Abstract
DNA methylation inhibitors such as 5-aza-2′-deoxycytidine (5-Aza-CdR) are currently used for the treatment of myelodysplastic syndrome. Although global DNA demethylation has been observed after treatment, it is unclear to what extent demethylation induces changes in nucleosome occupancy, a key determinant of gene expression. We use the colorectal cancer cell line HCT116 as a model to address this question and determine that <2% of regions demethylated by 5-Aza-CdR treatment assume an open configuration. Consolidating our findings, we detect nucleosome retention at sites of global DNA methylation loss in DKO1, an HCT116-derived non-tumorigenic cell-line engineered for DNA methyltransferase disruption. Notably, regions that are open in both HCT116 cells after treatment and in DKO1 cells include promoters belonging to tumor suppressors and genes under-expressed in colorectal cancers. Our results indicate that only a minority of demethylated promoters are associated with nucleosome remodeling, and these could potentially be the epigenetic drivers causing the loss of tumorigenicity. Furthermore, we show that the chromatin opening induced by the histone deacetylase inhibitor suberoylanilide hydroxamic acid has strikingly distinct targets compared with those of 5-Aza-CdR, providing a mechanistic explanation for the importance of combinatorial therapy in eliciting maximal de-repression of the cancer epigenome.
INTRODUCTION
There has been increasing support for the role of epigenetic aberrations in contributing to tumorigenesis over the past few decades (1,2). It is now clear that promoter CpG island-specific hypermethylation as well as global hypomethylation are widespread defects in tumors (3). Aside from aberrations in the DNA methylation marks, abnormal histone modifications, such as increased repression from the polycomb H3K27me3 mark, have been observed in tumors and confirmed to potentiate tumorigenesis independently of genetic alterations (1,3,4).
More recently, aberrations in nucleosome remodeling have also been noted in cancers (5). Nucleosome positioning regulates gene expression by modulating the accessibility of DNA to the transcriptional machinery (6,7). Nucleosome positioning is tightly controlled and maintained in cells by several factors, including DNA sequence, ATP-dependent chromatin remodelers and nucleosome modifications (8,9). In cancer, significant changes in nucleosome positioning result in aberrant compaction of chromatin structure and, hence, altered gene expression signatures (10,11). This can often be explained by malfunctioning of chromatin remodeling complexes, such as the SWI/SNF (SWItch/Sucrose NonFermentable) complex, which are frequently found to be mutated in cancer (4,5,8,12).
Although aberrant epigenetic patterns are heritable, their dynamic nature and potential reversibility through pharmacological interventions make them attractive therapeutic targets (13,14). The Federal Drug Administration has approved DNA methyltransferase inhibitors for the treatment of myelodysplastic syndrome (15). Drugs of this class have also been used successfully in other hematological malignancies (16). More recently, pre-clinical and clinical data have demonstrated that these inhibitors can be used to reduce the malignant potential of solid tumors as well (17). Even difficult-to-treat lung cancers have shown response to epigenetic modulators, improving patient outcomes (18). Histone deacetylase (HDAC) inhibitors are another class of drugs that allow for increased acetylation of histones, thereby permitting an open chromatin state. Suberoylanilide hydroxamic acid (SAHA) is a potent HDAC inhibitor that has been approved for the treatment of cutaneous T-cell lymphoma (13) and has shown response in acute myeloid leukemia patients (19). In a majority of cases, SAHA’s effectiveness as a mono-therapeutic agent has been found to be limited. Hence, it is often used in combination with DNA methyltransferase inhibitors (20,21). Although the need for combination epigenetic therapy has been widely recognized, much remains to be determined regarding the ideal combinations.
The study of the mechanism of these pharmacological agents in reversing epigenomic aberrations is still in its infancy. Although global DNA demethylation has been noted by treating cell lines, mouse models and patient samples with DNA methyltransferase inhibitors (17), it is not clear how this translates into phenotypic effects. It is crucial to understand how these inhibitors affect nucleosome positioning, given that it is a key determinant of gene expression. This understanding will allow for the discernment of functional DNA demethylation (22), those events that are accompanied by chromatin opening, from non-functional demethylation, following which the chromatin continues to remain in a repressed and an inaccessible state. By asking this question, one can define regions that are likely to open on demethylation. This will also allow for innovation to identify alternative methods to affect nucleosome repositioning of regions that are indifferent to DNA demethylation and to assess whether the usage of HDAC inhibitors will further aid in this process.
To address this important issue, we ask to what extent DNA demethylation induced by an inhibitor, 5-aza-2′-deoxycytidine (5-Aza-CdR), results in chromatin opening of a colorectal cancer cell line. We study this phenomenon globally using a novel assay that we have devised, using the CpG methyltransferase M.SssI (23) to infer chromatin accessibility using an Illumina methylation array. We probe this question further using a genetic model engineered for the disruption of DNA methyltransferases, DKO1 cells. Finally, we test the potential of the HDAC inhibitor SAHA to open other regions of chromatin.
MATERIALS AND METHODS
Cell culture
Culture of colorectal cancer cell lines HCT116 and DKO1 (a hypomethylated derivative of HCT116, DNMT1ΔE2–5/DNMT3B−/− double knockout) was carried using McCoy’s 5 A media with 10% fetal bovine serum (FBS). Human embryonic stem cell line H1 was carried under recommended conditions. Renal cancer cell line 786-O was cultured using Roswell Park Memorial Institute (RPM1)-1640 media + 10% FBS, and breast cancer cell line MCF7 (kindly provided by Cindy Zahnow and Huili Li from the Johns Hopkins School of Medicine) was carried in Minimum Essential Media (MEM) + 10% FBS + 0.01 mg/ml bovine insulin.
Drug treatments
HCT116, DKO1, 786-O and MCF7 cells were treated with either 0.3 µM 5-Aza-CdR for 24 h or 1 µM of SAHA for 24 h. Cells were harvested for study of chromatin accessibility 72 h post-treatment for 5-Aza-CdR and 24 h post-treatment for SAHA.
M.SssI treatment and DNA methylation array
For nuclei extraction, cells were trypsinized and centrifuged for 5 min at 200g, then washed with ice-cold PBS and resuspended in 1 ml ice-cold nuclei buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA and 0.5% NP-40, plus protease inhibitors) per 5 × 106 cells and incubated on ice for 5 min. Nuclei were recovered by centrifugation at 900g for 3 min and washed in nuclei wash buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1 mM EDTA containing protease inhibitors). Freshly prepared nuclei (2 × 105 cells) were resuspended in 1× M.SssI reaction buffer (NEB), and then treated with 50 U of M.SssI (NEB) in 15 µl 10× reaction buffer, 45 µl 1 M sucrose and 0.75 µl S-Adenosyl methionine (SAM) in a volume of 150 µl. Reactions were quenched by the addition of an equal volume of stop solution (20 nM Tris-HCl [pH 7.9], 600 mM NaCl, 1% SDS, 10 mM EDTA and 400 µg/ml proteinase K) and incubated at 55°C overnight (23). DNA was purified by phenol/chloroform extraction and ethanol precipitation. Bisulfite conversion was performed using the EZ DNA methylation kit (Zymo Research). All samples passed bisulfite conversion quality control and were subsequently processed for the Illumina Infinium DNA methylation platform (Human Methylation 450 Bead Chip). For validations of enzyme treatment and accessibility locally, molecules were cloned using the Topo TA Kit (Invitrogen) and sequenced.
Global DNA methylation and chromatin accessibility analysis
The Infinium HumanMethylation450 array (Illumina, Inc.) was used to analyze bisulfite-treated DNA. Briefly, the signal of methylation-specific probe over the sum of the signals of the methylation- and unmethylated-specific probes is calculated as the beta value, indicating full methylation of a specific CpG site (beta value = 1), absence of methylation (beta value = 0) and ranges in between (0 ≤ beta value ≤ 1). Probes with poor overall signals (P-value > 0.05) were removed from analysis (24). Gene promoters with probes that have beta value >0.7 were called methylated and <0.3 were called unmethylated, with everything in between being partially methylated. The accessibility scale was defined as the beta value of M.SssI-treated cells minus the beta value of the no-enzyme control (delta-beta), defined on a 0–1 scale, after removing the probes with delta-beta <0. Accessible probes were defined as delta-beta >0.3 using standard receiver-operating characteristic curve analysis. Heat maps were generated using R package gplots and the density plots using R package ggplot2.
Overlaps of DNAse, Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) and H3K4Me1 data for H1 and HCT116 were performed using the GRanges package.
Plots of Illumina probes and data on a genomic scale were produced using IlluminaHumanMethylation450kprobe and GViz packages through Bioconductor. Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis and visualization of gene expression. All statistical calculations were performed on R (Version 2.15.0).
Motif enrichment analysis was done de novo using the program Homer (25).
Network and ontology analysis was performed using MetaCore from GeneGo Inc.
Reagents and antibodies
Commercial primary antibodies used were anti-H3 (Abcam) and anti-AcH3 (Millipore Laboratories).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed according to the Upstate Biotechnology instructions. For each ChIP assay, 100 μg of DNA sheared by a sonicator was pre-cleared with salmon-sperm DNA-saturated protein A Sepharose and then precipitated by H3 and acetlayed-H3 antibodies. After immunoprecipitation, recovered chromatin fragments were subjected to real-time polymerase chain reaction. IgG control experiments were performed for all ChIPs and were accounted for in the IP/Input by presenting the results as (IP − IgG)/(Input − IgG). Primer sequences: TXNIP promoter: 5′-CAGCGATCTCACTGATTGGT, 3′-CCCAATTGCTGGAGAAAAGA; NEUROD2 promoter: 5′-CAGACGCATGCCAATCAC, 3′-CATTGTTCCCCCATCTTCAG; GAPDH promoter: 5′-CGGCTACTAGCGGTTTTACG, 3′-AAGAAGATGCGGCTGACTGT; STMN1 promoter: 5′-GTTGGAATGGGGAAGAAGGT, 3′-GGAACA AGGGCATCACTGAC.
Data access
ENCODE H1 DNAse-seq data set—
ENCODE H1 FAIRE-seq data set—
wgEncodeOpenChromFaireH1hescPk.narrowPeak.gz
ENCODE HCT116 DNAse-seq data set—
ENCODE HCT116 H3K4Me1 data set—
wgEncodeEH002874
Oncomine data sets—
TCGA colon adenocarcinoma versus normal
GEO AcceSssIble data set—
RESULTS
A majority of demethylated probes retain chromatin inaccessibility after 5-Aza-CdR treatment
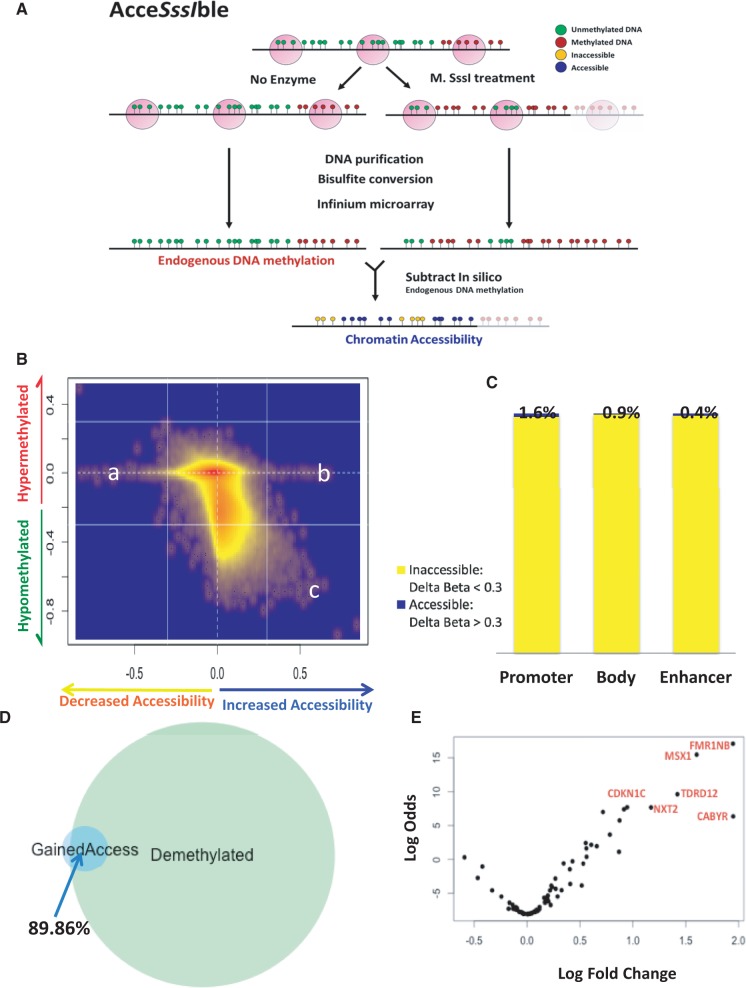
To understand, globally, whether demethylation after 5-Aza-CdR treatment results in increased accessibility of chromatin, we designed a novel method termed AcceSssIble (Figure 1A). This method involves comparing methylation of nuclei treated with a CpG methyltransferase enzyme, M.SssI (23), which specifically methylates unmethylated CpG sites on nucleosome-depleted unbound DNA, with a no-enzyme treated control using an Illumina HumanMethylation450 BeadChip. This platform queries >485 000 CpG sites across the genome, covering 99% of RefSeq genes, an average of 17 CpG sites per gene, with probes across promoters, gene bodies and enhancers (26). The accessibility or the lack thereof to M.SssI, determined by acquisition of methylation on enzyme treatment compared with the control, can be used to infer the positioning of nucleosomes or factors bound tightly to chromatin, at endogenously unmethylated loci (Figure 1A).
Figure 1.
Majority of probes demethylated by 5-Aza-CdR are inaccessible to CpG methyltransferase M.SssI after 5-Aza-CdR treatment. (A) Schematic of the AcceSssIble assay: after nuclei are treated with M.Sss1 (or a no-enzyme treatment control), DNA is extracted, bisulfite converted and assayed on the Illumina methylation 450 k beadchip. Endogenous DNA methylation status is obtained from the no-enzyme control (green indicating unmethylated and red methylated), and information on chromatin state for unmethylated probes is attained from the accessibility of the M.SssI methyltransferase to CpG dinucleotides (scaled from yellow to blue, with yellow as inaccessible and blue as highly accessible). (B) Kernel density scatter plot of delta-methylation (methylation in treated minus methylation in control) versus delta-accessibility (accessibility in treated minus accessibility in control) shows that majority of demethylated loci do not gain accessibility. Groups a and b are events that depict changes in chromatin accessibility independent of DNA methylation, whereas group c depicts regions that gain accessibility on DNA demethylation. (C) Bar graph depicting percentage of demethylated promoter- (9719), body- (1205) and enhancer-associated (28 348) probes that gain accessibility or remain inaccessible on demethylation. (D) Venn diagram of all promoter regions that are demethylated post-5-Aza-CdR treatment in HCT116 as well with those that gain accessibility. Results show that ∼90% of regions require demethylation to gain accessibility. (E) Volcano plot of genes whose promoters gain accessibility after treatment shows that most of these regions gain expression.
To compare our assay with results produced by other methods, we performed AcceSssIble on H1 human embryonic cells, for which there is publicly available genome-wide data on DNAse hypersensitivity and FAIRE sequencing (Supplementary Figure S1A). Methylation is quantified using beta values (27), and chromatin accessibility is quantified as the acquisition of methylation due to enzyme treatment using delta-beta values. To assess the performance of the AcceSssIble assay, we used a receiver-operating characteristic curve analysis of our data using DNAse-seq, FAIRE-seq as gold standards and empirically chose a delta-beta of 0.3 as the cutoff to define open/accessible regions (Supplementary Figure S1B). We also performed a comparative analysis of open transcription start sites (TSSs) specifically with DNAse-seq and/or FAIRE-seq data for H1 and colorectal cancer cell line, HCT116, and found a high degree of overlap (Supplementary Figure S1C and 1D). Together, these results confirm that our AcceSssIble assay can recapitulate the findings of comprehensive methods such as DNAse-seq and FAIRE-seq and identify regions of open chromatin in a rapid and cost-effective manner.
Having established the assay, we treated HCT116 cells with 0.3 µM of 5-Aza-CdR for 24 h and performed AcceSssIble 72 h post-treatment. As expected, endogenous DNA methylation decreased genome-wide with 5-Aza-CdR treatment (Supplementary Figure S2A). To characterize the spectrum of epigenetic changes using metrics of both DNA methylation and accessibility changes, we plotted delta-methylation (methylation in treated minus methylation in control) against delta-accessibility (accessibility in treated minus accessibility in control) as a kernel density scatter plot (Figure 1B). Interestingly, we observed that a majority of demethylation events were not associated with a corresponding gain of chromatin accessibility; only a small number of probes that lost methylation displayed accessibility changes indicative of chromatin opening (region c in Figure 1B). Further, only a small number of accessibility changes occurred independently of changes in DNA methylation (region a, b in Figure 1B). To validate our result in independent cell lines, we replicated the experiment in breast cancer cell line MCF7 and renal cancer cell line 786-O and found a similar pattern: majority of demethylation induced by 5-Aza-CdR is not accompanied by chromatin accessibility (Supplementary Figure S2B and C).
We then asked whether different genomic regions, the gene promoter, gene body and enhancer (defined by probes that lie within H3K4Me1 peaks, obtained from an ENCODE data set for HCT116) displayed similar patterns of changes in chromatin accessibility associated with demethylation (Figure 1C). Strikingly, only 1.6% of promoter-associated probes that became demethylated gained accessibility. Similarly, 0.9% of gene body-associated probes and 0.4% of enhancer-associated probes gained chromatin accessibility, indicative of the presence of a nucleosome-depleted region on demethylation. Of note, we observed a statistically significant enrichment of polycomb repressive complex 2 targets (28) in the group of genes that fail to gain accessibility, despite promoter demethylation, indicating that the polycomb machinery could be involved in maintaining the closed configuration by means of an epigenetic switch to the H3K27me3 mark (29,30). Overall, irrespective of gene region, a majority of probes that became demethylated did not assume an open chromatin configuration.
Next, we determined the converse: the number of promoter regions that gain accessibility after 5-Aza-CdR treatment that were dependent on DNA demethylation, and found that nearly 90% of probes did in fact require demethylation to open (Figure 1D). A minority of probes (∼10%) were unmethylated to begin with and switched from a repressed to an open state after treatment, potentially due to downstream indirect events. To determine the genes that gained expression on demethylation and chromatin opening at promoter regions after 5-Aza-CdR treatment, we performed a gene expression analysis for this group of genes (Figure 1E). We found that genes implicated in having anti-tumor effects, such as CDKN1C (31) and MSX1 (32), as well as testis antigen genes that could make the tumor more visible to the immune system (33), such as CABYR, FMR1NB, were up-regulated. Several other genes that had gained chromatin accessibility did not attain gene expression, indicating that although these genes are poised for expression with open chromatin, they may not have the necessary transcriptional machinery to drive transcription. Together, our results show that only a minority of demethylation events result in functional changes, wherein a gain of chromatin accessibility and gene expression accompanies the loss of methylation.
DKO1 promoters have retained inaccessible chromatin despite the loss of methylation
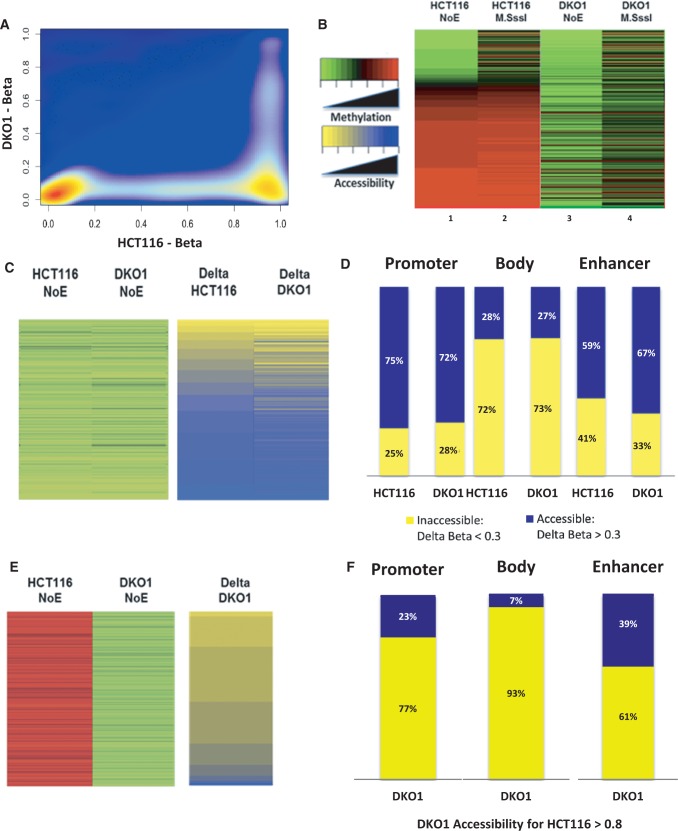
As we identified that demethylation does not induce widespread opening of chromatin after 5-Aza-CdR treatment, we asked whether this finding also applied to a cleaner model of DNA methylation loss. To address this and to deepen our understanding of nucleosome repositioning in response to demethylation, we used the HCT116-derived DKO1, which is a genetic model of permanent demethylation. DKO1 has been engineered for the genetic disruption of the DNA methyltransferases DNMT1ΔE2−5/DNMT3B−/− (34,35), resulting in the loss of ∼95% of the methylation seen in parental HCT116 cells (Figure 2A). Interestingly, the lack of functional methyltransferases resulted in the conversion of the tumorigenic HCT116 into the non-tumorigenic state (36).
Figure 2.
Unmethylated promoter regions shared by HCT116 and DKO1 are largely open; demethylated regions in DKO1 retain chromatin inaccessibility. (A) Density scatter plot between DKO1 and HCT116 shows that DKO1 is highly demethylated compared with HCT116. (B) A heat map of all probes on the Illumina platform depicting beta values from 0 to 1 (green to red, unmethylated to methylated) for HCT116 and DKO1 with and without M.SssI treatment. DKO1 has reduced endogenous methylation compared with HCT116, and they both show an increase in methylation on M.SssI treatment. (C) Heat maps depicting endogenous DNA methylation (left) and M.SssI accessibility (on a delta-beta scale from 0 to 1, with delta-beta = M.SssI beta value minus NoE beta value) for all TSS probes that are unmethylated in HCT116. DKO1 and the parental HCT116 have similar patterns of accessibilities to these probes. (D) Bar graphs of accessibilities calculated for all unmethylated probes in HCT116 and in DKO1 and analyzed across gene promoter regions (36 447 probes), bodies (11 695 probes) and enhancers (3696 probes). A majority of the unmethylated probes are highly accessible at the promoters and enhancers and primarily inaccessible at gene bodies. (E) Heat maps showing endogenous DNA methylation and M.SssI accessibility for promoter regions methylated in HCT116 but unmethylated in DKO1 indicate that DKO1 has maintained many of its promoters in an inaccessible state where it has lost DNA methylation. A small number of probes (4.7%) are highly accessible to M.SssI, despite being methylated in HCT116. (F) DKO1 probes that have lost methylation have decreased accessibility at promoters (9915 probes), bodies (35 786 probes) and enhancers (8966 probes), possibly due to the retention of nucleosomal occupancy.
First, we confirmed the methylation patterns of HCT116 and DKO1 obtained from the Illumina methylation array. Although HCT116 showed a high degree of methylation for >60% of probes queried by the platform, DKO1 displayed an expected genome-wide loss of methylation (Figure 2B, columns 1 and 3, NoE – no-enzyme control). Chromatin accessibility is visualized by the increase in methylation of unmethylated sites on enzyme treatment (Figure 2B, columns 2 and 4).
To establish the baseline patterns of accessibilities in the cell lines, we queried the chromatin states at promoter probes that are unmethylated in common between HCT116 and in DKO1. Indeed, a majority of these probes appeared to be highly accessible in both cell lines (Figure 2C), with similar patterns observable for the parental and the derivative cell lines. The inaccessible probes could be located in regions blocked by nucleosomes or transcription factors.
We selected two genes that displayed chromatin accessibility in both cell lines, GAPDH and GRP78, as well as two genes, FOS and STMN1, determined to be unmethylated and potentially nucleosome occupied. We confirmed these results using ChIP for histone H3, which can be used to infer the presence of nucleosomes, and validated that the inaccessible gene had higher presence of H3 than the accessible gene (Supplementary Figure S3A), and further confirmed the result using sequencing (Supplementary Figure S3B).
To determine whether unmethylated regions were similarly open at other genomic regions, such as the gene body or enhancer regions, we compared accessibilities in DKO1 and HCT116 at these regions. Although 75% of HCT116 unmethylated promoter probes and 59% of unmethylated enhancer probes were accessible (Figure 2D), only 28% of unmethylated gene body probes were accessible. A similar pattern of accessibility was seen for the identical probes in DKO1—72% of promoter probes and 67% of enhancer probes were accessible but only 27% of body sites were accessible. The data from both cell lines suggest that in an unmethylated state, promoter and enhancer regions have a higher percentage of accessible chromatin compared with gene body regions (Figure 2D). We also studied the CpG island-related probes and probes in promoters without islands and found that 1.7% of demethylated CpG island probes gained accessibility, whereas 1.4% of non-CpG island probes gained accessibility (Supplementary Figure S3C). On performance of a chi-squared test, we found that there is no statistically significant difference in the attainment of an open chromatin configuration due to the island status. We further looked at island-associated probes (shore, shelf) and found that 1.6% of probes gained accessibility on demethylation, not statistically different from the other categories (Supplementary Figure S3C).
Having established the accessibility patterns in regions unmethylated in both cell lines, we next examined probes that were methylated, initially, in the parental HCT116 cells but had lost methylation in the derivative DKO1 cells (Figure 2E). Interestingly, despite losing methylation, DKO1 cells showed patterns of inaccessibility across all studied regions, with a total accessibility of only 23% at the promoters, 7% at the body sites and 39% at the enhancers (Figure 2F), significantly lower levels of accessibility compared with unmethylated region in both cell lines (Figure 2D). This is similar to the results we observed in HCT116 post-5-Aza-CdR treatment, wherein demethylation seldom correlates with an increase in accessibility.
We took a closer look at the promoter of NEUROD2 using the ChIP assay and determined it to be closed, despite a loss of methylation in DKO1, with high levels of histone H3 and low ratio of acetylated-H3/H3 at the NEUROD2 promoters in both DKO1 and HCT116 cell lines. We further confirmed these results by sequencing TSSs of both genes with and without enzyme treatment and by visualizing all probes associated with the gene (Supplementary Figures S3D and E).
Our data suggest that, largely, DKO1 has retained nucleosomes across the different genomic loci to maintain the repressed state found in the parental HCT116. Overall, patterns of open or closed chromatin as seen in the parental cells have been maintained in DKO1, despite the loss of DNA methylation.
A majority of regions that gain chromatin accessibility on 5-Aza-CdR treatment overlap with de novo open regions in DKO1 cells and are associated with putative tumor suppressors
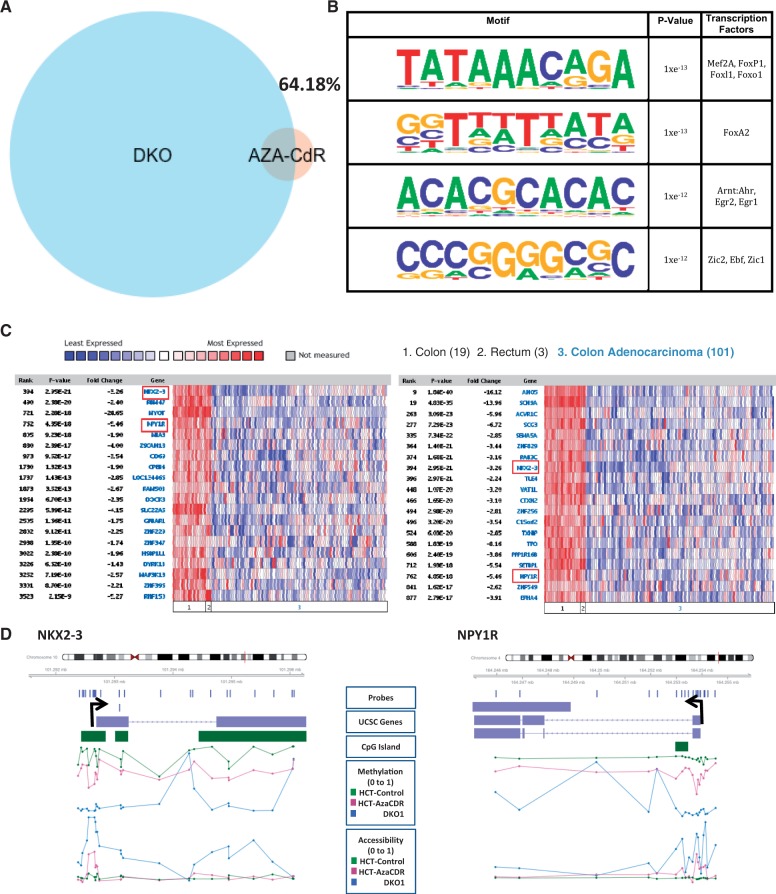
Aside from the group of genes with nucleosome retention, we found that a small portion of genes (4.7%) that showed endogenous hypermethylation in the parental HCT116 cells have gained accessibility by genetic disruption of DNA methyltransferases in DKO1 (Figure 2E). Interestingly, we noticed that this group contained several known tumor suppressor genes, such as GATA-4 (37), TXNIP (38) and NKX2-3 (39). A gene set enrichment analysis (40) of the genes corresponded significantly with genes methylated de novo in cancer (P-value = 1.3 e−6) (41). We also examined the accessibilities of genes identified as tumor suppressors and previously shown to be DNA-methylated and to lack gene expression in HCT116 but not in DKO1 (putative tumor suppressors) (42). We found that these genes have a significant increase in accessibility (P-value < 2.2 e−16) compared with the accessibility of all genes that have lost DNA methylation (Supplementary Figure S4A). Hence, these genes could potentially have tumor suppressive function in colorectal cancers and, therefore, regaining accessibility at these loci could contribute to the loss of tumorigenicity in DKO1 cells.
Given that only a minority of genes gain accessibility on 5-Aza-CdR treatment as well as in DKO1, despite a loss of methylation, we asked how these groups of genes compare. For this, we considered chromatin accessibility changes at the promoter regions and found that >64% of genes that open on 5-Aza-CdR treatment are also open in DKO1, despite the parental HCT116 being methylated (Figure 3A). We next asked if we could explain why the promoters of these groups of genes gained accessibility, despite this being the exception to the otherwise observed lack of chromatin accessibility after demethylation. To this end, we performed a motif enrichment analysis and found that the four motifs that were significantly enriched corresponded to the recognition sequence of several transcription factors that have been known to associate with chromatin remodelers (43–46) (Figure 3B). As we observed that five of the transcription factors were expressed in HCT116 and DKO1 cells to levels higher than the known negative control, PRND (Supplementary Figure S4B), it is plausible that the presence of these transcription factors and their associated chromatin remodelers caused the opening of chromatin at the observed promoters.
Figure 3.
Regions that gain accessibility in HCT116 on 5-Aza-CdR treatment and in DKO1 are associated with tumor suppressors. (A) Venn diagram of the overlap between DKO1 regions that are accessible despite methylation in HCT116 overlapped with regions that gain accessibility on HCT116 treatment with 5-Aza-CdR shows that ∼64% of regions that open on drug treatment are open in DKO1. (B) Motif enrichment analysis of the regions open in DKO1 when methylated in HCT116 and in HCT116 post 5-Aza-CdR treatment shows that motifs significantly enriched include binding sites for transcription factors with known chromatin remodeler associations. (C) Oncomine expression data for colon adenocarcinoma versus normal for genes from Figure 3A (left – promoters that gain accessibility in HCT116 post-5-Aza-CdR treatment; right – promoters that gain accessibility in DKO1 compared with methylated HCT116) show significant under-expression in the tumors. (D) Plots displaying all probes for NKX2-3 and NPY1R, arranged in the genomic context, show that both genes lose methylation and gain accessibility on 5-Aza-CdR treatment and are unmethylated and accessible in DKO1.
To assess the functional importance of these groups of genes that open on 5-Aza-CdR treatment and in DKO1, despite methylation in the parental, we surveyed the Oncomine™ database (Compendia Bioscience, Ann Arbor, MI). On studying gene expression for colorectal carcinoma samples compared with matched normal tissue (obtained from The Cancer Genome Atlas), we noticed that numerous genes in these categories were down-regulated in colorectal adenocarcinomas compared with the matched normal tissues (Figure 3C), some of which had gained accessibility in common under both conditions of demethylation, NKX2-3 and NPY1R. In summary, genes with promoter regions that open on treatment with 5-Aza-CdR in HCT116 and those that failed to retain nucleosomes to compensate for the lack of methylation at promoters regions in DKO1 cells include well-known tumor suppressors and genes frequently down-regulated in colorectal cancers. This could provide a plausible explanation for the epigenetic component behind the loss of tumorigenicity of HCT116 cells on 5-Aza-CdR treatment as well as in DKO1 cells due to the loss of methylation.
We examined the promoters of NKX2-3 (Figure 3D) and TXNIP (Supplementary Figure S4E), well-studied tumor suppressor genes, as well as NPY1R (Figure 3D) (aberrantly open in DKO1 cells as well as open after 5-Aza-CdR treatment in HCT116 cells) (Figure 3D). A close look at all probes for these genes revealed that all three are unmethylated in DKO1 cells at most sites around the start site, lose methylation on 5-Aza-CdR treatment at these sites compared with untreated HCT116 cells and show increased accessibility in this region for both conditions compared with the parental untreated HCT116 cells. This is a stark illustration of how the loss of methylation is associated with the creation of a nucleosome-depleted region specifically around the TSS in both contexts (Figure 3D). We confirmed results for TXNIP, wherein repressive histone patterns for HCT116 cells but a substantial acetylated-H3/H3 ratio for DKO1 cells were observed (Supplementary Figure S4C), and sequenced the TSS with and without enzyme treatment to validate the findings (Supplementary Figure S4D).
Our results from these genes illustrate important exceptions to the general pattern found of nucleosome retention compensating for the loss of methylation, which may suggest the functional need to restore the expression of certain genes to maintain the non-tumorigenic nature of DKO1 and to support the loss of tumorigenicity after 5-Aza-CdR treatment.
5-Aza-CdR and the HDAC inhibitor SAHA target a unique set of genes for chromatin opening
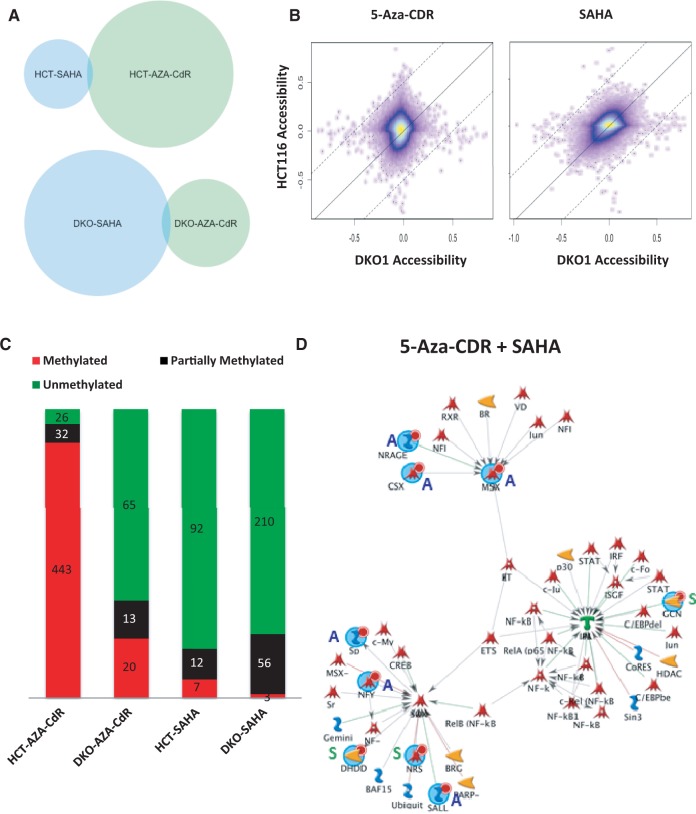
On observing that only 1.6% of promoter sites open after demethylation in HCT116 cells on 5-Aza-CdR treatment, we asked whether it is possible to further open the aberrantly repressed epigenome in cancers. To answer this question, we used DKO1 cells as a model system of demethylation and treated the cells with 0.3 µM of 5-Aza-CdR for 24 h to see if we can further open regions that may be bound by remnant DNA methyltransferases. We also treated DKO1 as well as HCT116 cells with 1 µM of the Food and Drug Administration-approved HDAC inhibitor SAHA for 24 h to determine the effects of increasing histone acetylation on chromatin opening.
Notably, we observed that the promoters that gain accessibility by each of the treatments in both cell lines show minimal overlap, indicating that the drugs have unique and independent targets (Figure 4A). We repeated the experiment in MCF7 and 786-O and confirmed that the two drugs drive chromatin opening in largely unique targets (Supplementary Figure S5A). To understand further the drug targets between the two cell lines, we compared the accessibility states for HCT116 and DKO1 cells treated with 5-Aza-CdR or with SAHA. We found that accessibility between HCT116 and DKO1 was non-linear on 5-Aza-CdR treatment. In contrast, SAHA treatment induced a highly linear relationship between the accessibilities of HCT116 and DKO1 post-treatment (Figure 4B). This suggests that 5-Aza-CdR targets different regions for the opening of chromatin in the two cell lines, whereas SAHA has shared targets. Given that the main difference between the cell lines is that of DNA methylation, this could explain the non-linearity of the accessibility patterns on 5-Aza-CdR treatment, which is specifically a methyltransferase inhibitor.
Figure 4.
Regions that display chromatin opening on 5-Aza-CdR or SAHA treatments are distinct. (A) Venn diagrams of regions that open in HCT116 and in DKO1 with 5-Aza-CdR and SAHA show that the drugs have distinct targets in both cells lines. (B) Kernel density scatter plot of accessibility of all probes in HCT116 and in DKO1 on 5-Aza-CdR treatment or SAHA treatment indicates that SAHA, unlike 5-Aza-CdR, has similar targets in both cell lines. (C) Quantification of the methylation status of probes that gain accessibility on treatment with 5-Aza-CdR or SAHA shows that 5-Aza-CdR targets regions of methylation even in a cell line severely retarded for methylation, DKO1, unlike SAHA. (D) Network enrichment diagram including seed genes that gain accessibility on either 5-Aza-CdR treatment (marked A) or on SAHA treatment (marked S). The symbols used are as seen on the Metacore Web site.
To further understand the relationship between the endogenous methylation state and drug response, we sought to determine the methylation status of all gene promoters that gain chromatin accessibility by each of the drugs. Our results demonstrate that ∼90% of 5-Aza-CdR reactivated genes in HCT116 and ∼20% in DKO1 have high degrees of methylation and ∼5 and ∼10% of them contain partial methylation, respectively, in the untreated control. The level of methylation in this group is much higher than the genome-wide average for each of the cell lines, especially for DKO1 cells which only have 5% remnant methylation (Figure 4C). This finding shows that 5-Aza-CdR targets methylated regions even in a cell line severely retarded for DNA methylation. On the other hand, the genes targeted for chromatin opening by SAHA contain few genes with high methylation in both cell lines, indicating that this drug likely targets regions that are independent of DNA methylation (Figure 4C). Similar patterns of methylation were seen for regions that gain accessibility in both MCF7 and 786-O cell lines treated with 5-Aza-CdR and SAHA, confirming the finding (Supplementary Figure S5B).
The minimal overlap between the genes that gain accessibility due to the two drugs strongly suggests that combinatorial treatment could result in the opening of promoter regions belonging to independent groups of genes, providing for a synergistic de-repression of the aberrant cancer epigenome. Interestingly, whereas the group of genes that open in HCT116 after treatment with 5-Aza-CdR or SAHA are enriched for specific functions such as cation/DNA binding (5-Aza-CdR) or channel activity (SAHA) (Supplementary Figure S5C), the combined list opens up a larger network of genes (Figure 4D). The combined network has significant enrichment for regulation of transcription (1.13 e−35), regulation of apoptosis (3.392e−16), immune response (9.135e−12) and chromatin remodeling (3.49e−7). This indicates that combinatorial treatment could functionally open a set of genes that could keep cell transcription in check, initiate apoptosis, make the cells more visible to the immune system and potentially initiate a cascading effect and open promoters of more genes that are abnormally repressed. In summary, genes that gain accessibility by both drugs in combination have the potential to activate multiple pathways that could work together to reduce the tumorigenicity of the cancer cells.
Together, our results show that both drugs, 5-Aza-CdR and SAHA, are capable of increasing the accessibility of chromatin on treatment, but have unique and non-overlapping targets, potentially due to different epigenetic aberrations that mark genes, methylation or histone deacetylation. This evidence provides a strong rationale for combination epigenetic therapy, to allow for the opening of different regions of aberrantly repressed chromatin.
DISCUSSION
Although several studies have established the occurrence of DNA demethylation after treatment with DNA methyltransferase inhibitor, the epigenetic consequences of such demethylation has not been carefully investigated (13,14). Previously, our group established, in a loci-specific manner, that the insertion of the histone variant H2A.Z by SRCAP is necessary for creating a nucleosome-depleted region after DNA demethylation induced by 5-Aza-CdR (47). In this study, we extend the analysis genome-wide in HCT116 post-treatment as well as in a genetic model of methyltransferase disruption, and find that demethylation is accompanied by the opening of only a minority of loci and, hence, is primarily a passenger event (22). We confirm our results in MCF7 and in 786-O cell lines. Interestingly, this group of genes is enriched for polycomb repressive complex 2 targets and, therefore, an epigenetic switch to the H3K27me3 mark could explain the maintenance of the closed configuration. The striking finding that most loci do not gain accessibility suggests that most demethylation events will not have a functional contribution to therapeutic efficacy. The areas that do open despite methylation in the parental or control cells appear to have a functional role in reducing tumorigenicity, corresponding with regions of well-studied and putative tumor suppressors. We also find combinatorial benefits to chromatin opening by treating cells with 5-Aza-CdR or SAHA, showing that each drug targets distinct and non-overlapping regions individually. Although the synergistic reactivation of specific genes using DNA methyltransferase inhibitors and HDAC inhibitors has been shown before (48), we now demonstrate that the drugs can open independent groups of genes, thereby increasing the extent of de-repression.
Our results have powerful implications, in that we have established in two colorectal cancer cell systems that specific loci have a predilection to gain chromatin accessibility after DNA demethylation. It would be interesting to expand this to a larger panel of primary colon cancers to understand whether the signature extends across the samples. We have observed similar patterns of chromatin opening on demethylation in a breast and renal cancer cell line as well. It would also be important to understand, from a mechanistic point of view, why some genes are more likely to gain chromatin accessibility after DNA demethylation than some others. Our preliminary findings suggest that specific transcription factor binding motifs are enriched in the regions that gain accessibility, and these transcription factors are expressed in the cell systems we studied. Given that these factors have been established to work in conjunction with known chromatin remodelers (43–46), this could result in the preferential targeting of remodelers to these sites and, hence, chromatin opening (9). This question needs to be explored further to understand how best to intervene pharmacologically to de-repress the epigenome in a targeted manner.
Our finding that <2% of demethylation induced by 5-Aza-CdR is functional, resulting in chromatin accessibility changes, forces us to re-evaluate the current design of epigenetic therapy. Although the use of DNA methyltransferase inhibitors has seen some success, it remains true that several patients receiving such therapeutic regimens do not see any benefit, often not correlating with observed demethylation (49). Our study allows for the identification of the functional events post-treatment, which could be patient and tumor-type specific. In a similar manner, it is crucial to study chromatin accessibility changes along with DNA demethylation and histone alterations in patients to better understand therapeutic response. One could then assess what combinatorial treatment would best benefit the patient.
Using our new methodology, AcceSssIble, we have developed an exciting new arena for studying the components of the epigenome in an integrated manner. We have shown that most DNA demethylation changes do not induce changes to the conformation of chromatin. Our results pave the way for future study to determine the ideal combination of epigenetic modulators that could best target the aberrant cancer epigenome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
National Institutes of Health (NIH) [RO1CA124518] to G.L. and the generous support of the Stand Up to Cancer [2000901485 to P.A.J. and S.B.B]. Funding for open access charge: NIH [RO1CA124518].
Conflict of interest statement. none declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the support of Dan Weisenberger in generating array data, and Hariharan Easwaran, Toshinori Hinoue and Tim Triche for help with data analysis. We thank Claudia Andreu-Vieyra and Jessica Charlet for helpful discussions.
REFERENCES
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr. Opin. Genet. Dev. 2012;22:50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreu-Vieyra CV, Liang G. Nucleosome occupancy and gene regulation during tumorigenesis. Adv. Exp. Med. Biol. 2012;754:109–134. doi: 10.1007/978-1-4419-9967-2_5. [DOI] [PubMed] [Google Scholar]
- 6.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 9.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinshelwood RA, Melki JR, Huschtscha LI, Paul C, Song JZ, Stirzaker C, Reddel RR, Clark SJ. Aberrant de novo methylation of the p16INK4A CpG island is initiated post gene silencing in association with chromatin remodelling and mimics nucleosome positioning. Hum. Mol. Genet. 2009;18:3098–3109. doi: 10.1093/hmg/ddp251. [DOI] [PubMed] [Google Scholar]
- 12.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 14.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 15.List AF, Vardiman J, Issa JP, DeWitte TM. Myelodysplastic syndromes. Hematology Am. Soc. Hematol. Educ. Program. 2004:297–317. doi: 10.1182/asheducation-2004.1.297. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths EA, Gore SD. Epigenetic therapies in MDS and AML. Adv. Exp. Med. Biol. 2013;754:253–283. doi: 10.1007/978-1-4419-9967-2_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Manero G. Demethylating agents in myeloid malignancies. Curr. Opin. Oncol. 2008;20:705–710. doi: 10.1097/CCO.0b013e328313699c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol. Oncol. 2010;116:195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blumenschein GR, Jr, Kies MS, Papadimitrakopoulou VA, Lu C, Kumar AJ, Ricker JL, Chiao JH, Chen C, Frankel SR. Phase II trial of the histone deacetylase inhibitor vorinostat (Zolinza, suberoylanilide hydroxamic acid, SAHA) in patients with recurrent and/or metastatic head and neck cancer. Invest. New Drugs. 2008;26:81–87. doi: 10.1007/s10637-007-9075-2. [DOI] [PubMed] [Google Scholar]
- 22.Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv. Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 27.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc. Natl Acad. Sci. USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 31.Larson PS, Schlechter BL, King CL, Yang Q, Glass CN, Mack C, Pistey R, de Las Morenas A, Rosenberg CL. CDKN1C/p57kip2 is a candidate tumor suppressor gene in human breast cancer. BMC Cancer. 2008;8:68. doi: 10.1186/1471-2407-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park K, Kim K, Rho SB, Choi K, Kim D, Oh SH, Park J, Lee SH, Lee JH. Homeobox Msx1 interacts with p53 tumor suppressor and inhibits tumor growth by inducing apoptosis. Cancer Res. 2005;65:749–757. [PubMed] [Google Scholar]
- 33.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 35.Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl Acad. Sci. USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi JM, Tsai HC, Glockner SC, Lin S, Ohm JE, Easwaran H, James CD, Costello JF, Riggins G, Eberhart CG, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094–8103. doi: 10.1158/0008-5472.CAN-07-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 38.Jin HO, Seo SK, Kim YS, Woo SH, Lee KH, Yi JY, Lee SJ, Choe TB, Lee JH, An S, et al. TXNIP potentiates Redd1-induced mTOR suppression through stabilization of Redd1. Oncogene. 2011;30:3792–3801. doi: 10.1038/onc.2011.102. [DOI] [PubMed] [Google Scholar]
- 39.Yu W, Lin Z, Pastor DM, Hegarty JP, Chen X, Kelly AA, Wang Y, Poritz LS, Koltun WA. Genes regulated by Nkx2-3 in sporadic and inflammatory bowel disease-associated colorectal cancer cell lines. Dig. Dis. Sci. 2010;55:3171–3180. doi: 10.1007/s10620-010-1138-0. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 42.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruffat H, Manet E, Sergeant A. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 2002;3:141–146. doi: 10.1093/embo-reports/kvf031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chokas AL, Trivedi CM, Lu MM, Tucker PW, Li S, Epstein JA, Morrisey EE. Foxp1/2/4-NuRD interactions regulate gene expression and epithelial injury response in the lung via regulation of interleukin-6. J. Biol. Chem. 2010;285:13304–13313. doi: 10.1074/jbc.M109.088468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung YM, Park SH, Tsai WB, Wang SY, Ikeda MA, Berek JS, Chen DJ, Hu MC. FOXO3 signalling links ATM to the p53 apoptotic pathway following DNA damage. Nat. Commun. 2012;3:1000. doi: 10.1038/ncomms2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. J. Biol. Chem. 2009;284:5936–5944. doi: 10.1074/jbc.M808103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Noushmehr H, Han H, Andreu-Vieyra C, Liang G, Jones PA. Gene reactivation by 5-aza-2′-deoxycytidine-induced demethylation requires SRCAP-mediated H2A.Z insertion to establish nucleosome depleted regions. PLoS Genet. 2012;8:e1002604. doi: 10.1371/journal.pgen.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 49.Oki Y, Aoki E, Issa JP. Decitabine–bedside to bench. Crit. Rev. Oncol. Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.