Abstract
Archaeal family-D DNA polymerase is inhibited by the presence of uracil in DNA template strands. When the enzyme encounters uracil, following three parameters change: DNA binding increases roughly 2-fold, the rate of polymerization slows by a factor of ∼5 and 3′–5′ proof-reading exonuclease activity is stimulated by a factor of ∼2. Together these changes result in a significant decrease in polymerization activity and a reduction in net DNA synthesis. Pol D appears to interact with template strand uracil irrespective of its distance ahead of the replication fork. Polymerization does not stop at a defined location relative to uracil, rather a general decrease in DNA synthesis is observed. ‘Trans’ inhibition, the slowing of Pol D by uracil on a DNA strand not being replicated is also observed. It is proposed that Pol D is able to interact with uracil by looping out the single-stranded template, allowing simultaneous contact of both the base and the primer-template junction to give a polymerase-DNA complex with diminished extension ability.
INTRODUCTION
Individual cells contain a varying repertoire of DNA polymerases, a subset of which is dedicated to genome replication (1). In all three domains of life, Bacteria, Eukarya and Archaea, chromosomes are copied by the replisome, a multi-protein replication machine (2,3). The bacterial replisome contains at least two molecules of DNA polymerase III, a family-C member which is responsible for copying the genetic material (4). Although the same molecular species is used to copy both the leading and lagging strands, individual molecules of DNA Pol III are believed to be arranged in an asymmetric fashion, compatible with the requirements needed to copy two dissimilar DNA strands (5). The eukaryotic replisome contains two family-B polymerases, δ and ε, shown in an elegant series of experiments to be responsible for lagging and leading strand replication, respectively (6–8). Which polymerases are responsible for duplicating the archaeal genome is not currently known with the same degree of certainty as the other two domains. All Archaea contain family-B polymerases, usually present as multiple members in the Crenarchaea and as a single exemplar in the Euryarchaea (3,9). These proteins are monomeric and contain both the polymerase and 3′–5′ proof-reading exonuclease active sites within the same polypeptide chain (10). In Crenarchaea, the only polymerases with properties compatible with DNA replication, i.e. interaction with proliferating cell nuclear antigen (PCNA) and rapid, accurate and processive DNA synthesis are the family-B enzymes which are widely assumed to fulfil this role (3,9,11). Whether distinct family members are individually responsible for leading and lagging strand replication or the same species copies both strands is presently unknown. Matters are more complicated in the Euryarchaea, which contain an unusual family-D enzyme (11–13) in addition to the replication-competent family-B polymerase. As well as being present in Euryarchaea, family-D polymerases are found in several emergent archaeal phyla (Thaumarchaea, Korarchaea and Nanoarchaea), which presently have few characterized members. However, this polymerase is noticeably absent in Crenarchaea.
Family-D polymerases exist as heterodimers, comprising a large (polymerase) and small (proof-reading exonuclease) subunit and may further assemble to give an L2S2 heterotetramer (11–15). The biochemical properties of the family-D enzymes are compatible with those required for a replicative polymerase: the presence of a proof-reading exonuclease activity should ensure accuracy (although fidelity has yet to be measured) and interaction with PCNA enables copying of long fragments of DNA (16–19). Both the family-B and -D polymerases have been shown to be essential for viability in a halophilic euryarchaeon using targeted gene deletion (20). Based on biochemical evidence it has been proposed that Pol D may act soon after initiation by primase and that at a later stage a switch occurs such that Pol B becomes responsible for leading strand replication, whereas Pol D continues to process the lagging strand (17,18). Both replicative DNA polymerases are also suspected to be involved in the resolution of RNA fragments in replicating cells (21). However, definitive genetic confirmation using experimental approaches such as those employed with the eukaryotic polymerases is awaited.
The archaeal family-B polymerases are unusual in recognizing uracil and hypoxanthine in DNA template strands and stalling replication when these bases are encountered (22–24). Tight and specific binding of the two deaminated bases is mediated by a pocket in the N-terminal domain (25,26). It is anticipated that stalling serves to prevent copying of uracil and hypoxanthine, which may arise by deamination of cytosine and adenine, respectively. The parent bases, cytosine and adenine, pair with guanine and thymine, respectively; however, their deamination products uracil and hypoxanthine are effective mimics of thymine and guanine and therefore code for adenine and cytosine. Thus, replication of deaminated bases results in a transition mutation (C:G → T:A, when uracil is copied; A:T → G:C, when hypoxanthine is copied) in 50% of progeny. Replicative polymerases from the bacterial and eukaryotic domains are unable to sense deaminated bases, despite the N-terminal domains of eukaryotic Pols ε and δ possessing considerable amino acid similarity with the corresponding region in the archaeal family-B enzymes (27). Very little is known about the response of the euryarchaeal family-D polymerases to deaminated bases. A brief report has indicated that they neither incorporate dUTP into extending strands nor copy uracil-containing templates (28). This publication presents a full characterization and shows that Pol D is indeed inhibited by template-strand uracil. While mechanistic details have not been completely characterized, it is apparent that the inhibition is markedly different from that previously observed with archaeal family-B polymerases.
MATERIALS AND METHODS
Enzymes
Wild-type and exonuclease-deficient (H451A) Pab-Pol D were produced and purified as previously described (29). These two variants of Pfu-Pol D (exo− = H445A, the equivalent amino acid change to that used in Pab-Pol D) were overexpressed in Escherichia coli BL21 (DE3) codon+ (RIL) cells using pWTD1 and pWTD2his, which encode the large and small subunits of the enzyme, respectively, with the small subunit having a (His)6 tag (30). Cells were suspended (50 ml buffer/g of cells) in 50 mM Tris-HCl (pH 8.0), 20 mM imidazole, 500 mM NaCl, 0.1 mM EDTA, 0.5 mM Dithiothreitol (DTT), 10% glycerol + 1 EDTA-free protease inhibitor tablet (Roche), sonicated (on ice) and subjected to DNaseI digestion at 37°C for 30 min. The lysed cell suspension was then incubated at 75°C for 20 min and denatured protein and cell debris pelleted by centrifugation. The supernatant was filtered and applied to a 5-ml HisTrap (GE Healthcare) column, equilibrated with the above buffer containing 50 mM imidazole and lacking the protease inhibitor. The column was extensively washed with this buffer and Pol D eluted using a 30 ml linear gradient of 50–500 mM imidazole. Final purification used gel filtration on a Superdex 200 10/300 GL column (GE Healthcare) with 20 mM Tris-HCl (pH 6.5), 400 mM NaCl and 1 mM DTT.
Primer extension assays
Pab-Pol D and Pfu-Pol D assays were carried out in 10 µl (20 μl for experiments with U+42, 70, 102 and 134 and replication fork mimics) of 10 mM Tris-HCl (pH 9), 50 mM KCl, 10 mM MgCl2 and 200 μM each of the dNTPs. With Pab-Pol D, 25 nM of fluorescent-labelled primed synthetic oligodeoxynucleotides and 30 nM of Pab-PolD exo+/exo− (unless otherwise specified) were used, and the reactions were performed at 55°C for 30 min. In the case of Pfu-Pol D reactions, 20 nM of primer template and 140 nM of polymerase were used and conducted at 50°C for the times indicated. With both enzymes, the reactions were quenched by the addition of an equal volume of stop buffer [95% formamide, 10 mM EDTA, 10 mM NaOH and 1 µM of ‘competitor oligonucleotide’ (an exact complement of the template strand under study)] (31). Samples were heated at 95°C for 5 min. The reaction products were resolved on 17% polyacrylamide, 8 M urea gels and visualized with a Mode Imager Typhoon 9400 or Typhoon FLA9500 (GE Healthcare) and quantified using Image Quant software. The percentage of extension was defined as the ratio: +1 to +n products (where n is the fully extended product)/total DNA.
DNA cutting by Pfu-Pol D
Pfu-Pol D exo+ or exo− (80 nM) was incubated with fluorescent oligodeoxynucleotides (single and double stranded; +/− uracil) (20 nM) at 50°C for 30 min in the buffer given above. As a positive control for strand cutting, a reaction was carried out with uracil-DNA glycosylase (0.5 units, Fermentas). After 30 min, the reactions were quenched by heating with stop buffer. This buffer contains NaOH resulting in an alkaline pH and heating at 95°C for 5 min is expected to cut DNA at any abasic sites generated by the polymerase. Analysis was performed by gel electrophoresis as discussed above.
Pfu-Pol D exonuclease assay and single dGTP incorporation
These were carried out as described above except that dNTPs were omitted for exonuclease assays and for dGTP incorporation only this triphosphate was added at concentrations that varied between 5 and 500 μM. The evaluation of the rate constant for the exonuclease reaction and the KD and kpol for incorporation of a single dGTP under single turnover conditions have been described earlier (26,29,32).
Binding of Pfu-Pol D to DNA
Determination of the KD values describing the binding of Pfu-Pol D to the oligodeoxynucleotides listed in Table 1 was performed using direct binding fluorescence anisotropy (with 5′-hexachorofluorescein labelled DNA) as previously described (33). Titrations were carried out in 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 1 mM DTT and 0.1 mg/ml of bovine serum albumin with 1 nM DNA at 25°C. Aliquots of Pol D (1–50 nM) were added and the data analysed to yield KD as described earlier (32).
Table 1.
Binding constants for the interaction of Pfu-Pol D with DNA
| DNA | X location | KD (nM) X = T | KD (nM) X = U |
|---|---|---|---|
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | Single stranded | 6.0 ± 1.6 | 3.4 ± 0.3 |
| 5′ AATAGGTCCTATAGGCGAATGGTCCAGCTGGAA | −1 | 9.3 ± 0.6 | 5.2 ± 1.3 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | |||
| 5′ AATAGGTCCTATAGGCGAATGGTCCAGCTGG | +1 | 9.1 ± 1.5 | 5.8 ± 0.4 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | |||
| 5′ AATAGGTCCTATAGGCGAATGGTCCAGC | +4 | 7.3 ± 1.2 | 4.8 ± 0.3 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | |||
| 5′ AATAGGTCCTATAGGCGAATGG | +10 | 8.4 ± 0.3 | 4.2 ± 0.5 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | |||
| 5′ AATAGGTCCTATAGGCGAATGGTCCAGCTGGAACCAGAAA | Double stranded | 27.9 ± 6.9 | 29.0 ± 7.7 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* | |||
| 5′ AATAGGTCCTAUAGGCGAATGGTCCAGCTGGAACCAGAAA | Double stranded | 29.4 ± 11.1 | 36.6 ± 2.4 |
| 3′ TTATCCAGGATATCCGCTTACCAGGTCGACCXTGGTCTTT-H* |
The KD values for the binding of Pfu-Pol D (average ± standard deviation from at least four determinations) to oligodeoxynucleotides containing uracil (thymine in controls) are given. The uracil is located in single strands, at various positions in a primer template and in double strands. H* = hexachlorofluorescein, used to determine the KD value using fluorescence anisotropy titration (33).
RESULTS
Uracil in DNA template strands inhibits extension by Pol D
Throughout this publication, Pol D has been used from two different Pyrococcus species, Pyrococcus abyssi and Pyrococcus furiosus (Pab- and Pfu-Pol D). The amino acid sequences of the two proteins are very similar with 86 and 77% identity seen for the large and small subunits, respectively (Supplementary Figure S1). Even when different amino acids are found, in ∼50% of cases the exchanges are conservative. The two polymerases behaved similarly enough in all the assays described in this publication to enable them to be used interchangeably.
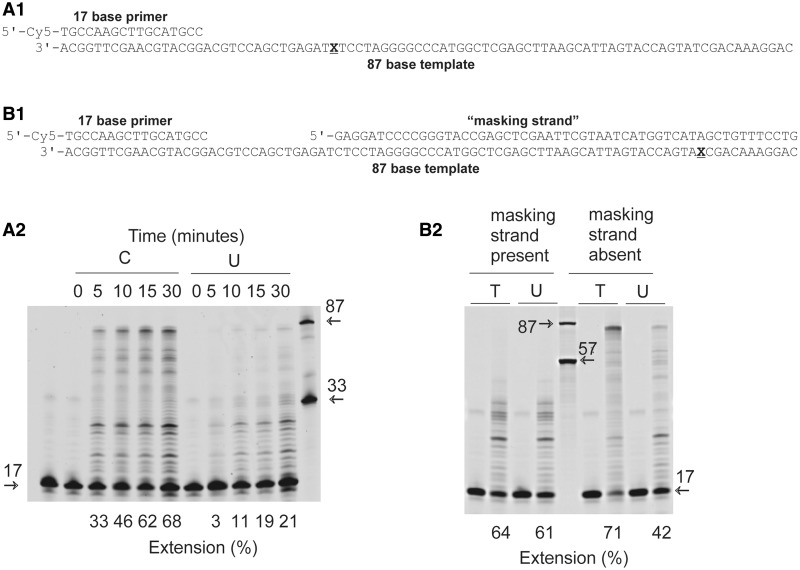
When Pab-Pol D was used to extend a primer template (Cy5-labelled primer, 17 bases long, annealed to a template 87 bases in length) containing a single uracil at +16 (i.e.16 bases ahead of the primer-template junction), a reduction in DNA synthesis was observed as compared with a control that lacked this base (Figure 1, A1 and A2). In general, the intensity of each extended band was reduced for the uracil-containing template and, in particular, much less full length product was observed. This figure shows the results obtained with wild-type Pab-Pol D (exo+); however, almost identical profiles were seen with a mutant lacking 3′–5′ exonuclease activity (exo−) (Supplementary Figure S2A). When the experiment was repeated with uracil further ahead of the primer-template junction at +59, inhibition was still observed (Figure 1, B1 and B2). However, when the uracil was situated in a double-stranded region by annealing a third ‘masking’ oligodeoxynucleotide to the primer template to give a gapped substrate, inhibition largely disappeared (Figure 1, B1 and B2). On this substrate, Pol D DNA synthesis was associated with pausing of the polymerase in the vicinity of the position of the nick, generating extension products in the 18–32 nucleotides (nt) size range. Although these experiments are not intended to be quantitative (kinetic parameters are determined more rigorously below), the gels were scanned to approximate the extent of polymerization. The extension percentages observed are consistent with uracil slowing polymerization by a factor of 2–5.
Figure 1.
Inhibition of Pab-Pol D (exo+) by template strand uracil. (A1 and B1) Primer templates used for the experiments shown in panels A2 and B2, respectively. (A2) Extension of the 17/87 primer-template A1 (X = C or U) by Pab-Pol for the times indicated. The numbers under the gel lanes represent the total percentage of extended product. Reference oligodeoxynucleotides of 17, 33 and 87 bases are indicated by the arrows. (B2) Influence of burying uracil in double stranded DNA on extensions by Pab-Pol D. The image shows the products formed with the 17/87 primer-template B1 (X = T or U) with either the ‘masking strand’ present or absent. Each reaction shows a zero and 30-min time point. The numbers under the gel lanes show the total percentage of extended product. Reference oligodeoxynucleotides of 17, 57 and 87 bases are indicated by the arrows.
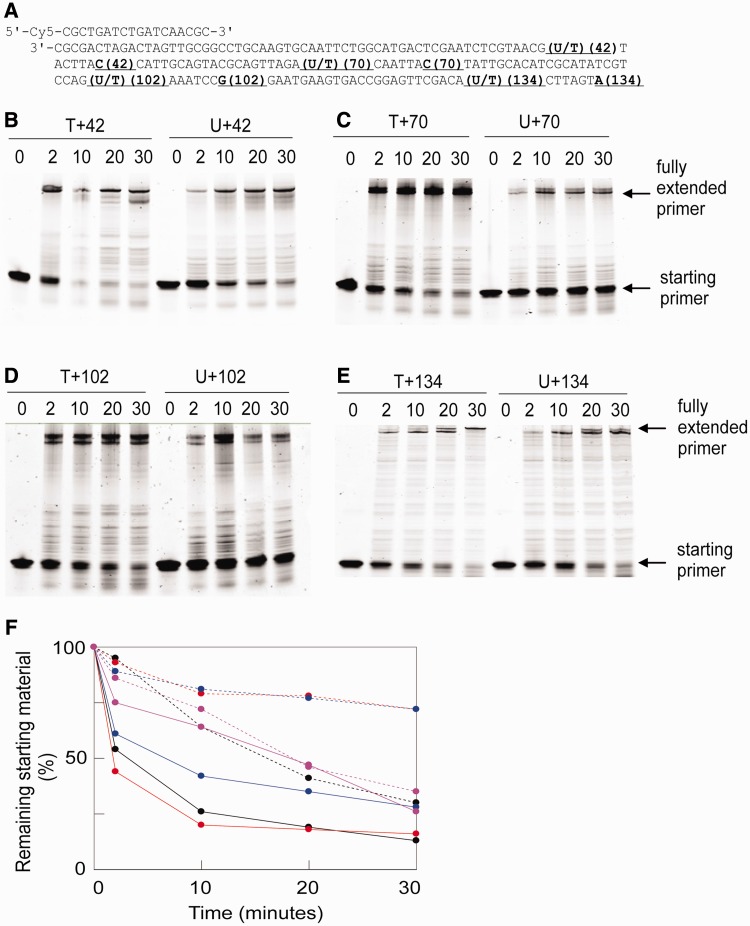
A parallel study was carried out with Pfu-Pol D (exo+) using a set of primer templates that position uracil increasingly further ahead of the primer-template junction (Figure 2A). Inspection of the extension gels obtained (Figure 2B–E) clearly shows less polymerization with uracil at +42, +70 and +102, compared with T controls. Inhibition is not obvious with uracil at +134. The impression is confirmed by Figure 2F, a summary schematic that shows the amount of primer template remaining at times of up to 30 min. In all cases, more starting material persists with U+42, 70 and 102 than with T-containing DNA. However, no difference is seen between U+134 and T+134.
Figure 2.
Extension by Pfu-Pol D (exo+) of primer templates that locate uracil at ever increasing distances ahead of the junction. (A) DNA substrates. Four different primer templates, consisting of a Cy5-labelled 18-mer primer annealed to four templates of increasing lengths, were used. These place uracil (control = thymidine) 42, 70, 102 and 134 bases ahead of the primer-template junction. The lengths of the templates are indicated using the same numbers, e.g. which places uracil at +42 terminates at the C labelled 42, etc. (B–E). Extension of the primer templates for the times shown above the gels (minutes) by Pfu-Pol D (exo+). The positions of the starting primer and full length products are indicated. (F) Summary of the data in panels (B–E) showing remaining primer template against time. Colour coding: black, +42; red, +70; blue, +102; magenta, +134. Solid lines, T; hatched lines, U.
A large number of extension reactions have been carried out, a selection of which are given in the Supplementary Figure S2. Strong inhibition was observed with uracil at +1 (Supplementary Figure S2B) and, surprisingly, a decrease in polymerization also occurred when uracil was located at −1, i.e. just within the double-stranded region, immediately behind the replication fork (Supplementary Figure S2C). In general, though, when uracil is well buried in double-stranded regions, either ahead of or behind the replication fork, no inhibition is observed (Figure 1B; Supplementary Figure S2D). However, exact boundaries, i.e. how far into a duplex uracil must be to prevent inhibition have not been mapped and a reduction in DNA synthesis was still noticeable using a derivative of the gapped substrate shown in Figure 1B, which locates uracil five bases into the duplex (Supplementary Figure S2E). Although the extensions described in the text and the Supplementary material use a very slight molar excess of Pab-Pol D and a 7-fold excess of Pfu-Pol D, inhibition by uracil with a 4-fold excess of Pab-Pol was also observed (Supplementary Figure S3).
Pol D incorporates dAMP opposite uracil
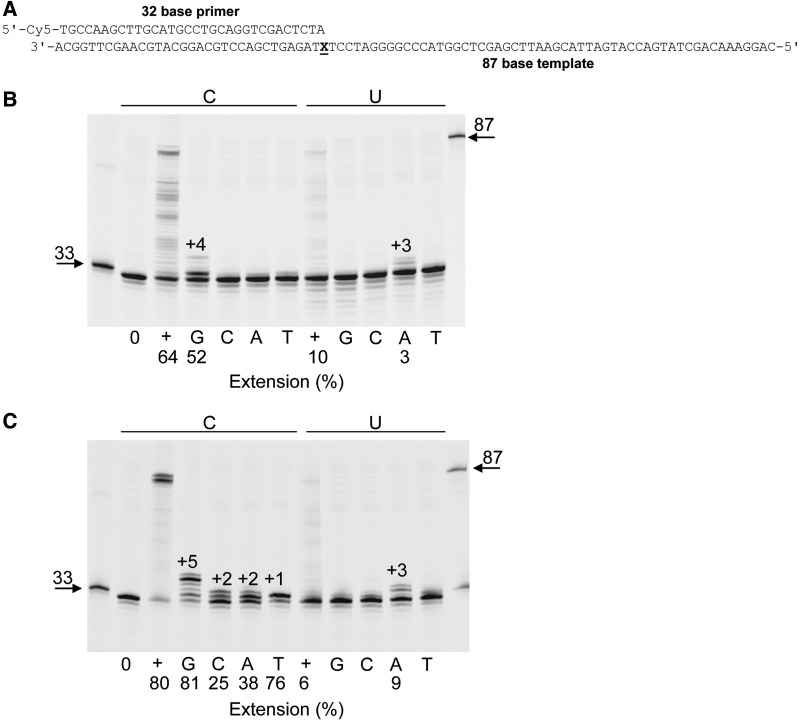
The gels shown in Figures 1, 2 and Supplementary Figure S2 indicate that although uracil inhibits primer extension by Pol D, copying beyond the deaminated base does take place. To determine which base is inserted opposite uracil, single additions of each of the four dNTPs were made to a solution containing Pab-Pol D and a primer template with uracil at +1 (Figure 3A). As can be seen in Figure 3B, Pab-Pol D exo+ only incorporated adenine opposite uracil, although at a reduced rate (∼3% incorporation) due to uracil inhibition. In a control experiment with cytosine at +1 guanine was inserted efficiently (∼52% incorporation), as expected. Similar results were seen with the exo− variant of Pol D, although multiple additions were observed due to the inability to excise incorrectly incorporated bases (Figure 3C). In agreement with Supplementary Figure S2B, when all four dNTPs were simultaneously added, strong reduction (∼6 to 13-fold depending on whether exo+ or exo− was used) in DNA synthesis was observed with the uracil-containing template.
Figure 3.
Base incorporated opposite template strand U by Pab-Pol D. (A) Primer templates used in these experiments (X = C or U). (B) and (C) Extensions using Pab-Pol D exo+ (B) or exo− (C) with either C or U at the +1 position (X), i.e. the first single-stranded template base. 0 = no dNTPs added; + = all four dNTPs added; G, C, A and T = only dGTP or dCTP or dATP or dTTP added, respectively. +1, +2, +3, +4 and +5 represent final products. The extension (%) for selected lanes is shown under the gels.
‘Trans’ inhibition of Pol D by uracil
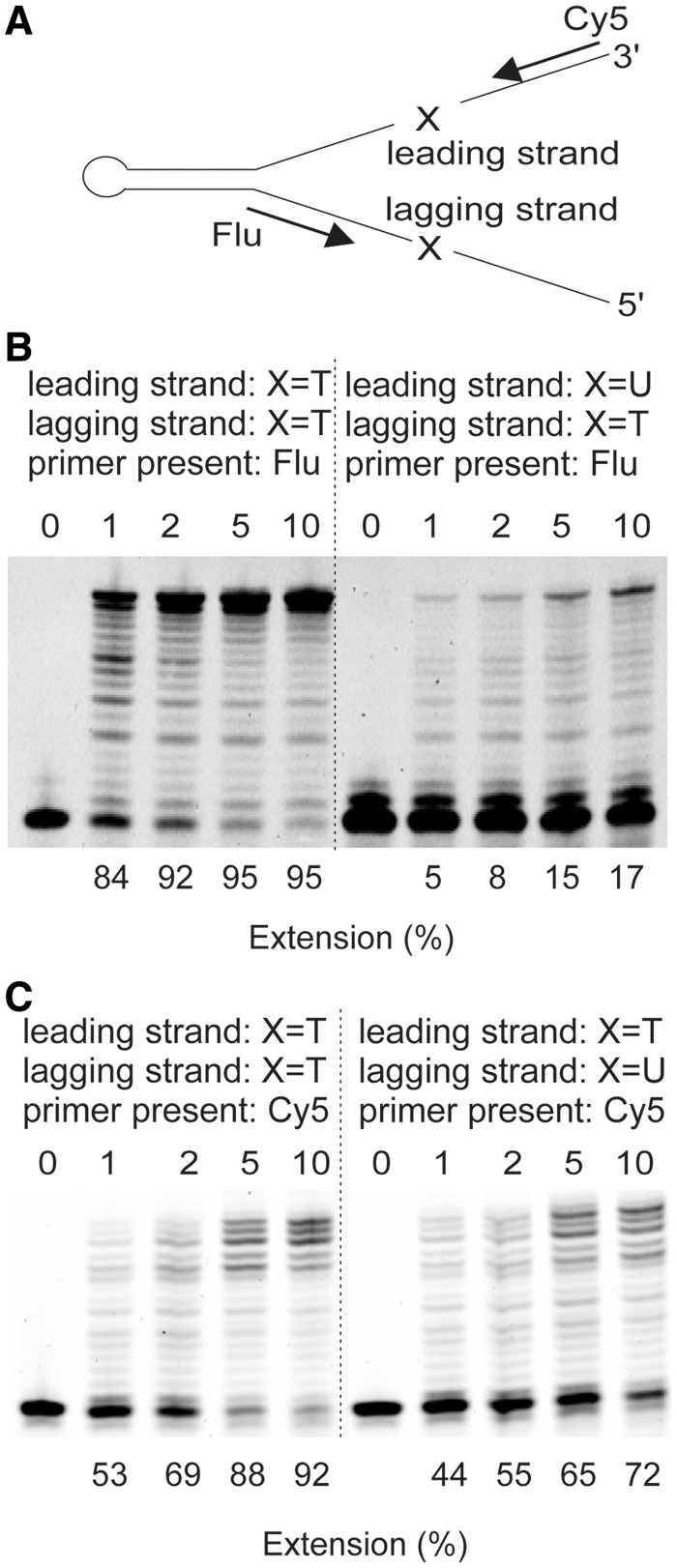
A replication fork mimic (Figure 4A; Supplementary Figure S4) has been used to assess the influence of translocated uracil. Following primer annealing, leading and lagging strand branches are created and uracil can be positioned at either location. Slowing of Pol D by uracil in the branch it is not copying is, strictly speaking, not true trans-inhibition; the design of the mimic means that leading and lagging strands uracil are actually in the same DNA molecule (Figure 4A; Supplementary Figure S4). However, when replicating the lagging strand Pol D will clearly be unable to copy any uracil present in the leading strand. During polymerization of the leading strand, if Pol D was able to melt the double-stranded region, it could conceivably access uracil located in the lagging strand. However, under the conditions used, Pfu-Pol D only copied the leading strand up to the fork junction and no further progression into the double-stranded region was observed. Here, therefore, trans-inhibition is defined as slowing of Pol D by uracil in a DNA region it cannot access by polymerization. As shown in Figure 4B, when Pfu-Pol D was used to copy an unmodified lagging strand, uracil in the leading strand was strongly inhibitory (∼10-fold reduction in extension). When the leading strand was replicated, uracil positioned on the lagging strand also hindered progression (<2-fold reduction in extension), although to a less profound extent than observed for the opposite orientation (Figure 4C). On this substrate, Pol D DNA synthesis was likely associated with futile cycles of assembly/disassembly upon approaching the double-stranded DNA junction, as observed by the accumulation of multiple final products. Further information, more representative of conditions expected in the cell, can be obtained by using both primers to simultaneously copy the leading and lagging strands. The fluorophores used to label the primers (fluorescein and cyanine-5) have spectral properties that allow each strand to be individually monitored. The results (Supplementary Figure S4) are in close agreement with the studies using a single primer shown in Figure 4A; again, Pol D is inhibited by uracil situated in the opposite branch of the primer template to the strand in which polymerization is being observed.
Figure 4.
Inhibition of Pfu-Pol D by ‘trans’ located uracil. (A) Structure of the primer-template mimic (for full sequences, see Supplementary Figure S4). A long ‘snap-back’ oligodeoxynucleotide forms the backbone of the mimic. Annealing of primers (either singly or in pairs) produces leading and lagging strand branches which can contain a single uracil residue (indicated by X) four bases ahead of the primer-template junction. (B) Results seen when the lagging strand is copied (only Flu primer present) with uracil (thymine in controls) present in the leading strand. (C) Results seen when the leading strand is copied (only Cy5 primer present) with uracil (thymine in controls) present in the lagging strand. Here, only one primer was used per experiment. For the results observed using two primers for each experiment, see Supplementary Figure S4.
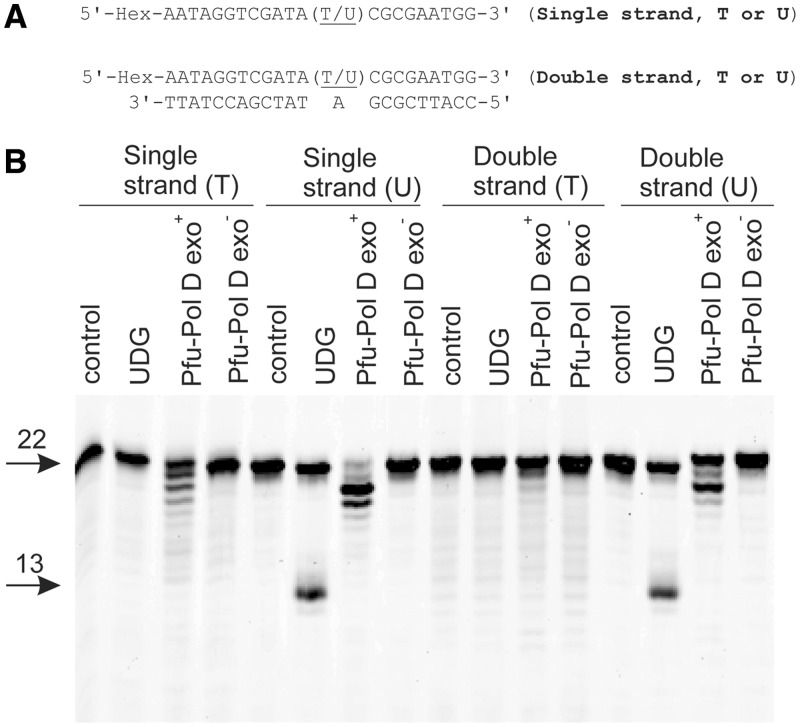
Pol D does not cut uracil-containing DNA
The gels shown in Figures 1–4 indicate that the presence of uracil in template strands reduces DNA synthesis by Pol D. The inhibition could conceivably arise because uracil-dependent degradation of the template reduces the amount of substrate available for extension. To address this possibility, hexachlorofluorescein-labelled oligodeoxynucleotides containing uracil in either single- or double-stranded regions (Figure 5A) were incubated with Pfu-Pol D exo+ or exo−. In all cases, the polymerase did not cut uracil-containing DNA in an endonucleotytic manner, i.e. by hydrolysis of the phosphodiester backbone at or near uracil (Figure 5B). The assay also scores for DNA-glycosylase action, as it involves a post-enzymatic heating step at alkaline pH which cleaves DNA at any abasic sites produced. Pfu-Pol showed no uracil-dependent DNA-glycosylase activity, in contrast to the expected positive result with uracil-DNA-glycosylase itself (Figure 5B). However, the presence of uracil does lead to a slight stimulation in 3′–5′ exonuclease activity, compared with thymidine-containing controls, which results in the removal of a few bases from the 3′ ends of the DNA substrates (Figure 5B). As discussed later in this publication, uracil-dependent stimulation of proof-reading exonuclease activity trims back primers and extending DNA strands and contributes to overall inhibition of polymerization. In principle, this exonuclease activity may remove bases from the 3′-termini of the template strands of the primer templates used in Figures 1–4. However, during polymerase assays, when dNTPs are present, these would be replaced. Therefore, template destruction cannot account for the inhibition of polymerization seen in the presence of uracil. A similar result was also found with Pab-Pol D (Supplementary Figure S5).
Figure 5.
Pol D does not cut uracil-containing DNA. (A) Hexachlorofluorescein-labelled oligodeoxynucleotides used in these experiments. (B) Results seen when Pfu-Pol D exo+ or exo− (80 nM) was incubated with the oligodeoxynucleotides (20 nM) at 50°C for 30 min followed by heating at 95°C for 5 min in the presence of NaOH. Control = no Pfu-Pol D added; UDG = addition of uracil-DNA glycosylase (positive control for strand cleavage at uracil). The starting oligodeoxynucleotides (22 bases) and the 13 base products expected for cleavage at uracil are shown arrowed. The ladders of products slightly reduced in length (seen with Pfu-Pol D exo+, most prominently when uracil is present) arise from 3′ to 5′ proof-reading exonuclease activity.
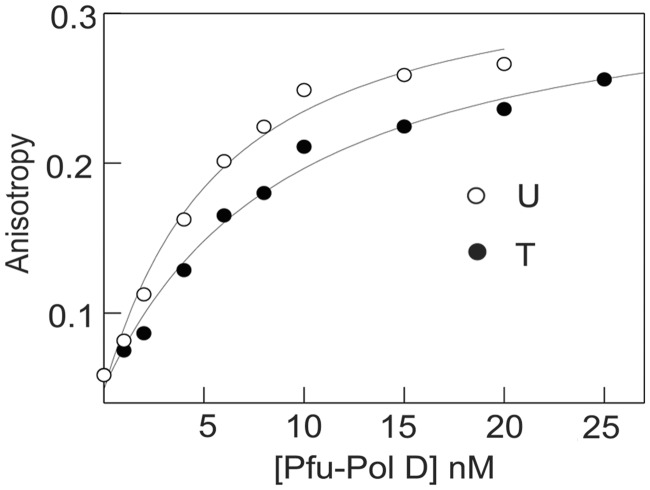
Pol D binds to uracil in single-stranded DNA
To determine if Pol D bound to uracil-containing DNA with greater affinity than to control substrates, fluorescence anisotropy binding titrations were used with hexachlorofluorescein-labelled oligodeoxynucleotides (33). A number of substrates were used (listed in Table 1), which place uracil in single-stranded DNA at a variety of locations in a primer template and in fully duplexed strands. In all cases where uracil was located in single strands, a slight increase, ∼2-fold, in binding was observed when compared with controls lacking this base. As a typical example, the results seen when Pfu-Pol D was added to a primer template containing uracil (or thymine as a control) at +4 are shown in Figure 6. The KD values found for a number of substrates are summarized in Table 1. Although the preference for uracil-DNA over controls is only about a factor of 2, the errors shown in Table 1, and the consistent results seen for a number of different DNA substrates, suggests significance. The slight selectivity for uracil is retained when the base is located at the −1 position of a primer template, just within the double-stranded region. The KD values given in Table 1 are apparent binding constants that represent the measured sum of any specific interaction with uracil plus the numerous non-specific DNA binding modes possible (for uracil-containing substrates) and non-specific binding (for controls). In cases, such as that described here, were specific binding is not significantly greater than non-specific modes, it was very difficult to extrapolate the true or intrinsic binding constant for specific binding to uracil from the measured KD. However, although the KDs shown in Table 1 are apparent, rather than true values, a small but significant preference for uracil in single-stranded regions is clear. Table 1 also demonstrates that Pol D binds more tightly to single than double-stranded DNA and that selectivity for uracil is lost when the base is buried in duplex DNA, away from the primer-template junction. Although binding constants are obtained at 25°C, preferential binding to uracil can be extrapolated to higher temperatures. This is supported by our recent study in which we show that increased temperature does not alter the binding modes of Pol D but likely accelerates the binding kinetics (association/dissociation) (34).
Figure 6.
Binding of Pfu-Pol D to primer templates containing T or U at the +4 template position (sequences given in Table 1). The polymerase was added to the hexachlorofluorescein-labelled primer templates and the increase in fluorescence anisotropy noted. The data were fitted to a 1:1 binding stoichiometry to give the titration curves shown. Binding constants are summarized in Table 1.
Uracil inhibits dNTP incorporation into primer templates
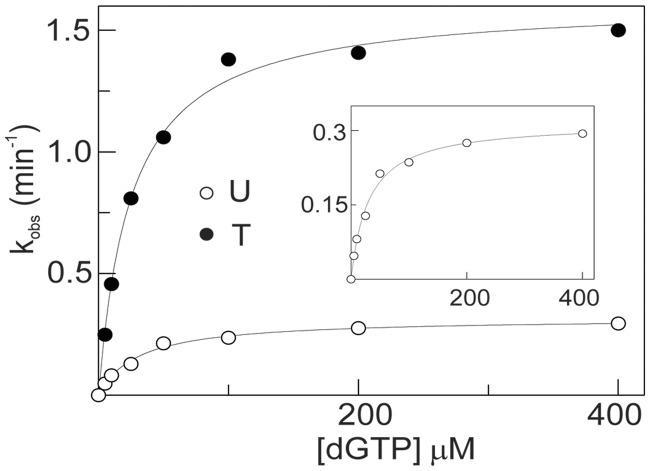
Although primer-template extensions (Figures 1–4) reveal very obviously that uracil inhibits Pol D, these experiments are, at best, semi-quantitative. In particular, it is difficult to accurately scan the multiple products formed, particularly those of low intensity. Therefore, for a more rigorous measure of the inhibitory influence of uracil, incorporation of a single dNTP under single turnover conditions has been used. This approach is widely used with DNA polymerases (35) and has previously been utilized to investigate the interaction of archaeal family-B enzymes with deaminated bases (32). For these experiments, Pol D exo− was used to enable investigation of just the polymerase activity free from any complications arising due to proof-reading exonuclease activity. In contrast to the archaeal Pol B which requires the addition of PCNA to ensure tight binding to primer templates (32), reactions were carried out in the presence of saturated Pol D exo− (140 nM) and primer templates (20 nM), respectively to the affinities for DNA reported in Table 1. The addition of 5–400 μM concentrations of dGTP to Pfu-Pol D and primer template resulted in extension of the primer by a single base, as assessed by gel electrophoresis, and enabled determination of kobs by fitting the amount of extended primer produced over a 30-min time course to a single exponential (Supplementary Figure S6). A secondary plot of kobs against dGTP concentration, fitted using the Michaelis–Menten equation, gave the results shown in Figure 7 and the kinetic constants summarized in Table 2. From these results it is clear that the presence of uracil inhibits polymerization, almost entirely by reducing kpol by a factor of ∼5 with little change in the KD for dGTP. This result is consistent with the slowdowns (between 2 - and 13-fold) measured, less accurately, using primer-template extension.
Figure 7.
Incorporation of a single dGTP into primer templates containing T or U at the +4 template position (sequences given in Table 2). The primer templates (20 nM) were mixed with Pfu-Pol D (140 nM) and the reaction initiated by adding appropriate amounts of dGTP (between 5 and 400 μM). Secondary fits to the Michaelis–Menten equation are given for both the T- and U-containing primer templates (the insert is an expansion of the U data). The kinetic parameters determined from this graph are summarized in Table 2.
Table 2.
Kinetic parameters for incorporation of a single dGTP into a primer templates containing dU or dT at +4, under single turnover conditions
| Base at +4 position of primer templatea | kpolb (min−1) | KDb (μM) | kpol/KDb (s−1 M−1) |
|---|---|---|---|
| dT | 1.6 ± 0.2 | 25 ± 6 | 3.8 ± 1.4 × 106 |
| dU | 0.3 ± 0.06 | 31 ± 9 | 0.6 ± 0.3 × 106 |
aThe primer-template used in this experiment was:
5′ Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA
3′ CCCCTAGGAGATCTCAGCTGGACGTCCCGTTCGTXCG AACAGAGG
where X = dU or dT.
bKinetic parameters are the averages (± standard deviation) from five experiments.
Uracil stimulates 3′–5′ proof-reading exonuclease activity
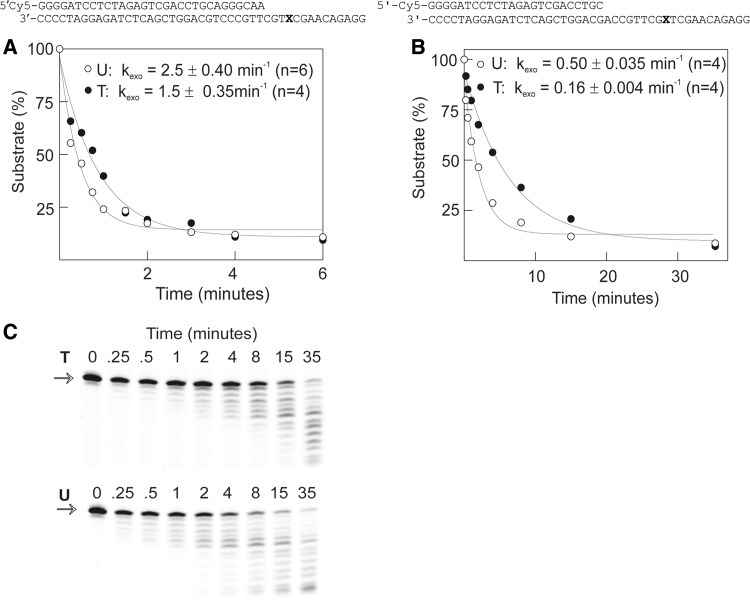
The influence of uracil on the proof-reading exonuclease activity of Pol D was determined by observing the degradation of the primer strand in a uracil-containing primer template. As with dNTP incorporation experiments, proof-reading exonuclease rates were measured with concentrations of primer template (20 nM) and Pol D (140 nM) that deliver single turnover conditions. Figure 8 shows the results obtained with two primer templates which contain either two A:T or two G:C base pairs at the primer-template junction. With the A:T primer template (identical to that used for the dNTP incorporation experiment), the presence of uracil at +4 increased the exonuclease rate slightly, by a factor of ∼1.7 as compared with thymidine-containing controls (Figure 8A). In the case of the G:C primer template, uracil at +9 resulted in an approximate 3-fold stimulation in the rate of 3′–5′ exonucleolysis (Figure 8B and C). Although the enhancements in exonuclease rates, when uracil is present in the template strand, are relatively small, they are consistently observed. The more rapid degradation of the A:T primer template, relative to G:C, is expected and arises from more facile strand separation; a requirement for proof-reading activity.
Figure 8.
Proof-reading exonucleolysis of primer templates containing either T or U (sequences given below) by Pfu-Pol D. The polymerase (140 nM) was added to the primer templates (20 nM) and the degradation of the Cy5-labelled primer measured. The amount of primer remaining over time was fitted to a single exponential to give the rate constants (kexo) for exonucleolysis. (A) and (B) Results found with the two primer templates illustrated at the top of each panels (X = T or U). The calculated kexo values are shown on the graph and represent the means (± standard deviation) for the number of experiments (n) given. (C) Gels used to generate the data shown in (B). The full-length starting primers are marked with an arrow.
DISCUSSION
The data presented in this publication provide compelling evidence that the occurrence of uracil in DNA template strands diminishes polymerization by euryarchaeal family-D enzymes. Uracil must be positioned in single-stranded DNA or, if in a double-stranded region, near the primer-template junction. When the base is well embedded in duplex DNA little inhibition is observed. It is most likely that Pol D can only interact specifically with uracil in single strands but is able to capture the base in double-stranded helical DNA, following transient melting of terminal regions. Preferential binding of uracil by Pol D is also only observed when the base is in single-stranded DNA, agreeing with the same requirement for inhibition of polymerization. When uracil is present in single-stranded templates, small changes to a number of parameters are observed: binding affinity is increased by a factor of ∼2, polymerization decreases roughly 5-fold (as measured using kpol) and 3′–5′ proof-reading exonuclease rates are about two to three times faster (as measured by kexo). Together these alterations result in markedly less DNA synthesis when uracil is present in template strands, as compared with controls. As DNA polymerization involves addition of multiple dNTPs, a relatively small change in the efficiency of incorporation for each single base (such as the 5-fold reduction in kpol seen in this study) may, cumulatively, lead to profound overall inhibition, especially when the accompanying increase in exonuclease activity is taken into account.
Template strand uracil has previously been observed to strongly suppress the activity of archaeal family-B polymerases, a feature assumed to reduce mutations that arise as a consequence of replicating DNA which has been subject to cytosine or adenine deamination (22,23,25,26) (uracil can also be directly incorporated into DNA from dUTP by a polymerase, but this is not mutagenic). It is most likely that the uracil-dependent inhibition of Pol D serves an identical function in protecting the genome from base deamination. The capacity of the two polymerases to detect and respond to uracil suggests they might both play a role in replication, as previously proposed (17–19). However, biochemical experiments can only tentatively identify a polymerase as replicative and the exact roles of euryarchaeal Pol B and Pol D in copying the genome await genetic investigation. The mechanisms by which the two polymerases interact with uracil appear to be different. Pol B has been well characterized and uses ‘read-ahead recognition’, where a running polymerase scans the template ahead of the replication fork for the presence of uracil, sensing the base using a pocket in the N-terminal domain (22–26). The hall-marks of ‘read-ahead’ recognition are strong and specific binding of uracil and stalling of replication at a tightly defined position, four bases prior to uracil encounter. On the rare occasions that the polymerase manages to pass beyond uracil, inhibition ceases. Pol D shows none of these features and, therefore, must use a novel mechanism to sense uracil. Binding of the deaminated base is far from profound and a unique truncated extension product, suggesting stalling at a defined position, is not seen. Instead most of the bands that represent polymerization products are diminished, particularly those of longer products. Furthermore, Pol D is able to interact with template strand uracil at positions well beyond the replication fork and also when uracil is situated in double-stranded DNA just behind the fork. Should Pol D progress beyond template-strand uracil inhibition transiently persists until further replication positions the base deep within the duplex, when DNA melting to produce single strands becomes unlikely. Inhibition is also observed when uracil is located on a DNA strand not being copied by Pol D, a pattern never observed with Pol B. Such trans-inhibition is relevant to replication as uracil located on the lagging strand is capable of inhibiting Pol D travelling on the leading strand, or vice versa. The DNA repair processes that follow uracil sensing by both Pol B and Pol D await elucidation but must involve the accurate replacement of uracil with cytosine, the parent base from which uracil is derived by deamination. The near complete halting of replication when Pol B encounters uracil represents a very efficient method of buying time to enable downstream repair. The slowing of polymerization, characteristic of Pol D, seems at first sight to be a less effective strategy. However, the ability of the enzyme to recognize uracil well in advance of the replication fork presumably ensures success. The continuing inhibition that follows copying of uracil is counterintuitive as the damage (i.e. incorporation of adenine opposite uracil) has already been done; however, as mentioned previously, such inhibition fades as extension progresses.
The molecular mechanism that gives rise to the uracil-dependent inhibition of replication seen with Pol D is far from clear but the enzyme must have a means of sensing the presence of this pro-mutagenic base. Perhaps a uracil binding site is present either near to or overlapping the polymerase active site. Such a site would be able to interact with uracil near the primer-template junction, e.g. at the −1 and +1 positions, following which polymerase activity becomes attenuated. Interaction with more remotely located uracil, e.g. up to 100 bases ahead of the primer-template junction will rely on looping out of a long stretch of flexible single stranded DNA to bring uracil and its recognition site into proximity. Similarly, uracil present in single-stranded regions of a replication fork may be captured by a polymerase travelling on the opposite strand using looping, accounting for trans-inhibition. Interaction with uracil activates the 3′–5′ proof-reading exonuclease activity and lowering the rate of replication. Simple partial blocking of the polymerase active site with uracil would make interaction with the exonuclease site more probable. Alternatively, as the two sites are well separated, allosteric activation of exonuclease activity on uracil binding is also a possibility. This publication shows that Pol D can interact with single stranded uracil up to 100 bases in front of the replication fork. The lack of inhibition seen with uracil at +134 may arise from the greater conformational space that must be searched to locate the uracil-binding pocket. In general, excessive lengths of single-stranded DNA are not exposed during replication. In Archaea, including P. abyssi, Okazaki fragments are up to 120 nt long (36), implying a similar length of single stranded DNA produced during lagging strand replication. In viruses (37), bacteria (38) and presumably Archaea (3,9), the movements of the replicative helicase and polymerase are tightly coupled, mediated by protein–protein interactions. Again this suggests that relatively short lengths of single-stranded DNA are formed when the leading strand is copied. Therefore, forward scanning by Pol D for uracil at extreme distances is unlikely to be necessary for detection of this base as it moves from double- to single-stranded regions. It should be noted that we have never seen uracil-dependent complete cessation of polymerization, in contrast to an earlier brief publication which reports full inhibition of Pol D by uracil (28). Pol D is a heterodimer (11,12) but it is presently unknown if the uracil sensing apparatus is located in the large (polymerase) or the small (exonuclease) subunit. The heterodimer has been reported to further assemble into a tetramer (14); again it is unresolved whether a single heterodimer interacts with both the primer-template junction and uracil or if two separate heterodimers present in the tetramer are used for individual recognition events. At present no high resolution crystal structure is available for the entire Pol D, although information is available for both the N-terminal regions of the small and large subunits (39,40). Inspection does not reveal any obvious uracil-binding pocket as seen with Pol B (24–26). Pol D contains a large number of cysteines arranged in groups of four and may be an iron-sulphur protein as observed for two archaeal DNA repair enzymes, uracil-DNA glycosylase and XPD (41,42). Very recently it has been determined that the eukaryotic family-B polymerases (Pols α, δ, ε and ζ) contain an Fe-S cluster (43). Isolation of these polymerases, after heterologous expression in E. coli, revealed high lability of the Fe-S cluster, with anaerobic purification required to preserve its integrity. It appears that the role of the Fe-S cluster is to facilitate correct folding and interaction with other subunits necessary for the assembly of the polymerase holoenzyme, rather than direct participation in catalysis. This publication also observes that archaeal Pol D contains a similar cysteine motif and so may also be an Fe-S protein. The samples of Pol D used in this investigation were overexpressed in E. coli and purified under aerobic conditions, giving no indication of the olive green hue that is characteristic of iron-sulphur clusters. It is therefore possible that these putative iron-sulphur clusters may have been lost and that these elements could be involved in the correct folding of the uracil sensing region. Further studies are currently in progress, including preparation of native Pol D under anaerobic conditions, to more thoroughly characterize the novel and enigmatic interaction of this protein with deaminated bases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
French National Research Agency [ANR-10-JCJC-1501-01 to G.H.]; T.T.R. and L.G. are UK BBSRC supported PhD students. Funding for open access charge: French National Research Agency (ANR).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The excellent technical assistance of Pauline Heslop throughout this project is greatly appreciated.
REFERENCES
- 1.Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 2.Pomerantz RT, O'Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Barry ER, Bell SD. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelman Z, O'Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J. Biol. Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 6.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelman Z, White MF. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cann IK, Komori K, Toh H, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cann IK, Ishino Y. Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uemori T, Sato Y, Kato I, Doi H, Ishino Y. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells. 1997;2:499–512. doi: 10.1046/j.1365-2443.1997.1380336.x. [DOI] [PubMed] [Google Scholar]
- 14.Shen Y, Musti K, Hiramoto M, Kikuchi H, Kawarabayashi Y, Matsui I. Invariant Asp-1122 and Asp-1124 are essential residues for polymerization catalysis of family D DNA polymerase from Pyrococcus horikoshii. J. Biol. Chem. 2001;276:27376–27383. doi: 10.1074/jbc.M011762200. [DOI] [PubMed] [Google Scholar]
- 15.Gueguen Y, Rolland JL, Lecompte O, Azam P, Le Romancer G, Flament D, Raffin JP, Dietrich J. Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 2001;268:5961–5969. doi: 10.1046/j.0014-2956.2001.02550.x. [DOI] [PubMed] [Google Scholar]
- 16.Cann IK, Ishino S, Hayashi I, Komori K, Toh H, Morikawa K, Ishino Y. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 1999;181:6591–6599. doi: 10.1128/jb.181.21.6591-6599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henneke G, Flament D, Hubscher U, Querellou J, Raffin JP. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005;350:53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 18.Castrec B, Rouillon C, Henneke G, Flament D, Querellou J, Raffin JP. Binding to PCNA in Euryarchaeal DNA Replication requires two PIP motifs for DNA polymerase D and one PIP motif for DNA polymerase B. J. Mol. Biol. 2009;394:209–218. doi: 10.1016/j.jmb.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 19.Rouillon C, Henneke G, Flament D, Querellou J, Raffin JP. DNA polymerase switching on homotrimeric PCNA at the replication fork of the Euryarchaea Pyrococcus abyssi. J. Mol. Biol. 2007;369:343–355. doi: 10.1016/j.jmb.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 20.Berquist BR, DasSarma P, DasSarma S. Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1. BMC Genet. 2007;8:31. doi: 10.1186/1471-2156-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henneke G. In vitro reconstitution of RNA primer removal in Archaea reveals the existence of two pathways. Biochem. J. 2012;447:271–280. doi: 10.1042/BJ20120959. [DOI] [PubMed] [Google Scholar]
- 22.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 24.Gouge J, Ralec C, Henneke G, Delarue M. Molecular recognition of canonical and deaminated bases by P. abyssi family B DNA polymerase. J. Mol. Biol. 2012;423:315–336. doi: 10.1016/j.jmb.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 25.Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA. Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J. Mol. Biol. 2008;381:529–539. doi: 10.1016/j.jmb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry. 2010;49:5772–5781. doi: 10.1021/bi100421r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wardle J, Burgers PM, Cann IK, Darley K, Heslop P, Johansson E, Lin LJ, McGlynn P, Sanvoisin J, Stith CM, et al. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res. 2008;36:705–711. doi: 10.1093/nar/gkm1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawai H, Nagashima J, Kuwahara M, Kitagata R, Tamura T, Matsui I. Differences in substrate specificity of C(5)-substituted or C(5)-unsubstituted pyrimidine nucleotides by DNA polymerases from thermophilic bacteria, archaea, and phages. Chem. Biodivers. 2007;4:1979–1995. doi: 10.1002/cbdv.200790165. [DOI] [PubMed] [Google Scholar]
- 29.Palud A, Villani G, L'Haridon S, Querellou J, Raffin JP, Henneke G. Intrinsic properties of the two replicative DNA polymerases of Pyrococcus abyssi in replicating abasic sites: possible role in DNA damage tolerance? Mol. Microbiol. 2008;70:746–761. doi: 10.1111/j.1365-2958.2008.06446.x. [DOI] [PubMed] [Google Scholar]
- 30.Tori K, Kimizu M, Ishino S, Ishino Y. DNA polymerases BI and D from the hyperthermophilic archaeon Pyrococcus furiosus both bind to proliferating cell nuclear antigen with their C-terminal PIP-box motifs. J. Bacteriol. 2007;189:5652–5657. doi: 10.1128/JB.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell HJ, Richardson TT, Emptage K, Connolly BA. The 3'-5' proofreading exonuclease of archaeal family-B DNA polymerase hinders the copying of template strand deaminated bases. Nucleic Acids Res. 2009;37:7603–7611. doi: 10.1093/nar/gkp800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emptage K, O'Neill R, Solovyova A, Connolly BA. Interplay between DNA polymerase and proliferating cell nuclear antigen switches off base excision repair of uracil and hypoxanthine during replication in archaea. J. Mol. Biol. 2008;383:762–771. doi: 10.1016/j.jmb.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Reid SL, Parry D, Liu HH, Connolly BA. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry. 2001;40:2484–2494. doi: 10.1021/bi001956p. [DOI] [PubMed] [Google Scholar]
- 34.Castrec B, Laurent S, Henneke G, Flament D, Raffin JP. The glycine-rich motif of Pyrococcus abyssi DNA polymerase D is critical for protein stability. J. Mol. Biol. 2010;396:840–848. doi: 10.1016/j.jmb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Joyce CM. Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsunaga F, Norais C, Forterre P, Myllykallio H. Identification of short ‘eukaryotic’ Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delagoutte E, von Hippel PH. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001;40:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 39.Yamasaki K, Urushibata Y, Yamasaki T, Arisaka F, Matsui I. Solution structure of the N-terminal domain of the archaeal D-family DNA polymerase small subunit reveals evolutionary relationship to eukaryotic B-family polymerases. FEBS Lett. 2010;584:3370–3375. doi: 10.1016/j.febslet.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 40.Matsui I, Urushibata Y, Shen Y, Matsui E, Yokoyama H. Novel structure of an N-terminal domain that is crucial for the dimeric assembly and DNA-binding of an archaeal DNA polymerase D large subunit from Pyrococcus horikoshii. FEBS Lett. 2011;585:452–458. doi: 10.1016/j.febslet.2010.12.040. [DOI] [PubMed] [Google Scholar]
- 41.Hinks JA, Evans MC, De Miguel Y, Sartori AA, Jiricny J, Pearl LH. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 2002;277:16936–16940. doi: 10.1074/jbc.M200668200. [DOI] [PubMed] [Google Scholar]
- 42.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 2011;8:125–132. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.