural +2 m/z isotopic abundance of ~0.7% of the endogenous signal can be observed in the circled chromatogram for the control TK6 cells.
Biomarkers of Exposure
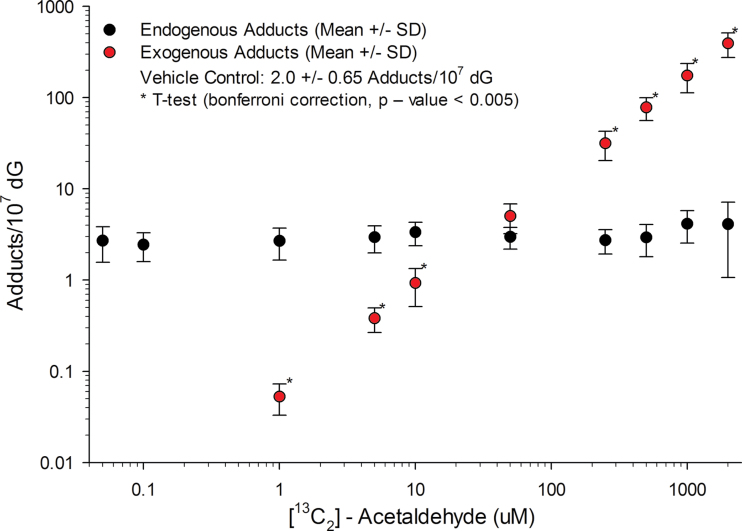
Human lymphoblastoid TK6 cells routinely maintained in conditions described in the Materials and Methods section were exposed to [13C2]-acetaldehyde for 12h at 10 concentrations ranging from 0.05 to 2000µM, along with nonexposed controls. Following completion of exposures, media containing [13C2]-acetaldehyde were removed, cells washed, and cell pellets frozen until DNA isolation and quantitation of DNA adducts by LC-MS/MS. The amounts of N2-ethyl-dG, [13C2]-N2-ethyl-dG, and sum of adducts (N2-ethyl-dG and [13C2]-N2-ethyl-dG) were determined from replicate samples (n = 5–6/exposure concentration) and are shown in Table 1. There were small changes in the endogenous adduct formation across the dose range though none were statistically significant compared with the controls. There was a clear dose-dependent increase in the formation of exogenous adducts with increasing [13C2]-acetaldehyde exposure concentration, whereas the endogenous level of N2-ethyl-dG remained nearly constant with 3.0±1.4 (mean ± SD) adducts/107 dG (Fig. 3). [13C2]-N2-Ethyl-dG was not detectable at exposure concentrations < 0.1µM due to the natural +2 m/z isotopic abundance (~0.7%) from the endogenous adduct. At 0.1µM, two samples had quantifiable amounts of exogenous adducts (0.02 and 0.06 adducts/107 dG). At the 1.0µM exposure concentration, a clear increase in [13C2]-N2-ethyl-dG was observed, with the exogenous peak equal to ~4% of the endogenous peak. This allowed for clear quantitation of the exogenous adducts. Exogenous adducts were significantly lower (Student’s t-test, p value < 0.005 threshold to account for 10 tests) than the corresponding endogenous adducts at [13C2]-acetaldehyde concentrations ≤ 10µM (Table 1). Exogenous adducts were significantly higher (Student’s t-test, p value < 0.005) than the corresponding endogenous adducts at [13C2]-acetaldehyde concentrations ≥ 250µM (Table 1).The sum of the adducts was significantly increased (Student’s t-test, Bonferroni p value < 0.005) from the average endogenous adducts at [13C2]-acetaldehyde concentrations ≥ 50µM (Table 1).
Table 1.
Endogenous, Exogenous, and Sum of N2-Ethylidene-dG Levels from [13C2]-Acetaldehyde Exposures in TK6 Cells
| [13C2]-Acetaldehyde (µM) | Endogenous adducts/107 dG (mean ± SD) | Exogenous adducts/107 dG (mean ± SD) | Sum adducts/107 dG (mean ± SD) | Exogenous versus endogenous, p value | Sum versus mean, p value | n |
|---|---|---|---|---|---|---|
| 0 | 2.0±0.65 | N.D. | 2.0±0.65 | — | — | 5 |
| 0.05 | 2.7±1.1 | N.D. | 2.7±1.1 | — | — | 6 |
| 0.1 | 2.4±0.84 | 0.2±0.06* | 2.5±0.85 | — | — | 6 |
| 1 | 2.7±1.0 | 0.05±0.02 | 2.7±1.0 | 1.52E−03 | 7.06E−01 | 6 |
| 5 | 3.0±1.0 | 0.38±0.17 | 3.3±1.0 | 1.19E−03 | 2.20E−01 | 6 |
| 10 | 3.4±0.96 | 0.93±0.42 | 4.2±1.3 | 8.71E−04 | 2.78E−02 | 6 |
| 50 | 3.0±0.79 | 5.0±1.8 | 8.0±2.4 | 6.22E−02 | 4.96E−03 | 5 |
| 250 | 2.8±0.81 | 32±11 | 34±12 | 1.44E−03 | 6.16E−04 | 6 |
| 500 | 2.9±1.1 | 78±22 | 81±22 | 3.94E−04 | 1.82E−04 | 6 |
| 1000 | 4.2±1.6 | 180±62 | 180±61 | 1.05E−03 | 4.33E−04 | 6 |
| 2000 | 4.1±3.0 | 390±120 | 400±120 | 4.84E−04 | 2.25E−04 | 6 |
Notes. The endogenous mean across all exposure concentrations and the control was 3.0±1.4 adducts/107 dG. The sum was calculated by adding the endogenous and exogenous adducts from each sample, and mean and SD were calculated. A statistical comparison of exogenous and endogenous adducts was conducted using a t-test with a Bonferroni multiple-testing threshold for an overall false-positive rate of 0.05, corresponding to 0.005 as the threshold for each test. A second comparison between the sum of adducts and the endogenous mean was conducted using the same Bonferroni correction. *For the 0.1µM concentration, two samples had quantifiable amounts of exogenous adducts (shown). The remaining four samples did not have detectable levels of the exogenous adducts.
Fig. 3.
Endogenous versus exogenous adducts. The endogenous and exogenous N2-ethylidene-dG adducts at each exposure concentration are plotted on a log versus log scale. Exogenous adducts from samples with no detectable amounts are not shown. The exogenous adducts from the two samples with detectable amounts for the 0.1µM concentration are not shown because an average and SD could not be calculated.
Exogenous [13C2]-N2-ethyl-dG increased with two distinct linear regions (Supplementary fig. SI-3), an initial linear increase in exogenous adducts from 1 to 50µM [13C2]-acetaldehyde (adducts = 0.102(x) − 0.091) and a second more steep increase from 250 to 2000µM [13C2]-acetaldehyde (adducts = 0.208(x) − 25.3). Using a linear regression from 1 to 50μM, with an intercept set at the origin, exogenous adducts were extrapolated using the following equation (exogenous adducts = 0.100 (acetaldehyde concentration)). The predicted values were 0.01, 0.005, and 0 for the 0.1, 0.05, and 0µM concentrations, respectively.
Biomarkers of Effect
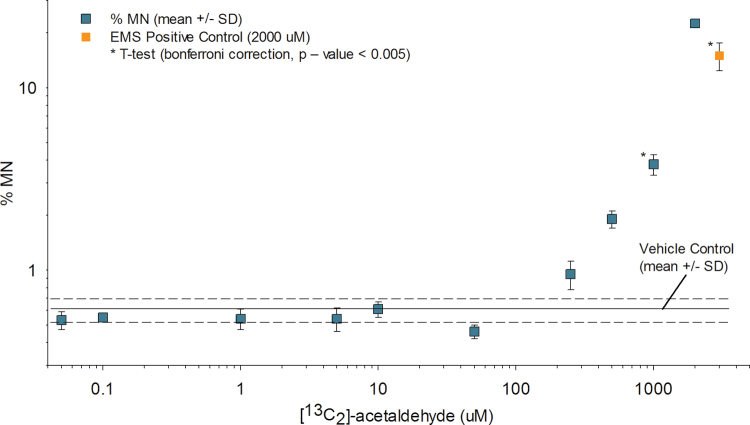
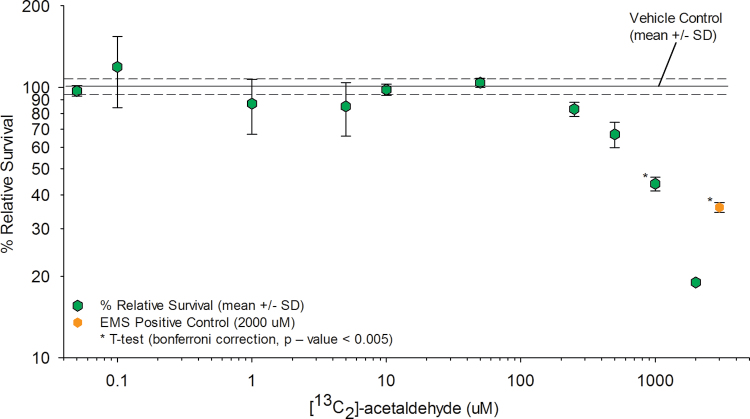
TK6 cells were exposed to the same 10 concentrations of [13C2]-acetaldehyde used for the determination of DNA adducts, though 12-well plates seeded at 8.0×105 cells/well were used for determination of biomarkers of effect. Following completion of the 12-h exposures, media were removed, cells were washed, and the formation of micronucleated cells and relative cell survival were assessed using flow cytometry (Figs. 4 and 5; Supplementary table SI-3). Samples were run in triplicate for each exposure concentration. Only one determination was made for the 2000µM exposure concentration due to poor sample quality. The frequency of MN formation was significantly greater than the vehicle control data at concentrations ≥ 1000µM [13C2]-acetaldehyde (Student’s t-test p value = 0.0038, with Bonferroni-corrected p value threshold = 0.0056). Statistically significant reductions in relative cell survival were observed at concentrations ≥ 1000µM [13C2]-acetaldehyde (Student’s t-test p value = 0.0004, with Bonferroni p value threshold at 0.0056). Increases in MN formation and decreases in cell survival were observed for concentrations < 1000µM though they were not statistically significant.
Fig. 4.
MN formation. The percent micronucleated cells (% MN) for each exposure concentration are shown (mean ± SD). The % MN for the vehicle control was 0.61±0.10 (mean ± SD).
Fig. 5.
Cell survival. The percent relative survival for each exposure concentration is shown (mean ± SD). The percent relative survival for the vehicle control was 100±6.8 (mean ± SD).
DISCUSSION
The [13C2]-acetaldehyde exposures in human TK6 lymphoblastoid cells showed clear changes to both biomarkers of exposure (DNA adducts) and effect (micronucleated cells and cell survival) in a dose-dependent fashion. The use of [13C2]-acetaldehyde allowed for the determination of both endogenous and exogenous DNA adducts and their relative contribution to the overall amount of adducted DNA. This information was used to determine thresholds for both biomarkers of exposure and effect.
Previous studies have commented on the possible artifactual formation of N2-ethyl-dG during sample processing, being unable to demonstrate its occurrence (Balbo et al., 2012a, b; Singh et al., 2009). In contrast, a small peak at the corresponding retention time of N2-ethyl-dG artifact was observed using this methodology though the amount was minimal with less than 0.25 adducts/107 dG found in 50 µg of DNA. Our detection of this small coeluting peak is likely due to the greater sensitivity achieved with the nanospray source employed in this method. The presence of the peak was monitored in reagent blanks and if present, subtracted from the endogenous peak areas to correct for the relatively small amount of increased signal.
The stability of the N2-ethylidene-dG adduct in DNA and as a nucleoside was determined, with a large difference in stability observed between the two forms of the adduct. At the nucleoside level, N2-ethylidene-dG rapidly degraded back to dG with a t 1/2 of 20min, whereas it was markedly more stable in intact calf thymus DNA with a t 1/2 of 69.3h. The > 200× difference in stability illustrates the importance of the reduction step prior to enzymatic digestion of the DNA. These data do not illustrate the possible rates of degradation and repair in biological systems but do provide information on the stability during processing of the samples. The rapid degradation of the nucleoside correlates well with previous reports on the stability of the N2-ethylidene-dG adduct (5min at 37°C) and shows the importance of the reduction step to convert the unstable adduct to the more stable N2-ethyl-dG allowing for analysis using LC-MS/MS (Wang et al., 2000). The slower t 1/2 may be temperature dependent, with degradation at 37°C occurring at a faster rate than at 15°C though additional studies are needed to confirm this relationship. Recent studies have shown that N2-ethylidene-dG adducts are rapidly formed and removed in buccal and white blood cells following ethanol consumption in human volunteers (Balbo et al., 2012a, b). Additional in vitro studies have shown that the N2-ethylidene-dG adducts are repaired in HL60 cells with a t1/2 of 35h (Hori et al., 2012). Given these studies, it is likely that active repair of the N2-ethylidene-dG adduct occurs in humans. Further studies investigating the in vivo half-life and possible accumulation of N2-ethylidene-dG using stable isotope exposures are needed to better understand the repair or removal of exogenous acetaldehyde-derived DNA adducts. We have recently quantified N2-ethylidene-dG from nonexposed rat nasal mucosa with in vivo samples having higher endogenous adducts (~10 adducts/107 dG), illustrating that our methodology is capable of generating results similar to results of other groups.
There is considerable variation in the amount of endogenous N2-ethylidene-dG values reported in the literature, with differences observed between different tissues and in both in vitro and in vivo samples (Abraham et al., 2011; Balbo et al., 2012a, b; Chen et al., 2007; Fang and Vaca, 1997; Garcia et al., 2011; Hori et al., 2012; Matsuda et al., 2007; Nagayoshi et al., 2009; Oyama et al., 2010; Singh et al., 2009, 2012; Vaca et al., 1998; Wang et al., 2006). As acetaldehyde is a ubiquitous environmental compound, there may be considerable variation in human exposures due to several factors including lifestyle choices (food/alcohol consumption and smoking) and occupation. There are additional genetic components that may influence acetaldehyde’s potential toxicity following exposures in humans. ALDH2 is a well-characterized polymorphic enzyme primarily responsible for the clearance of acetaldehyde. Individuals with the ALDH2−/+ and ALDH2−/− genotyope (SNP genotype reference = ALDH2*2 rs671) are highly susceptible to the effects of acetaldehyde with markedly decreased rates of clearance compared with those with ALDH2+/+ genotype. This has been shown to be an additional risk factor for ethanol-induced carcinogenesis (IARC Monographs on the Evaluation of Carciongenic Risks to Humans, 2010). The TK6 cell line is ALDH2+/+ although neither gene expression data nor protein expression data were not collected in this study. Acetaldehyde is also endogenously produced, with myeloperoxidase likely playing a key role in the endogenous production (Hazen et al., 1998; Shin et al., 2009). Myeloperoxidase has also been implicated in inflammation-induced nitrous and chlorination-derived DNA damage (Mangerich et al., 2012). The TK6 cell line does not express myeloperoxidase in large amounts (Ji et al., 2009), which may help explain the lower endogenous adducts compared with the in vivo samples.
There was a clear increase in the formation of exogenous N2-ethylidene-dG adducts at [13C2]-acetaldehyde concentrations above 1µM. Although the exogenous adducts increased with increasing exposure concentrations, the endogenous adducts remained nearly constant. An upward trend to higher endogenous adducts was observed though not statistically significant (p = 0.24). The slight increase may be due to saturation of repair or metabolic clearance of endogenous acetaldehyde. The relatively constant amount of endogenous N2-ethylidene-dG adducts observed in human TK6 cells was lower than the reported values obtained from human tissues including lung, liver, buccal cells, and lymphocyte samples (Balbo et al., 2008, 2012a, b; Chen et al., 2007; Singh et al., 2009; Wang et al., 2006). This is not surprising given that the TK6 cells have a much faster rate of replication and the exposure to both endogenous and exogenous (food and environmental) sources is much lower in cell culture.
The lowest exposure concentration with a detectable amount of exogenous adducts was 0.1µM although it was not detectable in all the samples. Samples had a detectable peak in the 298.1 → 182.1 m/z transition; however, the relative percentage of that peak to the endogenous peak was very close to the natural isotopic abundance of 0.7%, making low-level detection of exogenous N2-ethyl-dG more difficult. At the 1.0µM exposure concentration, a clear increase in exogenous N2-ethyl-dG was observed, with the exogenous peak equal to ~4% of the endogenous peak, which allowed for clear quantitation of exogenous adducts. The small mass difference (+2Da) found in the exogenous adducts is limited by acetaldehyde having only two carbons that can be stable-isotope labeled with 13C. The use of deuterium labels would result in larger differences in mass (+ 1Da/deuterium), which may allow for detection of exogenous adducts at lower exposure concentrations, but would be limited by possible differences in rates of metabolism of acetaldehyde and hydrogen-deuterium exchange. Acetaldehyde’s small size offers a limited number of positions for the incorporation of stable or radioactive isotopes to increase the mass difference between endogenous and exogenous adducts. It can be problematic to increase the mass difference above the natural isotopic abundance using stable isotopes. This limitation can be observed through other small endogenous compounds or exogenous compounds that form lesions identical to those of endogenous compounds (Swenberg et al., 2011).
The formation of exogenous adducts had two distinct linear segments over the exposure concentrations, with a change in slope occurring between 50 and 250µM [13C2]-acetaldehyde (Supplementary fig. SI-3). The dose-response relationship for the formation of exogenous adducts suggests a sublinear relationship, with a higher rate of formation of exogenous adducts occurring at higher acetaldehyde concentrations. The reason for the two distinct slopes is not known although we hypothesize that it may be due to either the saturation of DNA repair or saturation of metabolism of acetaldehyde by the cells allowing for the increase in exogenous adducts (Swenberg et al., 2008). It has been shown that several DNA repair pathways are critical to the repair of DNA lesions caused by acetaldehyde, with FANCD2 playing an important role (Garaycoechea et al., 2013; Langevin et al., 2011; Ridpath et al., 2007). Acetaldehyde is primarily cleared following metabolism to acetic acid by ALDH2, which is a known polymorphic enzyme (IARC Monographs on the Evaluation of Carciongenic Risks to Humans, 2010). The metabolism of acetaldehyde to acetate was not monitored in this study, and the metabolic capabilities of the TK6 cell line for acetaldehyde are not well characterized. It is critical to note that acetaldehyde is primarily metabolized by ALDH2 to acetate in humans (Brooks et al., 2009a; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, 1999, 2010). ALDH2 polymorphisms have been indicated as an additional risk factor for ethanol-induced cancers in primarily northern Asians (Brooks et al., 2009a, b; Yukawa et al., 2012). Accordingly, this susceptible population should be taken into account for the assessment of risk associated with acetaldehyde exposures in humans.
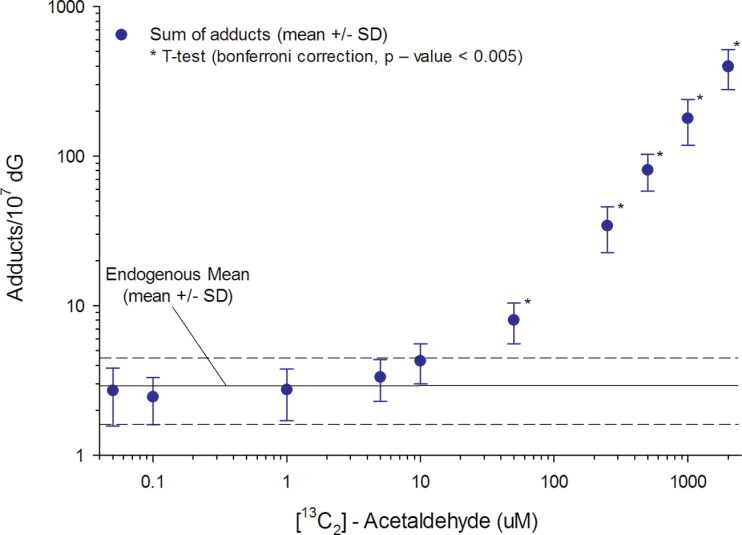
The use of the stable isotope–labeled acetaldehyde allowed for the determination of the relative contribution of exogenous and endogenous adducts to the total amount of acetaldehyde adduction over more than 4 orders of magnitude. As there is no biological difference between endogenous and exogenous adducts, the sum of adducts was plotted (Fig. 6). A t-test (Bonferroni correction, p < 0.005) was used to determine whether the sum of adducts was statistically different from the endogenous background, with significant increases over background occurring at 50µM (p = 0.0050) and more dramatic increases occurring at higher concentrations. Using our combined approach of nano-LC-MS/MS and stable isotope exposures, we were able to quantitate the formation of exogenous adducts at ~50× lower concentration (1.0µM) than if nonlabeled acetaldehyde were utilized in the exposures. Additional information on the formation of exogenous adducts at lower exposures may be gained through the use of 14C, 3H, or 2H to increase the mass difference between endogenous and exogenous adducts. Using both the sum of and the difference between endogenous and exogenous adducts, we clearly show that at low exposures (< 50µM), the additional [13C2]-acetaldehyde has no impact on the overall amount of adducted DNA.
Fig. 6.
Sum of adducts. The mean of the endogenous adducts was 3.0 adducts/107 dG, and the sum of the adducts at each exposure concentration is shown (mean ± SD). The sum of the adducts for the control is not shown (2.0±0.65 adducts/107 dG, mean ± SD).
The biological relevance of N2-ethylidene-dG in acetaldehyde-induced carcinogenesis is not fully characterized. Acetaldehyde can form other DNA lesions (DNA-protein-crosslinks, 1,N2-propano-dG) that are known to be more bulky and mutagenic compared with the N2-ethyl-dG. Additional information of the relevance of the N2-ethylidene-dG adducts and other acetaldehyde-derived DNA lesions in carcinogenesis is needed. It is clear that acetaldehyde is a direct-acting genotoxic agent capable of forming a number of DNA lesions. These lesions may help explain the cytogenetic effects that have been observed following acetaldehyde exposures.
Biomarkers of exposure, including cell survival and MN formation, were monitored across 10 exposure concentrations, along with nonexposed controls. The increased MN formation was well correlated with increased cytotoxicity (cell death). Statistically significant changes in MN formation and cell survival occurred at 1000µM, respectively. It should be noted that decreases in cell survival and increases in MN formation occurred at lower concentrations (250 and 500µM) though they were not statistically significant. Previous studies have reported cell survival and MN data in other cell lines, with statistically significant changes occurring at similar concentrations as observed from our results in the TK6 cell line (Bird et al., 1982; IARC Monographs on the Evaluation of Carciongenic Risks to Humans, 2010; Migliore et al., 1996). The changes in cell survival and MN formation over the vehicle controls occurred at a 20× higher concentration (1000µM) than the increase in the sum of the adducts (50µM) over the endogenous background. This suggests that the internal cellular concentration of acetaldehyde, as measured by adduct formation, requires ~20× higher concentrations to have impact on this study’s biomarkers of effect. This relationship can be further explored by the ~1000× difference in exogenous acetaldehyde concentrations required to show statistically significant changes in the biomarkers of effect (1000µM) and the formation of exogenous adducts (1µM). The use of other cell lines with different metabolic capacities for the clearance of acetaldehyde may provide different dose-response relationships.
Assessing the dose-response relationship across a wide range of exposure conditions is critical to advancing the understanding of biomarkers of exposure and effects of genotoxic compounds to better discern the potential implications for human health. This study further advances such knowledge for the health effects of acetaldehyde by providing high-quality data for both endogenous and exogenous biomarkers of exposure, as well as biomarkers of effect across a wide range of concentrations. There are several critical points illustrated by the data described in this article. First, the use of [13C2]-acetaldehyde allowed for the discrimination between endogenous and exogenous DNA adducts across a wide dose range. The data clearly show that exogenous adducts can be extrapolated through zero. This confirms findings of other direct-acting genotoxic compounds (Pottenger et al., 2009). Second, at low exogenous exposure conditions, the amount of endogenous DNA adducts dominates over the exogenous adducts. This is evident by looking at the sum of endogenous and exogenous adducts, which shows a clear threshold (> 50µM) at which additional exogenous acetaldehyde causes increases in the sum of acetaldehyde-derived DNA adducts. Third, both biomarkers of effect showed clear thresholds (> 1000µM) at which statistically significant changes from the nearly constant background values occurred. Fourth, integrating the results from the biomarkers of exposure and effect clearly shows that there are differences between the concentrations at which these thresholds were observed. This is important in advancing the use of mode of action data for the assessment of risk of compounds such as acetaldehyde that have identical endogenous exposures. These data further question the use of default linear extrapolations and strongly support the use of such data for science-based regulation of genotoxic chemicals (Gollapudi et al., 2012; Pottenger et al., 2009; Pottenger and Gollapudi, 2009; Swenberg et al., 2008, 2011).
CONCLUSIONS
The dose-response relationships for biomarkers of exposure (N2-ethylidene-dG adducts) and effect (cell survival and MN formation) were determined across 4.5 orders of magnitude using [13C2]-acetaldehyde exposures in human lymphoblastoid TK6 cells for 12h. The results suggest that a threshold exists for both biomarkers of exposure and effect with a ~ 20-fold difference in the exposure concentrations required to achieve statistically significant changes over background levels between the measured biomarkers. Similar results are likely to be observed in animal models and human subjects following acetaldehyde exposures though additional studies would be needed to determine the possible human health relevance.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (P42 ES005948, P30 ES010126 to J.A.S., T32 ES007126 to B.C.M.); Vinyl Acetate Council (J.A.S. and L.R.). BCM was also supported by the Leon and Bertha Golberg Postdoctoral Fellowship.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge Patricia Upton for assistance in manuscript preparation. They would also like to acknowledge the support of Leonard Collins and Valeriy Alfonin for their assistance with LC-MS/MS instrumentation and the isolation of DNA from cells, respectively. The authors would like to acknowledge Dr Carol Swartz DVM, PhD, for the preparation of the formulation used for [13C2]-acetaldehyde exposures. The authors would like to acknowledge Dr Xu Tian for his assistance in determination of the ALDH2 genotype in TK6 cells.
REFERENCES
- Abraham J., Balbo S., Crabb D., Brooks P. J. (2011). Alcohol metabolism in human cells causes DNA damage and activates the Fanconi anemia-breast cancer susceptibility (FA-BRCA) DNA damage response network. Alcohol. Clin. Exp. Res. 35, 2113–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S., Hashibe M., Gundy S., Brennan P., Canova C., Simonato L., Merletti F., Richiardi L., Agudo A., Castellsagué X., et al. (2008). N2-ethyldeoxyguanosine as a potential biomarker for assessing effects of alcohol consumption on DNA. Cancer Epidemiol. Biomarkers Prev. 17, 3026–3032 [DOI] [PubMed] [Google Scholar]
- Balbo S., Meng L., Bliss R. L., Jensen J. A., Hatsukami D. K., Hecht S. S. (2012a). Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol. Biomarkers Prev. 21, 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S., Meng L., Bliss R. L., Jensen J. A., Hatsukami D. K., Hecht S. S. (2012b). Time course of DNA adduct formation in peripheral blood granulocytes and lymphocytes after drinking alcohol. Mutagenesis. 27, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird R. P., Draper H. H., Basrur P. K. (1982). Effect of malonaldehyde and acetaldehyde on cultured mammalian cells. Production of micronuclei and chromosomal aberrations. Mutat. Res. 101, 237–246 [DOI] [PubMed] [Google Scholar]
- Bogdanffy M. S., Valentine R. (2003). Differentiating between local cytotoxicity, mitogenesis, and genotoxicity in carcinogen risk assessments: The case of vinyl acetate. Toxicol. Lett. 140-141, 83–98 [DOI] [PubMed] [Google Scholar]
- Brooks P. J., Enoch M. A., Goldman D., Li T. K., Yokoyama A. (2009a). The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 6, e50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P. J., Goldman D., Li T. K. (2009b). Alleles of alcohol and acetaldehyde metabolism genes modulate susceptibility to oesophageal cancer from alcohol consumption. Hum. Genomics. 3, 103–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks P. J., Theruvathu J. A. (2005). DNA adducts from acetaldehyde: Implications for alcohol-related carcinogenesis. Alcohol. 35, 187–193 [DOI] [PubMed] [Google Scholar]
- Bryce S. M., Avlasevich S. L., Bemis J. C., Lukamowicz M., Elhajouji A., Van Goethem F., De Boeck M., Beerens D., Aerts H., Van Gompel J., et al. (2008). Interlaboratory evaluation of a flow cytometric, high content in vitro micronucleus assay. Mutat. Res. 650, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce S. M., Bemis J. C., Avlasevich S. L., Dertinger S. D. (2007). In vitro micronucleus assay scored by flow cytometry provides a comprehensive evaluation of cytogenetic damage and cytotoxicity. Mutat. Res. 630, 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang M., Villalta P. W., Luo X., Feuer R., Jensen J., Hatsukami D. K., Hecht S. S. (2007). Quantitation of an acetaldehyde adduct in human leukocyte DNA and the effect of smoking cessation. Chem. Res. Toxicol. 20, 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Guengerich F. P. (2006). Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase iota. J. Biol. Chem. 281, 12315–12324 [DOI] [PubMed] [Google Scholar]
- Fang J. L., Vaca C. E. (1995). Development of a 32P-postlabelling method for the analysis of adducts arising through the reaction of acetaldehyde with 2’-deoxyguanosine-3’-monophosphate and DNA. Carcinogenesis. 16, 2177–2185 [DOI] [PubMed] [Google Scholar]
- Fang J. L., Vaca C. E. (1997). Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 18, 627–632 [DOI] [PubMed] [Google Scholar]
- Garaycoechea J. I., Crossan G. P., Langevin F., Daly M., Arends M. J., Patel K. J. (2013). Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 489, 571–575 [DOI] [PubMed] [Google Scholar]
- Garcia C. C., Angeli J. P., Freitas F. P., Gomes O. F., de Oliveira T. F., Loureiro A. P., Di Mascio P., Medeiros M. H. (2011). [13C2]-Acetaldehyde promotes unequivocal formation of 1,N2-propano-2’-deoxyguanosine in human cells. J. Am. Chem. Soc. 133, 9140–9143 [DOI] [PubMed] [Google Scholar]
- Gollapudi B. B., Johnson G. E., Hernandez L. G., Pottenger L. H., Dearfield K. L., Jeffrey A. M., Julien E., Kim J. H., Lovell D. P., Macgregor J. T., et al. (2012). Quantitative approaches for assessing dose-response relationships in genetic toxicology studies. Environ. Mol. Mutagen. 54, 8–18 [DOI] [PubMed] [Google Scholar]
- Grafström R. C., Dypbukt J. M., Sundqvist K., Atzori L., Nielsen I., Curren R. D., Harris C. C. (1994). Pathobiological effects of acetaldehyde in cultured human epithelial cells and fibroblasts. Carcinogenesis. 15, 985–990 [DOI] [PubMed] [Google Scholar]
- Hazen S. L., Hsu F. F., d’Avignon A., Heinecke J. W. (1998). Human neutrophils employ myeloperoxidase to convert alpha-amino acids to a battery of reactive aldehydes: A pathway for aldehyde generation at sites of inflammation. Biochemistry. 37, 6864–6873 [DOI] [PubMed] [Google Scholar]
- He S. M., Lambert B. (1990). Acetaldehyde-induced mutation at the hprt locus in human lymphocytes in vitro. Environ. Mol. Mutagen. 16, 57–63 [DOI] [PubMed] [Google Scholar]
- Hecht S. S., McIntee E. J., Cheng G., Shi Y., Villalta P. W., Wang M. (2001a). New aspects of DNA adduct formation by the carcinogens crotonaldehyde and acetaldehyde. Adv. Exp. Med. Biol. 500, 63–71 [DOI] [PubMed] [Google Scholar]
- Hecht S. S., McIntee E. J., Wang M. (2001b). New DNA adducts of crotonaldehyde and acetaldehyde. Toxicology. 166, 31–36 [DOI] [PubMed] [Google Scholar]
- Hori K., Miyamoto S., Yukawa Y., Muto M., Chiba T., Matsuda T. (2012). Stability of acetaldehyde-derived DNA adduct in vitro. Biochem. Biophys. Res. Commun. 423, 642–646 [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. Monographs on the Evaluation of Carcinogenic Risks to Humans. Re-evaluation of some organic chemicals, hydrazine and hydrogen peroxide. 71, 319–335 1999. [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. Monographs on the Evaluation of Carcinogenic Risks to Humans. Alcohol consumption and ethyl carbamate. 96 2010. [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Zhang L., Guo W., McHale C. M., Smith M. T. (2009). The benzene metabolite, hydroquinone and etoposide both induce endoreduplication in human lymphoblastoid TK6 cells. Mutagenesis. 24, 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R. A., Hanaoka T., Sugimura H., Toda H., Matsui S., Matsuda T. (2006). Development of the adductome approach to detect DNA damage in humans. Antioxid. Redox Signal. 8, 993–1001 [DOI] [PubMed] [Google Scholar]
- Kuykendall J. R., Bogdanffy M. S. (1992a). Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 283, 131–136 [DOI] [PubMed] [Google Scholar]
- Kuykendall J. R., Bogdanffy M. S. (1992b). Reaction kinetics of DNA-histone crosslinking by vinyl acetate and acetaldehyde. Carcinogenesis. 13, 2095–2100 [DOI] [PubMed] [Google Scholar]
- Kuykendall J. R., Bogdanffy M. S. (1994). Formation and stability of acetaldehyde-induced crosslinks between poly-lysine and poly-deoxyguanosine. Mutat. Res. 311, 49–56 [DOI] [PubMed] [Google Scholar]
- Langevin F., Crossan G. P., Rosado I. V., Arends M. J., Patel K. J. (2011). Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 475, 53–58 [DOI] [PubMed] [Google Scholar]
- Mabuchi R., Kurita A., Miyoshi N., Yokoyama A., Furuta T., Goda T., Suwa Y., Kan T., Amagai T., Ohshima H. (2012). Analysis of N(ε) -ethyllysine in human plasma proteins by gas chromatography-negative ion chemical ionization/mass spectrometry as a biomarker for exposure to acetaldehyde and alcohol. Alcohol. Clin. Exp. Res. 36, 1013–1020 [DOI] [PubMed] [Google Scholar]
- Mangerich A., Knutson C. G., Parry N. M., Muthupalani S., Ye W., Prestwich E., Cui L., McFaline J. L., Mobley M., Ge Z., et al. (2012). Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U.S.A. 109, E1820–E1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Matsumoto A., Uchida M., Kanaly R. A., Misaki K., Shibutani S., Kawamoto T., Kitagawa K., Nakayama K. I., Tomokuni K., et al. (2007). Increased formation of hepatic N2-ethylidene-2’-deoxyguanosine DNA adducts in aldehyde dehydrogenase 2-knockout mice treated with ethanol. Carcinogenesis. 28, 2363–2366 [DOI] [PubMed] [Google Scholar]
- Migliore L., Cocchi L., Scarpato R. (1996). Detection of the centromere in micronuclei by fluorescence in situ hybridization: Its application to the human lymphocyte micronucleus assay after treatment with four suspected aneugens. Mutagenesis. 11, 285–290 [DOI] [PubMed] [Google Scholar]
- Millonig G., Wang Y., Homann N., Bernhardt F., Qin H., Mueller S., Bartsch H., Seitz H. K. (2011). Ethanol-mediated carcinogenesis in the human esophagus implicates CYP2E1 induction and the generation of carcinogenic DNA-lesions. Int. J. Cancer. 128, 533–540 [DOI] [PubMed] [Google Scholar]
- Nagayoshi H., Matsumoto A., Nishi R., Kawamoto T., Ichiba M., Matsuda T. (2009). Increased formation of gastric N(2)-ethylidene-2’-deoxyguanosine DNA adducts in aldehyde dehydrogenase-2 knockout mice treated with ethanol. Mutat. Res. 673, 74–77 [DOI] [PubMed] [Google Scholar]
- National Toxicology Program Report on Carcinogens. 2011. [Google Scholar]
- O’Brien P. J., Siraki A. G., Shangari N. (2005). Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit. Rev. Toxicol. 35, 609–662 [DOI] [PubMed] [Google Scholar]
- OECD (2010). Test No. 487: In Vitro Mammalian Cell Micronucleus Test. 7-23-2010. OECD Publishing; Paris. [Google Scholar]
- Oyama T., Nagayoshi H., Matsuda T., Oka M., Isse T., Yu H. S., Pham T. T., Tanaka M., Kagawa N., Kaneko K., et al. (2010). Effects of acetaldehyde inhalation in mitochondrial aldehyde dehydrogenase deficient mice (Aldh2-/-). Front. Biosci. (Elite Ed). 2, 1344–1354 [DOI] [PubMed] [Google Scholar]
- Pottenger L. H., Gollapudi B. B. (2009). A case for a new paradigm in genetic toxicology testing. Mutat. Res. 678, 148–151 [DOI] [PubMed] [Google Scholar]
- Pottenger L. H., Schisler M. R., Zhang F., Bartels M. J., Fontaine D. D., McFadden L. G., Gollapudi B. B. (2009). Dose-response and operational thresholds/NOAELs for in vitro mutagenic effects from DNA-reactive mutagens, MMS and MNU. Mutat. Res. 678, 138–147 [DOI] [PubMed] [Google Scholar]
- Ridpath J. R., Nakamura A., Tano K., Luke A. M., Sonoda E., Arakawa H., Buerstedde J. M., Gillespie D. A., Sale J. E., Yamazoe M., et al. (2007). Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 67, 11117–11122 [DOI] [PubMed] [Google Scholar]
- Sako M., Kawada H., Hirota K. (1999). A convenient method for the preparation of N(2)-ethylguanine nucleosides and nucleotides. J. Org. Chem. 64, 5719–5721 [DOI] [PubMed] [Google Scholar]
- Setshedi M., Wands J. R., Monte S. M. (2010). Acetaldehyde adducts in alcoholic liver disease. Oxid. Med. Cell. Longev. 3, 178–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. W., Umber B. J., Meinardi S., Leu S. Y., Zaldivar F., Blake D. R., Cooper D. M. (2009). Acetaldehyde and hexanaldehyde from cultured white cells. J. Transl. Med. 7, 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Gromadzinska J., Mistry Y., Cordell R., Juren T., Segerbäck D., Farmer P. B. (2012). Detection of acetaldehyde derived N2-ethyl-2’-deoxyguanosine in human leukocyte DNA following alcohol consumption. Mutat. Res. 737, 8–11 [DOI] [PubMed] [Google Scholar]
- Singh R., Sandhu J., Kaur B., Juren T., Steward W. P., Segerbäck D., Farmer P. B. (2009). Evaluation of the DNA damaging potential of cannabis cigarette smoke by the determination of acetaldehyde derived N2-ethyl-2’-deoxyguanosine adducts. Chem. Res. Toxicol. 22, 1181–1188 [DOI] [PubMed] [Google Scholar]
- Speit G., Fröhler-Keller M., Schütz P., Neuss S. (2008). Low sensitivity of the comet assay to detect acetaldehyde-induced genotoxicity. Mutat. Res. 657, 93–97 [DOI] [PubMed] [Google Scholar]
- Stein S., Lao Y., Yang I. Y., Hecht S. S., Moriya M. (2006). Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat. Res. 608, 1–7 [DOI] [PubMed] [Google Scholar]
- Swenberg J. A., Fryar-Tita E., Jeong Y. C., Boysen G., Starr T., Walker V. E., Albertini R. J. (2008). Biomarkers in toxicology and risk assessment: Informing critical dose-response relationships. Chem. Res. Toxicol. 21, 253–265 [DOI] [PubMed] [Google Scholar]
- Swenberg J. A., Lu K., Moeller B. C., Gao L., Upton P. B., Nakamura J., Starr T. B. (2011). Endogenous versus exogenous DNA adducts: Their role in carcinogenesis, epidemiology, and risk assessment. Toxicol. Sci. 120(Suppl. 1)S130–S145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T., Kawai T., Morimoto K. (1997). Elevated levels of hemoglobin-associated acetaldehyde related to alcohol drinking in the atypical genotype of low Km aldehyde dehydrogenase. Cancer Res. 57, 1241–1243 [PubMed] [Google Scholar]
- Tanaka F., Shiratori Y., Yokosuka O., Imazeki F., Tsukada Y., Omata M. (1996). High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology. 23, 234–239 [DOI] [PubMed] [Google Scholar]
- Terashima I., Matsuda T., Fang T. W., Suzuki N., Kobayashi J., Kohda K., Shibutani S. (2001). Miscoding potential of the N2-ethyl-2’-deoxyguanosine DNA adduct by the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I. Biochemistry. 40, 4106–4114 [DOI] [PubMed] [Google Scholar]
- Upton D. C., Wang X., Blans P., Perrino F. W., Fishbein J. C., Akman S. A. (2006a). Replication of N2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem. Res. Toxicol. 19, 960–967 [DOI] [PubMed] [Google Scholar]
- Upton D. C., Wang X., Blans P., Perrino F. W., Fishbein J. C., Akman S. A. (2006b). Mutagenesis by exocyclic alkylamino purine adducts in Escherichia coli. Mutat. Res. 599, 1–10 [DOI] [PubMed] [Google Scholar]
- Vaca C. E., Nilsson J. A., Fang J. L., Grafström R. C. (1998). Formation of DNA adducts in human buccal epithelial cells exposed to acetaldehyde and methylglyoxal in vitro. Chem. Biol. Interact. 108, 197–208 [DOI] [PubMed] [Google Scholar]
- Wang M., McIntee E. J., Cheng G., Shi Y., Villalta P. W., Hecht S. S. (2000). Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 13, 1149–1157 [DOI] [PubMed] [Google Scholar]
- Wang M., Yu N., Chen L., Villalta P. W., Hochalter J. B., Hecht S. S. (2006). Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem. Res. Toxicol. 19, 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woutersen R. A., Appelman L. M., Feron V. J., Van der Heijden C. A. (1984). Inhalation toxicity of acetaldehyde in rats. II. Carcinogenicity study: Interim results after 15 months. Toxicology. 31, 123–133 [DOI] [PubMed] [Google Scholar]
- Woutersen R. A., Appelman L. M., Van Garderen-Hoetmer A., Feron V. J. (1986). Inhalation toxicity of acetaldehyde in rats. III. Carcinogenicity study. Toxicology. 41, 213–231 [DOI] [PubMed] [Google Scholar]
- Woutersen R. A., Feron V. J. (1987). Inhalation toxicity of acetaldehyde in rats. IV. Progression and regression of nasal lesions after discontinuation of exposure. Toxicology. 47, 295–305 [DOI] [PubMed] [Google Scholar]
- Yang I. Y., Chan G., Miller H., Huang Y., Torres M. C., Johnson F., Moriya M. (2002). Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 41, 13826–13832 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Huang I. Y., Ikawa M. (1984). Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc. Natl. Acad. Sci. U.S.A. 81, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa Y., Muto M., Hori K., Nagayoshi H., Yokoyama A., Chiba T., Matsuda T. (2012). Combination of ADH1B*2/ALDH2*2 polymorphisms alters acetaldehyde-derived DNA damage in the blood of Japanese alcoholics. Cancer Sci. 103, 1651–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.