Abstract
The potent anticancer drug actinomycin D (ActD) functions by intercalating into DNA at GpC sites, thereby interrupting essential biological processes including replication and transcription. Certain neurological diseases are correlated with the expansion of (CGG)n trinucleotide sequences, which contain many contiguous GpC sites separated by a single G:G mispair. To characterize the binding of ActD to CGG triplet repeat sequences, the structural basis for the strong binding of ActD to neighbouring GpC sites flanking a G:G mismatch has been determined based on the crystal structure of ActD bound to ATGCGGCAT, which contains a CGG triplet sequence. The binding of ActD molecules to GCGGC causes many unexpected conformational changes including nucleotide flipping out, a sharp bend and a left-handed twist in the DNA helix via a two site-binding model. Heat denaturation, circular dichroism and surface plasmon resonance analyses showed that adjacent GpC sequences flanking a G:G mismatch are preferred ActD-binding sites. In addition, ActD was shown to bind the hairpin conformation of (CGG)16 in a pairwise combination and with greater stability than that of other DNA intercalators. Our results provide evidence of a possible biological consequence of ActD binding to CGG triplet repeat sequences.
INTRODUCTION
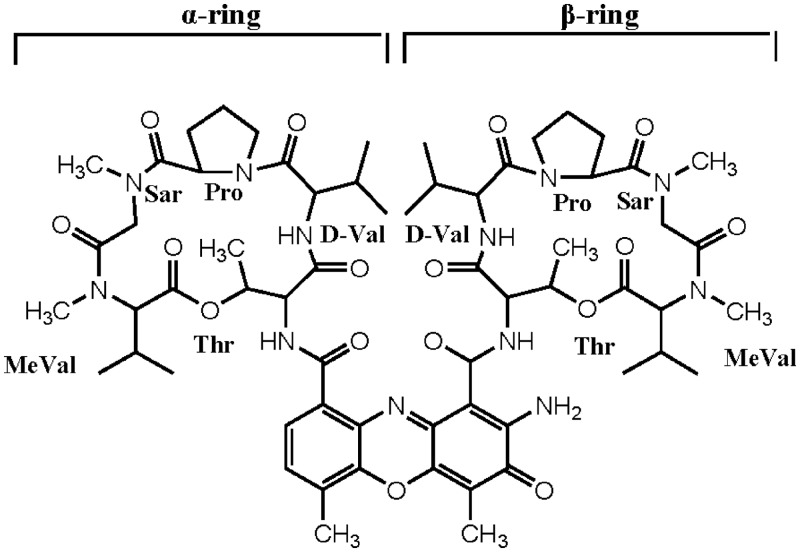
Actinomycin D (ActD) (Figure 1) is a potent anticancer drug. ActD binds to DNA by intercalating its phenoxazone ring at a GpC step such that the two cyclic pentapeptides of the drug are located in the DNA minor groove (1,2). The biological activity of ActD may be related to its binding to DNA, which interferes with replication and transcription (3). The GpC sequence specificity of ActD, which has been analysed extensively using a variety of biochemical and biophysical methods (2,4), is attributed to the strong hydrogen bonds between the NH/C=O groups of threonines in ActD and the corresponding N3/N2 sites of adjacent guanine bases of the GpC step (5). The binding affinity of ActD to GpC sites is also influenced by flanking sequences due to their interactions with the cyclic pentapeptide lactone rings of ActD (5–8).
Figure 1.
Chemical structure of ActD. The cyclic pentapeptides attached to the quinonoid and benzenoid rings of the phenoxazone group are labelled α and β, respectively.
CGG triplet repeat expansions (TREs) within genes are associated with various neurological diseases (9,10). The correlations of these unusual repetitive sequences with the aetiology of these diseases and the mechanism by which those repeats are expanded during replication have been extensively studied. The massive CGG expansion found in neurodegenerative disorders may be caused by the slipped register of the DNA complementary strands along with the transient formation of hairpins during DNA replication (11,12). During this process, DNA polymerase pauses with relocation of the primer, which results in the elongation of integral numbers of triplets to produce TREs. The transient intrastrand hairpin structure containing the mismatched base pairs has been proposed to promote DNA slippage and is a causative factor for DNA expansion (13–15). For example, the CGG TREs in the coding sequence of the FMR1 (Fragile mental retardation 1) gene result in the production of an aberrant protein that has a critical role in the pathogenesis of these diseases (16).
The binding of ActD to non-canonical sequences has been previously studied. It has been reported that two equivalents of ActD bind to the partially overlapping G–C sites on the G–C–G–C segment, and a bend is induced by crowding of the neighbouring pentapeptide (17). In addition, the molecular basis for the tight binding of ActD to a -TGCT- sequence has been reported based on the structure of the ActD–GATGCTTC complex in which the N-methyl group of N-methylvaline (MeVal) was shown to fit snugly in a cavity, which is created by the T:T mismatched base pair, during the TpG step (8). ActD was also able to bind to certain single-stranded sequences with a high affinity (18). Interestingly, when (CGG)n triplet sequences adopt a hairpin arm (as part of a cruciform) or duplexes form between antiparallel CGGs, they contain many GpC ActD-binding sites that alternate with G:G mismatches. Here, we solved the crystal structure of the ActD–ATGCGGCAT complex and analysed the properties of ActD binding to understand the structural basis of the tight binding of ActD. This complex was studied because the -GCGGC- region, which is a part of (CGG)n triplet repeat sequences, creates two GpC sites separated by a G:G mismatch. We characterized the stabilizing and structural effects of ActD on CGG triplet sequences using biophysical analysis. These results provided valuable information regarding the action of ActD on the CGG triplet sequence associated with neurological disease.
MATERIALS AND METHODS
Drug and oligonucleotides
The synthetic DNA oligonucleotides were purified by gel electrophoresis. ActD was purchased from Sigma Chemical Co. (St. Louis, MO, USA). The concentrations of the ActD solutions were determined based on the optical density (ε224 nm = 35 280 M−1 cm−1). The oligonucleotide ATGCGGCA[br5U] was used for the MAD experiments. The oligomer extinction coefficients were calculated according to tabulated values of the monomer and dimer extinction coefficients, with reasonable assumptions. The standard buffer used in the ultraviolet (UV) and circular dichroism (CD) studies contains 50 mM NaCl buffered with 20 mM Tris–HCl buffer at pH 7.3.
Melting temperature measurements
The Tm values of the DNA duplexes were analysed as previously described using a JASCO UV/VIS spectrophotometer by monitoring the sample absorption (O. D.) at 260 nm (19,20). The experiments were performed by increasing the temperature from 0°C to 100°C at a rate of 0.5°C/min. The temperature was recorded every 30 s. The Tm values were determined based on the polynomial fitting of the observed curves and were considered to be the temperatures corresponding to the half-dissociation of the DNA duplexes. The first derivative of the absorption with respect to the temperature (dA/dT) of the melting curve was computed and used to determine the Tm.
CD spectroscopy
CD spectra were collected between 325 and 200 nm at 1 nm intervals using a JASCO-815 spectropolarimeter. The temperature was controlled using a circulating water bath. All of the spectra were calculated as the average of three runs. The methods used for the CD spectral analyses have been previously described (21,22).
Surface plasmon resonance–binding analysis
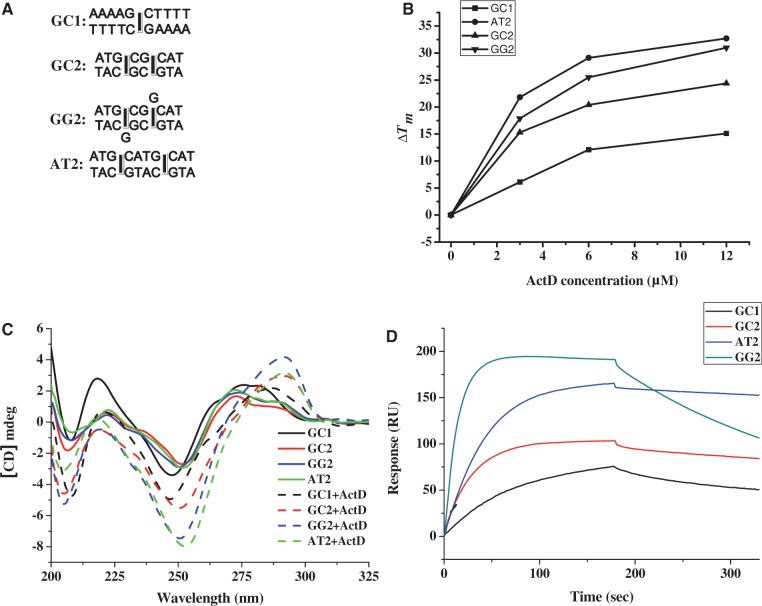
The affinity between the drug and the DNA duplexes was measured using a BIAcore 3000 A surface plasmon resonance (SPR) instrument (Pharmacia, Uppsala, Sweden) with a SensorChip SA5 (Pharmacia) by monitoring the change in the refractive index of the sensor chip surface. In general, these changes are assumed to be proportional to the mass of the molecules bound to the chip and are recorded in resonance units (RU) (23). The 5′-biotin-labelled hairpin DNA duplexes used in the SPR experiments were purified by polyacrylamide gel electrophoresis. To control the amount of DNA that bound to the streptavidin SA chip surface, the biotinylated oligomer was manually immobilized on the chip surface. ActD was prepared in a solution of 20 mM Tris–HCl buffer (pH 7.3) and 50 mM NaCl. Different concentrations of the drug were passed over the chip surface for 180 s at a flow rate of 70 µl min−1 to reach equilibrium. One of the flow cells was used as a blank control. Then, the blank buffer solution was passed over the chip to initiate the dissociation reaction, and this flow was continued for an additional 300 s to complete the reaction. Next, the surface was recovered by washing with 10 μl of 10 mM HCl. Sensorgrams for the interactions between the hairpin DNA duplexes and the drugs were analysed using the BIA evaluation software, version 3. Because 5′-AAAGCTTTTGTAAAGCTTT-3′ (GC1) provides one site for ActD binding, the SPR-binding constants of ActD bound to GC1 were calculated using a 1:1 Langmuir binding model as described below:
In addition, 5′-ATGCGCATTGTATGCGCAT-3′ (GC2), 5′-ATGCGGCATTGTATGCGGCAT-3′ (GG2) and 5′-ATGCATGCATTGTATGCATGCAT-3′ (AT2) provided two sites for ActD binding. The SPR-binding constants of ActD bound to GC2, GG2 and AT2 were calculated using bivalent ligand model, which describes the two sequential binding events for ActD binding to DNA duplex as described below:
This model usually takes cooperative effects into account. The fit was considered acceptable when the chi-square values were <3.
Crystallography
Crystals were obtained from solutions containing 1.0 mM single-stranded DNA, 1.5 mM ActD, 40 mM sodium-cacodylate buffer (pH 6.0), 3 mM MgCl2, 5 mM CaCl2, 10 mM spermine and 8% 2-Methyl-2,4-pentanediol (MPD) solution. The solutions were previously equilibrated at 4°C with 500 μl of 50% MPD using the vapour diffusion method (24). The crystallographic statistics are listed in Supplementary Table S1. The DNA in the complex is numbered A1 to T9 in one strand and A10 to T18 in the complementary strand. The two ActD molecules are numbered ActD1 and ActD2. Diffraction data for the ActD–ATGCGGCA[br5U] complex crystal were measured at 100 K at the National Synchrotron Radiation Research Center 13B1 beamline using an ADSC Q315r detector. The crystallographic data integration and reduction was performed using the software package HKL2000 (25). The MAD data were collected at four wavelengths using bromine as the anomalous scattering atoms. The phases were calculated as implemented in the crystallographic suite Crystallography & NMR System (CNS) (26). The figure-of-merit values were 0.50 and 0.74 before and after density modification, respectively. The resulting MAD electron density map at 2.8 Å resolution was used to build the initial models using the program Mifit (http://code.google.com/p/mifit/). The structure was refined using the simulated annealing procedure incorporated in CNS (26). The DNA force field parameters reported by Parkinson et al. (27) were used. The force field for ActD was generated using the atomic coordinates of its high-resolution crystal structure. The torsion angles were calculated using the CURVES v5.3 program (28).
RESULTS
Overall crystal structure of the ActD–DNA complex
To understand the structural basis of (CGG)n triplet repeat binding by ActD, we solved the crystal structure of ActD bound to ATGCGGCAT at 2.8 Å resolution using the multiple wavelength anomalous diffraction method with 5-bromo-U-DNA. The DNA is embedded with the (CGG)n triplet sequence signature. The final refined electron density map at 2.6 Å resolution was of good quality. Supplementary Figure S1 shows an electron density map indicating the phenoxazone plus the cyclic pentapeptide ring in the minor groove of DNA.
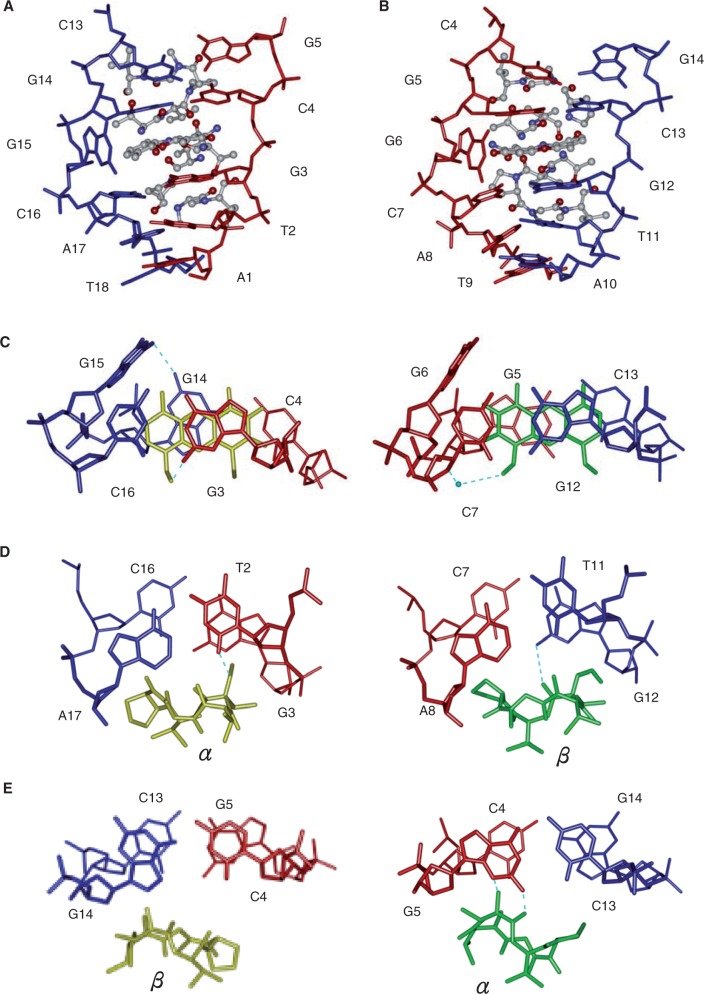
The asymmetric unit of the unit cell contains only one duplex complex consisting of the nonamer palindromic DNA sequence, d(ATGCGGCAT)2, bound by two ActD drug molecules (labelled ActD1 and ActD2) (Figure 2A and B). A symmetrical duplex containing a 2-fold axis running though the centre of the duplex was observed in our structural analysis. The structure revealed many unexpected features. In the complex, two ActD molecules were observed to bind at adjacent overlapping GpC sites, and two guanines are looped out and perpendicular to the long axis of the flanking G:C base pairs (Figure 2A). The general mode of ActD binding to each of the GpC sites resembles that of previously reported crystal structures of ActD–DNA complexes (2). Previous crystallographic analyses showed that CGG/CGG contain C:G and G:C Watson–Crick base pairs with a central non-canonical G:G pair in which one guanosine is always in the syn conformation, whereas the other is in the anti conformation (29). In this study, on ActD binding, we found that the 3′ guanosine in the central CGG triplet is flipped out of the duplex DNA and the 5′ cytosine that was originally paired with it forms a base pair with the guanosine from the destructed G:G mispair in d(ATGCGGCAT). The phenoxazone rings of ActD1 (yellow) and ActD2 (green) intercalates into the G3-C16/C4-G14 and G5-C13/C7-G12 base pairs, respectively, from the minor groove side. The G6 and G15 positions, which are no longer base paired within the duplex, are flipped out and are perpendicular to the long axis of the flanking Watson–Crick base pairs (Figure 2B).
Figure 2.
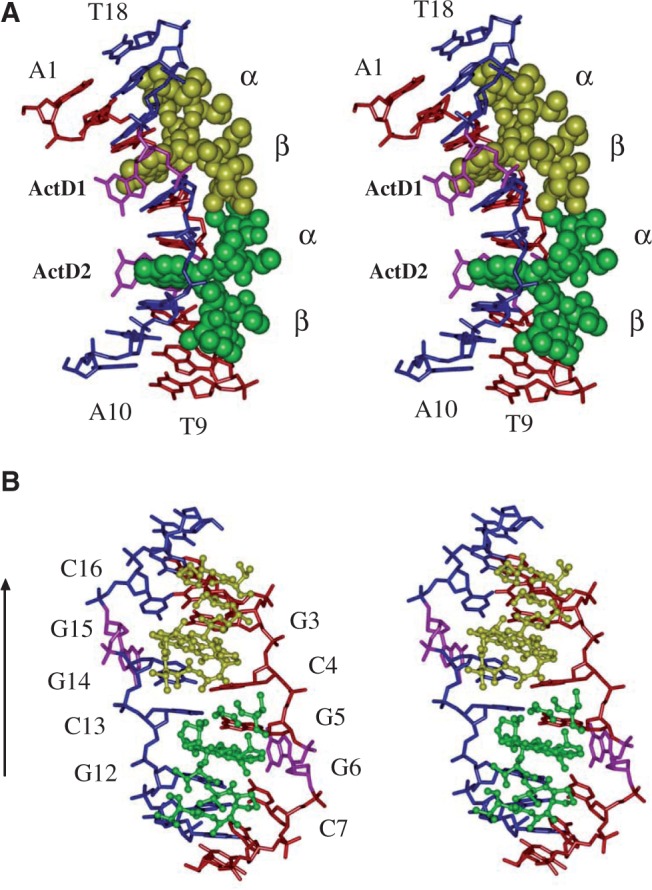
(A) Stereoscopic drawings of the crystal structure of the 2:1 ActD–(ATGCGGCAT)2 complex (ActD is shown as a van der Waals drawing, and DNA is shown using a skeletal line drawing). Two ActD molecules, ActD1 (yellow) and ActD2 (green), which are related by a 2-fold symmetry operation, bind to DNA by intercalating its phenoxazone ring at the two GpC steps with cyclic pentapeptide moieties located in the minor groove, and two guanines (G6 and G15) are looped out and perpendicular to the long axis of the flanking G:C base pairs. (B) Stereoscopic drawings of the ActD–DNA complex (ActD molecules are shown as balls and sticks, and DNA in shown as a skeletal line drawing) viewed from the minor groove direction.
The cyclic pentapeptide lactone rings of individual ActD molecules project in the same direction as the intercalated phenoxazone ring in the ActD–d(ATGCGGCAT) complex (Supplementary Figure S2). The proximity of the two ActD molecules in the minor groove results in contacts between the β and α peptide rings. The bulky MeVal side chain from the α ring of ActD2 is in van der Waals contact with the cyclic side chain of the Pro of the β ring of ActD1. Similarly, crowding occurs between MeVal of the β ring of ActD1 and the cyclic side chain of the Pro of the α ring of ActD2 and between the N-methyl group of the Sar of the α ring of ActD2 and the N-methyl group of the Sar of the β ring of ActD1. Such extensive hydrophobic interactions provide stabilization forces between the two ActD molecules. The contacts between the β and α peptide rings result in a sharp bend of the nonamer DNA duplex in which the two halves are tilting towards the direction of the major groove (Figure 2A). The helix axis of the top half (with an A1-T18 terminal base pair) is bent with an angle of ∼87.8° away from that of the bottom half (with an A10-T9 terminal base pair) according to the CURVE results. The bending angle is far greater than those observed in the current structure of the DNA–drug complex. To relieve the steric conflict between the inwardly pointing bulky peptide rings associated with the two ActD molecules bound at partially overlapping sites on the duplex, the two bulky peptide rings expand the minor groove width at the central (C4-G14)·(G5-C13) step in the ActD–d(ATGCGGCAT) complex and force the two backbones to unwind to generate additional space (Figure 2B).
Structure of the binding site
Figure 3 shows a close-up view of the ActD–DNA complex at the ActD-intercalation site. The two halves of the complex show global similarities as well as local differences with an r.m.s.d. between the two halves (DNA + ActD) of 1.33 Å (Figure 3A and B). In the complex, ActD1 and ActD2 maintain a pseudo 2-fold symmetry. However, there is a rotation (∼180°) between ActD1 and ActD2. The phenoxazone ring of ActD1 (and ActD2) is intercalated at the G3-C16/C4-G14 base pairs (and G5-C13/C7-G12 base pairs) from the minor groove side by pushing out G6 (and G15) (Figure 3A and B). The strong G–C preference of ActD binding can even extrude nucleotides located between a 5′-G and a 3′-C (5,30). There are extensive stacking interactions between the phenoxazone ring and the guanine bases from both sides of the ring, which were also observed in the X-ray (2,31) and nuclear magnetic resonance (NMR) structures (8,32). The flipped-out G6 and G15 sites, which are not disordered, form a perpendicular π-ring/C-H interaction with the chromophore groups 4-CH3 and 6-CH3 in ActD2 and ActD1, respectively (Figure 3C). One hydrogen bond was detected for G14 and G15. In addition, the flipped-out G6 and G15 also form hydrogen bonds and mutual stacking interaction with the flipped out G6 and G15 of other symmetry-equivalent duplexes (Supplementary Figure S3). In the ActD1–DNA complex, G3 and C16 maintain a standard Watson–Crick base pair (Figure 3C). However, G14 has adopted a syn conformation that may maximize the stacking interaction between the purine and the phenoxazone ring leading to the formation of Hoogesteen-type base pairing with C4. Strong intermolecular hydrogen bonds form between G3-N3 and the NH of Thr(α) of ActD1 and G3-N2 and the O′ of the phenoxazone ring of ActD1 (Supplementary Figure S4A and B). In the ActD2–DNA complex, G12 and C7 also maintain standard Watson–Crick base pairing. The G5-C13 base pair has Watson–Crick geometry with two full hydrogen bonds (Figure 3C). Strong intermolecular hydrogen bonds are formed between G12-N2 and the CO of ActD2 Thr(β) and G5-N2/N3 and the CO/NH of ActD2 Thr(α) (Supplementary Figure S4A and C). In addition to these hydrogen bonds, one bridging water molecule also plays an important role in mediating the interactions between ActD2 and the DNA duplex (Supplementary Figure S4B).
Figure 3.
Enlarged side view of the ActD–ATGCG region (ActD is shown as a ball-and-stick model, and DNA is shown as a skeletal model). The two phenoxazone rings of ActD1 (left) and ActD2 (right) are individually intercalated into the G3pC4 step in A and the G12pC13 step in B, respectively C–E. The detailed conformation showing the stacking interactions in the ActD–DNA complex at various base pair steps of the refined structure. Hydrogen bonds are shown as blue dotted lines. Water is shown as a blue sphere.
The outward outer edges of the peptide rings (i.e. the Sar-MeVal dipeptide part) from ActD reach beyond the flanking A:T base pairs in the minor groove. The N-methyl group of the MeVal from the ActD2 β and ActD1 α rings are wedged between the T11pG12 and T2pG3 steps, respectively (Figure 3D). In addition, the methyl group of the MeVal from the ActD1 β and ActD2 α ring from the inwardly outer edges of the peptide rings is wedged between the C13pG14 and C4pG5 steps, respectively, which causes unwinding in this step (Figure 3E). ActD2 has a similar overall conformation as that of free ActD (r.m.s.d. = 0.81) (Supplementary Figure S5) even though ActD1 has a different conformation compared with free ActD (r.m.s.d. = 1.52), especially at the proline residue (33). The proline side in the β and α peptide rings of ActD2 pushes the cytosine of the C7:G12 and G5:C13 base pairs towards the major groove side, which reduces the base stacking on the DNA strand closest to the proline (Figure 3D and E). However, the proline side of the peptide ring of ActD1 moves away from the DNA surface.
DNA backbone torsion angles and helical parameters of the DNA duplex
The entire DNA structure is distorted by the binding of two ActD molecules. The binding of ActD to the central -GCGGC- causes considerable changes in the DNA backbone torsion angles and helical parameters relative to those of B-DNA (Supplementary Tables S2 and S3). The glycosidic torsion angles, χ, of most of the residues fall into an anti conformation (160° to ∼300°) with the exception of G14, which adopts a syn conformation (χ = 108°). The majority of sugar puckers preserve C2′-endo or closely related C3′-exo structures, suggesting alternating C2′-endo/C3′-endo conformations, which is a configuration frequently found in intercalator-dinucleotide monophosphate complexes, are not required for ActD intercalation (34). In addition, flipped-out G6 and G15 are most evident at torsion angles (ξ) of 102.4° and 78.6°, which differ by ∼289° and 265°, respectively, from those observed in A-DNA and B-DNA. C4 and G14, which adopt syn (G14) and anti (C4) conformations, form Hoogesteen base pairs that exhibit a higher propeller twist angle and stretch distance than other G:C base pairs.
An increase of 6.7 and 8.2 Å for the two GpC steps at the intercalation sites in the ActD–DNA complex was observed and is similar to changes observed in the structures of DNA oligomers on DNA intercalator binding (34–36). The DNA helical twist at the ActD1 intercalation site at the GpC step is 62.5° indicates that this step is overwound (Supplementary Figure S6A). However, the helix unwinds by 30° at the ActD2 intercalation site at the G–C step (twist angle 5.8°). Interestingly, the duplex is unwound at the central CpG step to generate a left-handed helix with a twist angle of −43.45°, which is characteristic of the Z-DNA structure. We propose that this action may relieve the steric conflict between the inwardly pointing bulky peptide rings associated with the two ActD molecules and absorbs the torsional stress caused by over-winding at the GpC step on ActD1 binding. In addition, there is a significant increase in the width of the minor groove in the DNA–ActD complex in the 17–18 Å range relative to the minor groove widths of 6 and 11 Å in B-DNA and A-DNA, respectively. Large positive roll and tilt angles were also detected in the three central steps of d(ATGCGGCAT) on ActD binding, which results in a sharp bending of the DNA helix towards the major groove (Supplementary Figure S6B and C). Another distortion induced by the ActD is a slide displacement in the five central steps of d(ATGCGGCAT). These DNA conformational changes result in an opening and widening of the minor groove to accommodate two intercalated ActD molecules bound at partially overlapping sites on the DNA.
Crystal packing of ActD–DNA complexes
There are two different major packing interactions that occur between the two asymmetric units in the crystal lattice (P6522) (Supplementary Figure S7A), i.e. end-to-end and end-to-side, that are mediated by π–π stacking and van der Waals contact, respectively (Supplementary Figure S7B). At one interface of the end-to-end packing, the end of one DNA duplex molecule is packed against the end of another molecule with a base-stacking distance of ∼3.4 Å between the contacting terminal residues of the ActD–DNA complex (A1-T18) of the independent asymmetric units. The other interface in the end-to-side packing is stabilized by the formation of intramolecular van der Waals contact between the terminal residues of the ActD–DNA complex (A8-T9) of the independent asymmetric units.
UV melting, CD and SPR analysis of ActD bound to DNA duplexes
We characterized the stabilizing and structural effects of ActD on d(ATGCGGCAT) (GG2) by UV melting, CD and SPR analysis. For comparison, the DNA duplexes, GC2 and AT2, which contain two related GpC sites that are either adjacent or non-overlapping, respectively, and GC1, which contains one central GC step site, were used as the reference sequences in this study. These duplexes were expected to be intercalated at the central GpC location by an ActD molecule (Figure 4A). The melting temperatures of the DNA duplexes with or without ActD were determined by recording their A260 values at different temperatures (Table 1). In the absence of ActD, GG2 with a central G:G mispair showed a lower Tm value than GC2 and AT2. In a concentration-dependent manner, ActD can stimulate the stability of DNA duplexes. However, the ΔTm values of GG2 and AT2 are more than those of GC1 and GC1 after the addition of ActD at various drug/DNA ratios (Figure 4B). These data suggest that ActD can show a high preference for stabilizing two GC sites flanking a non-canonical G:G pair or two standard AT base pairs. The Tm values of the GC2 and GG2 duplexes with ActD are lower than that of AT2 due to crowding between two neighbouring cyclic peptides of ActD at two overlapping GpC sites, which results in a severe bending of the helix.
Figure 4.
(A) The DNA duplexes including GC1, GC2, AT2 and GG2 were used in this study (squares represents the binding sites for the phenoxazone ring of ActD). (B) The effect of ActD at various concentrations on the ΔTm values of GC1, GC2, GG2 and AT2 (3 μM) in standard buffer. (C) The CD spectra of GC1, GC2, AT2 and GG2 (4 μM) in standard buffer alone and with 10 μM ActD are shown. The CD spectra of ActD–DNA complexes were obtained by subtracting out the spectra of ActD. (D) A sensorgram of the ActD–DNA interaction between immobilized hairpin DNA and the target ActD obtained by subtracting the reference control is shown. The concentration of the drug was 2 µM in 50 mM NaCl buffered with 20 mM Tris–HCl buffer at pH 7.3.
Table 1.
The effects of ActD at various concentrations on the melting temperature of various DNA duplexes at 3 μM
| DNA forms | 0 μM ActD | 3 μM ActD | 6 μM ActD | 12 μM ActD |
|---|---|---|---|---|
| GC1 | 28.1 | 34.2 | 40.2 | 43.3 |
| GC2 | 36.9 | 52.5 | 57.6 | 61.3 |
| GG2 | 28.8 | 46.3 | 54.6 | 60.1 |
| AT2 | 37.4 | 59.2 | 67.6 | 70.2 |
Figure 4C shows the CD spectra of the DNA duplexes including GC1, GC2, GG2 and AT2 with and without ActD. The CD spectra of the DNA duplexes showed the same profiles (i.e. a band of negative and positive peaks at 250 and 275 nm that are typical of B-DNA). After binding to ActD, GC1, GC2, GG2 and AT2 showed an increase in the intensity of their CD spectra at ∼250 nm and a red shift from 275 to 290 nm, which revealed a conformational change in the DNA on ActD binding. It is interesting to note that GG2 exhibited the largest change in its CD intensity at 290 nm in the presence of ActD indicating the largest conformational change of GG2 on ActD binding compared with other DNA duplexes.
To characterize the binding affinity of ActD for GG2, ActD was allowed to interact with biotin-labelled hairpin DNA duplexes at various concentrations, and the results were monitored by SPR (Supplementary Figure S8). The tested DNA formed self-complementary hairpin duplexes using 5′-TGT-3′ trinucleotides as a loop region. In Figure 4D, the association between the hairpin DNA duplexes and ActD is shown by an increase in the RU values, whereas the dissociation of these two species is indicated by a decrease in the same trace. According to the SPR sensorgram, the interactions between GG2 and ActD exhibited the highest DNA-binding capacity (∼202 RU) compared with those between other DNA duplexes and ActD.
Kinetic experiments were performed by measuring the parameters of the binding between ActD and its target DNA duplexes (Table 2). The kinetic constants of association (ka in M−1s−1) and dissociation (kd in s−1) for ActD binding were measured based on the calculations from the association and dissociation phases of the SPR traces, respectively. Because GC1 provided one site for ActD binding, only one ka1 and kd1 values that contribute towards the respective association constants (Ka1) were observed in the ActD–GC1 complex. In addition, GC2, GG2 and AT2 provide two sites for ActD binding. The ka1, kd1 and ka2, kd2 values contribute towards the respective association constants (Ka1 and Ka2) for the two binding modes obtained for the ActD–GC2, ActD–GG2 and ActD–AT2 interactions. GG2 was shown to have a higher ka1 and ka2 compared with other duplexes containing regions with two related GpC sites. For the dissociation rate constants, GG2 had a higher dissociation rate constant (kd1 = 5.71 × 10−3 s−1) relative to GC2 and AT2 (kd = 1.15 × 10−3 and 0.46 × 10−3 s−1, respectively). The kd2 values of GC2, GG2 and AT2 were essentially the same. The association constants (Ka) were calculated as ka/kd (M−1) and listed in Table 2. AT2 showed the highest Ka1 value, which is ∼2-fold and 4-fold larger than the Ka1 of GC2 and GG2, respectively, whereas GC1 exhibited the lowest Ka1 among all of the duplexes. The Ka2 (1.55 × 106) of GG2 was slightly higher than the Ka2 values of AT2 and GC2. In addition, GG2 had a higher Ka1 value that was ∼2-fold greater than the Ka2 of GG2, suggesting that the binding of two ActDs to GG2 may exhibit negative cooperative binding.
Table 2.
Binding parameters for ActD and hairpin DNA duplexes determined by SPR
| DNA forms | ka1 (M−1S−1) | ka2 (M−1S−1) | kd1 (S−1) | kd2 (S−1) | Ka1 (M−1) | Ka2 (M−1) |
|---|---|---|---|---|---|---|
| GC1 | 3.51 × 103 | 1.53 × 10−3 | 2.29 × 106 | |||
| GC2 | 6.98 × 103 | 0.94 × 103 | 1.15 × 10−3 | 0.94 × 10−3 | 6.07 × 106 | 1.01 × 106 |
| GG2 | 1.76 × 104 | 1.70 × 103 | 5.71 × 10−3 | 1.14 × 10−3 | 3.08 × 106 | 1.55 × 106 |
| AT2 | 5.86 × 103 | 1.02 × 103 | 0.46 × 10−3 | 0.98 × 10−3 | 12.87 × 106 | 1.04 × 106 |
Binding of ActD to CGG repeats
The oligo (CGG)16 was used to further characterize the action of ActD on the CGG repeats. This sequence forms intrastrand hairpins consisting of mismatched base pairs that promote primer–template slippage during DNA replication. The structure of (CGG)16 offers many GC sites flanking a G:G mispair for ActD binding. To explore the ActD interactions and any subsequent structural perturbations to the (CGG)16, CD spectra were monitored in the presence of various concentrations of ActD. Figure 5A shows the CD spectra (320–200 nm) of (CGG)16 on addition of ActD. The CD spectra of (CGG)16 alone showed a band with a negative peak at 260 nm and two small shoulders appearing at 276 and 240 nm, which is a pattern that is typical of undefined structures. After binding of ActD, (CGG)16 exhibited a concomitant increase in a positive peak at 295 nm and two negative peaks at 205 and 246 nm. These results suggest that long CGG repeats also show substantial DNA conformational change on addition of ActD. The occurrence of one isobestic point at 270 nm, which remains largely unchanged, demonstrates the presence of one-mode conformation alternation of DNA by binding to ActD.
Figure 5.
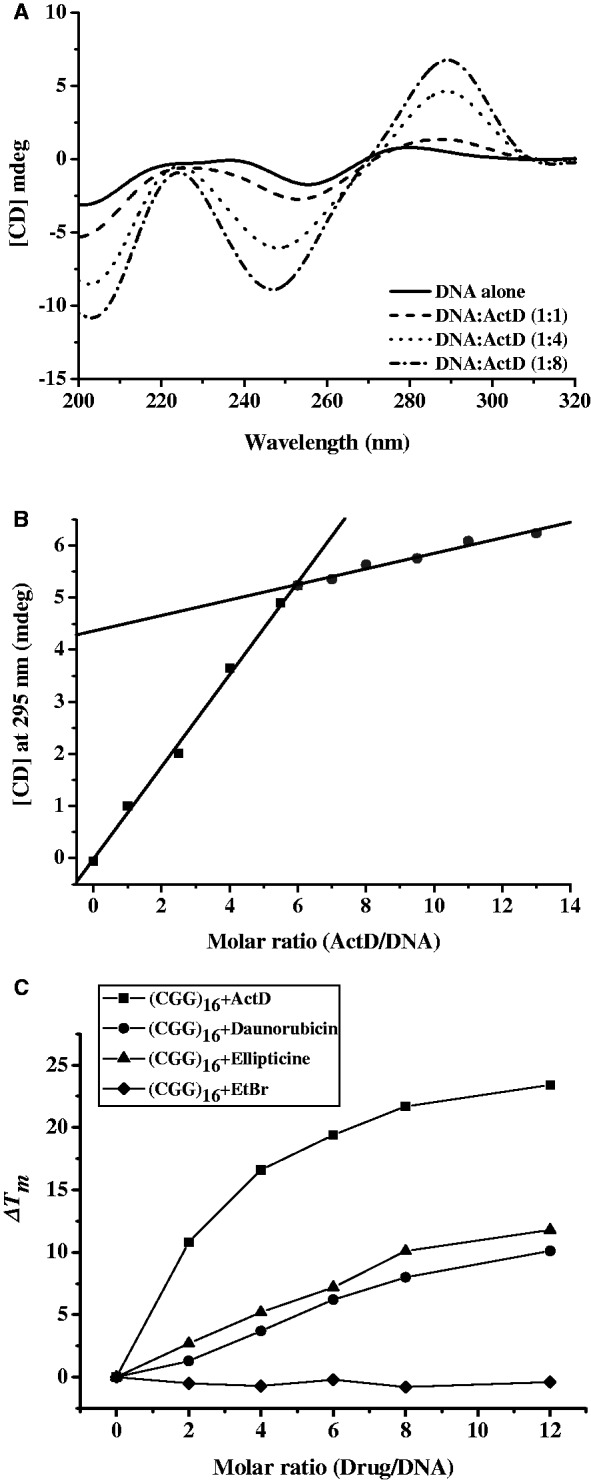
(A) CD spectra of (CGG)16 (1 μM) with ActD at various concentrations. The CD spectra of ActD–DNA complex were obtained by subtracting out the spectra of ActD. (B) CD titration of (CGG)16 against ActD with varied CD intensities at 295 nm as a function of the molar equivalents of ActD–DNA at 25°C. A DNA concentration of 1 μM, which was buffered by 20 mM sodium-cacodylate buffer at pH 7.3, was used. The two solid lines represent the initial binding curve of ActD to DNA and the curve that reaches the plateau. (C) ΔTm values of (CGG)16 (1 μM) in standard buffer induced by various DNA intercalators at different drug/DNA ratio.
To determine the stoichiometry between ActD and (CGG)16, the CD intensity of DNA at 295 nm with increasing concentrations of ActD was performed at 25°C. A plot of the CD intensity of the signal at 275 nm as a function of the concentration of added ActD showed that a plateau was reached when six equivalents of ActD had been added. The points of intersection of two extrapolated lines (initial and final), which represent the saturation levels, indicate a 6:1 stoichiometry between ActD and DNA (Figure 5B).
To determine the stabilizing effects of ActD on the thermodynamics of the (CGG)16 hairpin formation, the melting temperatures of the DNA duplexes in the presence of ActD at various concentrations were determined following their complex formation by recording the A260 values at different temperatures (Table 3). Several DNA intercalators, including ethidium bromide, daunorubicin and ellipticine, were used as reference compounds. Daunorubicin and ellipticine are GC-specific intercalators. Daunorubicin at 2, 4, 6, 8 and 12 μM elevated the Tm value of (CGG)16 by 1.3°C, 3.7°C, 6.2°C, 8°C and 10.1°C, respectively (Figure 5C). The Tm value of (CGG)16 was estimated to increase by 3.7°C, 5.2°C, 7.2°C, 10.1°C and 11.8°C on treatment with 2, 4, 6, 8 and 12 μM ellipticine, respectively. Ethidium bromide at various concentrations was unable to stabilize the hairpin structure of (CGG)16. The addition of ActD to (CGG)16 at 2, 4, 6, 8 and 12 μM resulted in a dramatic increase in the Tm value to 10.8°C, 16.6°C, 19.4°C, 21.7°C and 23.4°C, respectively, demonstrating that ActD exhibits higher DNA-stabilizing effects towards (CGG)16 than do the other compounds.
Table 3.
The effects of drugs at various concentrations on the melting temperature (°C) of (CGG)16 at 1 μM
| Drugs | 0 μM | 2 μM | 4 μM | 6 μM | 8 μM | 12 μM |
|---|---|---|---|---|---|---|
| EtBr | 70.6 | 70.1 | 69.9 | 70.4 | 69.8 | 70.2 |
| Ellipticine | 70.6 | 73.3 | 75.8 | 77.8 | 80.7 | 82.4 |
| Daunorubicin | 70.6 | 71.9 | 74.3 | 76.8 | 78.6 | 80.7 |
| ActD | 70.6 | 81.4 | 87.2 | 90.0 | 92.3 | 94.0 |
DISCUSSION
Small-molecule ligands that bind to CXG repeats could provide potent biological applications. For example, a naphthyridine–azaquinolone conjugate and a naphthyridine carbamate dimer that target (CAG)n and (CGG)n, respectively, and bind to A:A and G:G mismatches, respectively, were developed to probe the length of a repeat (37–39). Previous studies reported that DNA-acting drugs, such as mitomycin C, doxorubicin and chromomycin A3, can prevent the expansion of the CXG triplet repeats (40,41). The (CXG)n triplet repeat sequence has extensive GpC sequence clustering, which provides a potent ActD-binding target. Our current studies demonstrate that ActD can tightly bind to consecutive GpC sites that are separated by a G:G mismatch. Certain significant conformational distortions including a particular nucleotide loop-out, a sharp bend of the helix and an unwinding to generate a left-handed DNA twist were observed. The NMR structure of the ActD bound to a 5′-GGC/CCG-5′ site was also reported to contain looped-out nucleotides in the DNA strand with one of the looped-out bases perpendicular to the ActD chromophore (30). A similar conformational feature (nucleotide loop-out) was observed in the complex of trioxacarcin bound to a DNA duplex (42). In addition, the looped-out bases play an important role in certain DNA–protein interactions (43). For example, the DNA repair enzyme (i.e. DNA glycosylase) flips the damaged base out of the double helix into an active site pocket to excise it.
There are parallels in the observed mode of DNA-binding in the current study of the ActD–d(ATGCGGCAT) complex and an NMR study from Patel's group on the ActD–d(AAGCGCTT) complex (two drugs per duplex) (17). For both complexes, two ActD molecules bind at adjacent overlapping GpC sites. The contacts between the peptide rings of two different ActD molecules result in a bend of the DNA duplex and an unwinding at the central GC step in structures as well as a widening and opening up of the minor groove accompanied by tilting towards the major groove. Of particular interest is the bend angle (∼88°) in the ActD–d(ATGCGGCAT) complex, which is greater than the bend angle of ∼20° derived from the NMR solution structure. The sharp bend associated with minor groove complex formation has been independently observed in the structures of the complexes formed between the tetraazaphenanthrene ligand and chromomycin A3 and their DNA target sites (44,45). Both interactions are accompanied by a widening of the minor groove and unwinding of the helix. In addition, the left-handed helix at the central GpC step of d(ATGCGGCAT) was induced by ActD binding, which has not been previously reported in other DNA–drug complex structures.
Previous studies have shown that the GpC site is the preferred intercalation site for ActD where A or T residues adjacent to the intercalation site are favoured (2). In agreement with the Tm results that showed ActD formed a more stable complex with AT2 than the other duplexes, our SPR results clearly showed that the binding affinity of the first ActD to the DNA duplexes follows the following order: AT2 > GC2 > GG2 > GC1. However, ActD was shown to bind to GG2 quickly. The CGG repeat helices exhibited a smooth bend caused by preferential stacking interactions at the guanine bases. The bend is produced by a roll in the base pairs along their long axes in the direction that compresses the wide major groove of the double helix. Therefore, it is likely that this major-groove compression and the concomitant widening of the minor groove partially contribute to the fast binding of the ActD to the CGG triplet repeats. In addition, the rates of dissociation of the first ActD (ka1) from the DNA duplexes with two related GC sites were significantly modulated by the inserted base pairs flanked by the two GC sites. For example, the first ActD dissociates from GG2 faster compared with AT2 and GC2. According to the crystal structure of the ActD–d(ATGCGGCAT) complex, the severe bending of the helix is most likely responsible for this observation. Our biophysical studies showed that the binding of ActD to d(ATGCGGCAT) occurs via a pairwise binding model. The SPR results indicating that the binding constant for one of the binding sites is 2-fold greater than the value for the other site, which can be explained by the structural differences between the two binding sites as previously described. For example, the GpC step for the ActD2 binding is tightly wound such that the minor groove becomes deeper and the proline residue of ActD2 fits into a hydrophobic pocket in this groove. In contrast, the GpC step for ActD1 binding is unwound such that the minor groove becomes shallow and wide and the proline residue of ActD1 remains distant from the DNA surface without a significant interaction. In addition, there are more hydrogen bonds in the DNA–ActD2 complex than in the DNA–ActD1 complex.
In the (CGG)16 triplet repeats, there is an extensive clustering of GpC sequences separated by a mismatched G:G base pair. This clustering provides an excellent ActD-binding target. Although (CGG)16 contains seven GC sites, ActD binds to (CGG)16 with a 6:1 drug/strand binding stoichiometry supporting the concept that binding to the (CGG)n repeat proceeds in a pairwise combination (Supplementary Figure S9), which has been demonstrated for the ActD–d(ATGCGGCAT) complex. We also provided evidence that ActD can modulate the stability and conformation of an intrastranded hairpin structure in CGG-repeat sequences. Currently, no treatments exist for neurological diseases associated with DNA TRE. The length of the repeat is the most important determinant of these neurological diseases. Chemicals or drugs that reduce the repeat length may be used to delay onset and reduce the severity of diseases linked to DNA TRE. Our results provide a possible biological consequence of ActD binding to CGG triplet repeat sequences associated with neurological disease.
ACCESSION NUMBERS
The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 4HIV).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–9.
FUNDING
Funding for open access charge: NSC [100-2113-M-005-004-MY3 to M.-H.H.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Andrew H.-J. Wang and Lou-Sing Kan (Academia Sinica) for their help in making this research possible. We are grateful for the assistance of the beamline staff at the 13B1 beamline, NSRRC, Hsinchu, Taiwan.
REFERENCES
- 1.Sobell HM, Jain SC. Stereochemistry of actinomycin binding to DNA. II. Detailed molecular model of actinomycin-DNA complex and its implications. J. Mol. Biol. 1972;68:21–34. doi: 10.1016/0022-2836(72)90259-8. [DOI] [PubMed] [Google Scholar]
- 2.Kamitori S, Takusagawa F. Crystal structure of the 2:1 complex between d(GAAGCTTC) and the anticancer drug actinomycin D. J. Mol. Biol. 1992;225:445–456. doi: 10.1016/0022-2836(92)90931-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang SY, Lee YL, Lai YH, Chen JJ, Wu WL, Yuann JM, Su WL, Chuang SM, Hou MH. Spermine attenuates the action of the DNA intercalator, actinomycin D, on DNA binding and the inhibition of transcription and DNA replication. PLoS One. 2012;7:e47101. doi: 10.1371/journal.pone.0047101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen FM. Kinetic and equilibrium binding studies of actinomycin D with some d(TGCA)-containing dodecamers. Biochemistry. 1988;27:1843–1848. doi: 10.1021/bi00406a008. [DOI] [PubMed] [Google Scholar]
- 5.Robinson H, Gao YG, Yang X, Sanishvili R, Joachimiak A, Wang AHJ. Crystallographic analysis of a novel complex of actinomycin D bound to the DNA decamer CGATCGATCG. Biochemistry. 2001;40:5587–5592. doi: 10.1021/bi002859z. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Chen FM. Actinomycin D binds strongly and dissociates slowly at the dGpdC site with flanking T/T mismatches. Biochemistry. 1996;35:16346–16353. doi: 10.1021/bi961060d. [DOI] [PubMed] [Google Scholar]
- 7.Chen FM. Binding of actinomycin D to DNA oligomers of CXG trinucleotide repeats. Biochemistry. 1998;37:3955–3964. doi: 10.1021/bi972110x. [DOI] [PubMed] [Google Scholar]
- 8.Lian C, Robinson H, Wang AHJ. Structure of actinomycin D bound with (GAAGCTTC)2 and (GATGCTTC)2 and its binding to the (CAG)n:(CTG)n triplet sequence as determined by NMR analysis. J. Am. Chem. Soc. 1996;118:8791–8801. [Google Scholar]
- 9.Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 10.Warren ST, Ashley CT., Jr Triplet repeat expansion mutations: the example of fragile X syndrome. Annu. Rev. Neurosci. 1995;18:77–99. doi: 10.1146/annurev.ne.18.030195.000453. [DOI] [PubMed] [Google Scholar]
- 11.Pluciennik A, Iyer RR, Parniewski P, Wells RD. Tandem duplication. A novel type of triplet repeat instability. J. Biol. Chem. 2000;275:28386–28397. doi: 10.1074/jbc.M000154200. [DOI] [PubMed] [Google Scholar]
- 12.Parniewski P, Staczek P. Molecular mechanisms of TRS instability. Adv. Exp. Med. Biol. 2002;516:1–25. doi: 10.1007/978-1-4615-0117-6_1. [DOI] [PubMed] [Google Scholar]
- 13.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 15.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Liu X, Patel DJ. DNA bending and unwinding associated with actinomycin D antibiotics bound to partially overlapping sites on DNA. J. Mol. Biol. 1996;258:457–479. doi: 10.1006/jmbi.1996.0262. [DOI] [PubMed] [Google Scholar]
- 18.Wadkins RM, Vladu B, Tung CS. Actinomycin D binds to metastable hairpins in single-stranded DNA. Biochemistry. 1998;37:11915–11923. doi: 10.1021/bi9809730. [DOI] [PubMed] [Google Scholar]
- 19.Hou MH, Lu WJ, Huang CY, Fan RJ, Yuann JM. Effects of polyamines on the DNA-reactive properties of dimeric mithramycin complexed with cobalt(II): implications for anticancer therapy. Biochemistry. 2009;48:4691–4698. doi: 10.1021/bi900092w. [DOI] [PubMed] [Google Scholar]
- 20.Hsu CW, Chuang SM, Wu WL, Hou MH. The crucial role of divalent metal ions in the DNA-acting efficacy and inhibition of the transcription of dimeric chromomycin A3. PLoS One. 2012;7:e43792. doi: 10.1371/journal.pone.0043792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YM, Chen CK, Hou MH. Conformational changes in DNA upon ligand binding monitored by circular dichroism. Int. J. Mol. Sci. 2012;13:3394–3413. doi: 10.3390/ijms13033394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu WJ, Wang HM, Yuann JM, Huang CY, Hou MH. The impact of spermine competition on the efficacy of DNA-binding Fe(II), Co(II), and Cu(II) complexes of dimeric chromomycin A(3) J. Inorg. Biochem. 2009;103:1626–1633. doi: 10.1016/j.jinorgbio.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Yuann JM, Tseng WH, Lin HY, Hou MH. The effects of loop size on Sac7d-hairpin DNA interactions. Biochim. Biophys. Acta. 2012;1824:1009–1015. doi: 10.1016/j.bbapap.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Wang AHJ, Gao Y-G. Crystallization of oligonucleotides and their complexes with antitumor drugs. Methods. 1990;1:91–99. [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson G, Vojtechovsky J, Clowney L, Brunger AT, Berman HM. New parameters for the refinement of nucleic acid-containing structures. Acta Crystallogr. D Biol. Crystallogr. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 28.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 29.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–7315. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou SH, Chin KH, Chen FM. Looped out and perpendicular: deformation of Watson-Crick base pair associated with actinomycin D binding. Proc. Natl Acad. Sci. USA. 2002;99:6625–6630. doi: 10.1073/pnas.102580399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou MH, Robinson H, Gao YG, Wang AHJ. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002;30:4910–4917. doi: 10.1093/nar/gkf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Chen H, Patel DJ. Solution structure of actinomycin-DNA complexes: drug intercalation at isolated G-C sites. J. Biomol. NMR. 1991;1:323–347. doi: 10.1007/BF02192858. [DOI] [PubMed] [Google Scholar]
- 33.Ginell S, Lessinger L, Berman HM. The crystal and molecular structure of the anticancer drug actinomycin D–some explanations for its unusual properties. Biopolymers. 1988;27:843–864. doi: 10.1002/bip.360270511. [DOI] [PubMed] [Google Scholar]
- 34.Lisgarten JN, Coll M, Portugal J, Wright CW, Aymami J. The antimalarial and cytotoxic drug cryptolepine intercalates into DNA at cytosine-cytosine sites. Nat. Struct. Biol. 2002;9:57–60. doi: 10.1038/nsb729. [DOI] [PubMed] [Google Scholar]
- 35.Niyazi H, Hall JP, O'Sullivan K, Winter G, Sorensen T, Kelly JM, Cardin CJ. Crystal structures of Lambda-[Ru(phen)(2)dppz](2)(+) with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes. Nat. Chem. 2012;4:621–628. doi: 10.1038/nchem.1397. [DOI] [PubMed] [Google Scholar]
- 36.Song H, Kaiser JT, Barton JK. Crystal structure of Delta-[Ru(bpy)(2)dppz](2)(+) bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation. Nat. Chem. 2012;4:615–620. doi: 10.1038/nchem.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatani K, Hagihara S, Goto Y, Kobori A, Hagihara M, Hayashi G, Kyo M, Nomura M, Mishima M, Kojima C. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat. Chem. Biol. 2005;1:39–43. doi: 10.1038/nchembio708. [DOI] [PubMed] [Google Scholar]
- 38.Hagihara M, Nakatani K. Inhibition of DNA replication by a d(CAG) repeat binding ligand. Nucleic Acids Symp. Ser. (Oxf) 2006:147–148. doi: 10.1093/nass/nrl073. [DOI] [PubMed] [Google Scholar]
- 39.Hagihara M, He H, Kimura M, Nakatani K. A small molecule regulates hairpin structures in d(CGG) trinucleotide repeats. Bioorg. Med. Chem. Lett. 2012;22:2000–2003. doi: 10.1016/j.bmcl.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 40.Hashem VI, Pytlos MJ, Klysik EA, Tsuji K, Khajavi M, Ashizawa T, Sinden RR. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 2004;32:6334–6346. doi: 10.1093/nar/gkh976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YW, Hou MH. The binding of the Co(II) complex of dimeric chromomycin A3 to GC sites with flanking G:G mismatches. J. Inorg. Biochem. 2013;121:28–36. doi: 10.1016/j.jinorgbio.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Pfoh R, Laatsch H, Sheldrick GM. Crystal structure of trioxacarcin A covalently bound to DNA. Nucleic Acids Res. 2008;36:3508–3514. doi: 10.1093/nar/gkn245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc. Natl Acad. Sci. USA. 2008;105:8890–8895. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall JP, O'Sullivan K, Naseer A, Smith JA, Kelly JM, Cardin CJ. Structure determination of an intercalating ruthenium dipyridophenazine complex which kinks DNA by semiintercalation of a tetraazaphenanthrene ligand. Proc. Natl Acad. Sci. USA. 2011;108:17610–17614. doi: 10.1073/pnas.1108685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou MH, Robinson H, Gao YG, Wang AHJ. Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion. Nucleic Acids Res. 2004;32:2214–2222. doi: 10.1093/nar/gkh549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.