Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression through RNA interference. Human miRNAs are generated through a series of enzymatic processing steps. The precursor miRNA (pre-miRNA) is recognized and cleaved by a complex containing Dicer and several non-catalytic accessory proteins. HIV TAR element binding protein (TRBP) is a constituent of the Dicer complex, which augments complex stability and potentially functions in substrate recognition and product transfer to the RNA-induced silencing complex. Here we have analysed the interaction between the RNA-binding region of TRBP and an oncogenic human miRNA, miR-155, at different stages in the biogenesis pathway. We show that the region of TRBP that binds immature miRNAs comprises two independent double-stranded RNA-binding domains connected by a 60-residue flexible linker. No evidence of contact between the two double-stranded RNA-binding domains was observed either in the apo- or RNA-bound state. We establish that the RNA-binding region of TRBP interacts with both pre-miR-155 and the miR-155/miR-155* duplex through the same binding surfaces and with similar affinities, and that two protein molecules can simultaneously interact with each immature miRNA. These data suggest that TRBP could play a role before and after processing of pre-miRNAs by Dicer.
INTRODUCTION
The majority of human micro-RNAs (miRNAs) are produced from long RNA transcripts in a two-step process (1,2). The initial primary miRNA (pri-miRNA) transcript can be 1-2 kilobases (kb) in length. Within the pri-miRNA, a short, imperfect stem-loop structure is recognized and excised by the nuclear ‘Microprocessor’ complex. The product is a 60–70-nucleotide (nt) precursor miRNA (pre-miRNA), which is exported from the nucleus by an Exportin-5/Ran-GTP complex. In the cytoplasm, the pre-miRNA is further processed by the Dicer complex into a 20–25-base-pair (bp) duplex, which contains the final miRNA and its reverse complement, termed miRNA*. The imperfect miRNA/miRNA* duplex is then transferred to the RNA-induced silencing complex (RISC), which is the mediator of miRNA-directed RNA interference in humans.
The multidomain human TAR element binding protein (TRBP) is a component of the Dicer complex and has been implicated in the processing of pre-miRNAs (3,4). TRBP was initially discovered as a protein that associates with the HIV TAR RNA (5) and was later demonstrated to interact with and regulate interferon-induced protein kinase R (PKR), a kinase involved in the cellular response to viral infection (6,7). In the context of miRNA biogenesis, TRBP associates with Dicer, the RNase III enzyme that catalyses the removal of the apical loop of pre-miRNAs (8) and processes small interfering RNAs [siRNAs; (9)]. TRBP has been shown to enhance dicing and augment Dicer stability (3,4,10), although the molecular mechanisms for these activities have not been elucidated. Downstream of Dicer, TRBP is also a component of the RISC-loading complex (RLC), the protein assembly responsible for transferring the miRNA/miRNA* duplex from Dicer to RISC (11,12). TRBP contains three double-stranded (ds) RNA-binding domains (dsRBDs), but only the first two dsRBDs interact with dsRNA. The third dsRBD is believed to mediate protein/protein interactions and bind to other proteins in the Dicer complex. The contribution of TBRP to the function of either the Dicer complex or RLC is not well-understood. The presence of two types of dsRBD supports the hypothesis that TRBP mediates both protein/RNA and protein/protein contacts, although the targets, location and consequences of these interactions in the context of the Dicer complex or RLC remain to be elucidated.
Dicer homologues are found in Drosophila melanogaster and Caenorhabditis elegans, and they associate with proteins with a similar domain composition to TRBP. Proteins such as R2D2 (13) and Loquacious (14,15) in D. melanogaster and RDE-4 (16) in C. elegans all contain two N-terminal canonical RNA-binding dsRBDs as well as a non-canonical C-terminal dsRBD, which is implicated in protein–protein interactions with the respective Dicer homologue. In plants, Dicer-like 1 (DCL1) is found in complex with HYPONASTIC LEAVES 1 (HYL1), an accessory protein, which contains only two dsRBDs (17). HYL1 interacts with pre-miRNA and miRNA/miRNA* duplexes primarily through its first dsRBD (18,19). Human Dicer has also been shown to associate with a TRBP homologue, Protein Activator of PKR [PACT; (20,21)]. A complex containing Dicer, TRBP and PACT can be purified from human cells (21). PACT also contains three dsRBDs and, like TRBP, is implicated in the regulation of PKR. However, unlike TRBP, PACT activates PKR (22).
miRNAs regulate gene expression at the post-transcriptional level (1,2). To date, ∼1500 human miRNAs have been documented in the database of miRNAs, miRbase (23). miRNAs are believed to regulate the expression of upwards of 30% of human genes by enabling RISC to recognize complementary sequences in target messenger RNAs. Many miRNAs have oncogene- or tumor-suppressor-like behaviour (24). Mis-expression or mutation of miRNAs can have considerable cellular consequences (25). For example, miR-155 has been implicated in lung and breast cancer as well as different leukaemias and lymphomas (24). Mutations in the proteins involved in the processing or regulation of miRNA biogenesis have also been linked to different cancers. For example, certain frame-shift mutations implicated in colorectal or endometrial cancers result in truncated forms of TRBP that lack dsRBDs 2 and 3 (26). Furthermore, TRBP is phosphorylated by the mitogen-activated protein kinase Erk (27), and it has been suggested to interact with the anti-cancer drug enoxacin (28). These recent studies have provided further evidence of the importance of TBRP in miRNA biogenesis.
Over the past decade, there has been a drive to understand the structural biology of miRNA biogenesis (29,30). Nuclear magnetic resonance (NMR) spectroscopic and X-ray crystallographic studies of TRBP have elucidated the isolated 3D structures of the first two dsRBDs (31) as well as the second dsRBD in complex with a short siRNA duplex (18). Negative-stain electron microscopy studies of miRNA biosynthesis complexes containing TRBP have also been reported; however, the level of resolution is not yet sufficient to accurately describe the location of TRBP in these complexes or determine how it interacts with other components (32,33). Many biochemical studies have sought to explore the mechanism for RNA recognition by TRBP or examine how TRBP associates with Dicer (3,4,10–12,34,21). However, despite these efforts, the exact role of TRBP in immature miRNA recognition and processing remains unclear.
Here we present an in-depth biophysical analysis of the complex formed between the RNA-binding region of TRBP and biosynthetic precursors of the oncogenic human miRNA miR-155. We establish that the two dsRBDs that form the RNA-binding region function independently in apo- and RNA-bound forms. We show that this region of TRBP interacts with both pre-miR-155 and miR-155/miR-155* with affinities in the low micromolar range and that it can form a large complex in which four dsRBDs interact with a single immature miRNA molecule. These data, which concern functional regions of the protein and their interactions with relevant biological targets, further enhance our understanding of the role of TRBP in miRNA biogenesis.
MATERIALS AND METHODS
Protein expression and purification
Residues 19–99 (TRBP-D1; monomeric molecular weight: 8.7 kDa), 157–228 (TRBP-D2; 8.2 kDa) or 19–228 (TRBP-D12; 22.6 kDa) of TRBP were sub-cloned into pLX06 (Protein'eXpert, France) using a codon-optimized TRBP2 (AAA36765) cDNA template (GeneArt). Each construct was expressed with an N-terminal 6-histidine purification tag and a tobacco etch virus cleavage site. Protein constructs were expressed and purified as previously described (35).
Large-scale production of pre-miR-155 and miR-155/miR-155*
Pre-miR-155 (5'-GGAAU GCUAA UCGUG AUAGG GGUUU UUGCC UCCAA CUGAC UCCUA CAUAU UAGCA UUCCC A-3') or miR-155/miR-155* (miR-155: 5'-GGAAU GCUAA UCGUG AUAGG CCU-3'; miR-155*: 5'-GGCCU ACAUA UUAGC AUUCC CA-3') were produced by in vitro transcription using in-house produced T7 RNA polymerase and fully complementary ds DNA oligonucleotides containing a T7 promoter. As each translated RNA sequence was required to start with a GG dinucleotide, it was necessary to modify the native RNA sequences. Additional sequence changes were required to preserve the Watson–Crick base-pairing. Non-native sequence nucleotides are underlined in the sequences provided previously.
Template oligonucleotides (Eurogentec) were annealed by incubation at 95°C for 5 minutes. The transcription reaction was performed in a standard T7 RNA polymerase buffer (40 mM Tris-HCl pH 8.1, 1 mM spermidine, 5 mM dithiothreitol, 0.1 % Triton X-100, 40 µg/ml bovine serum albumin, 80 mg/ml polyethylene 8000, 4 mM of each nucleotide triphosphate, 15-20 mM MgCl2, 35 µg/ml DNA matrix, 0.1 mg/ml T7 RNA polymerase and 1 U/ml pyrophosphatase). The reaction mixture was incubated at 37°C for 3 hours. RNA products were purified by polyacrylamide gel electrophoresis, dialyzed multiple times against water and lyophilized. The folding of each RNA was achieved by heating each sample in water at 96°C for 5 minutes, followed by rapid cooling induced by the addition of ice-cold 2× NMR buffer. All RNA samples were analysed by 1D (1H) NMR spectroscopy.
NMR spectroscopy
(1H,13C,15N) backbone resonance assignments of TRBP constructs in complex with either pre-miR-155 or miR-155/miR-155* were obtained using a combination of 3D BEST triple resonance NMR experiments (36) and titration experiments. (1H,13C,15N) backbone resonance assignments of apo-TRBP-D12 were taken from our previous work [BioMagResBank code: 18324; (35)]. Assignments of TRBP-D12 in complex with pre-miR-155 were obtained by analysing 3D (1H,13C,15N) HNCA and HNCO experiments, which were acquired using a 230 µM [U-2H,13C,15N]-labelled sample of TRBP-D12 and a 3.5-fold excess of unlabelled pre-miR-155. The results of titration experiments (see later in the text) were also used to aid assignment of TRBP-D12 in complex with pre-miR-155 or in complex with miR-155/miR-155*.
Apo- and RNA-bound states of the different TRBP constructs were analysed using 2D BEST (1H,15N) TROSY experiments at various protein concentrations (0.4 to 0.8 mM) and RNA/protein ratios. Titration experiments were performed with the following RNA/protein ratios: (i) TRBP-D12 with 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.75, 1.0, 1.25, 1.5, 1.75, 2.0 and 2.3 molar equivalents of pre-miR-155 or miR-155/miR-155*; (ii) TRBP-D1 with 0, 0.1, 0.22, 0.29, 0.33, 0.4, 0.5, 0.75, 1.0 and 1.5 molar equivalents of pre-miR-155; or (iii) TRBP-D2 with 0, 0.1, 0.5, 0.75, 1.0 and 1.5 molar equivalents of pre-miR-155. The time required to collect individual titration points depended on the protein concentration and the signal-to-noise ratio.
All NMR experiments were performed at 25°C using a Varian (Agilent) DirectDrive 800 MHz spectrometer equipped with a triple resonance cryogenic probe. NMR spectra were processed using NMRpipe (37) and analysed using CCPNMR Analysis, version 2.1.5 (38). Compound chemical shift differences, Δδ, between free protein and RNA-bound protein were calculated using the following equation: Δδ = [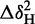 + (ΔδN/6.5)2]½, where ΔδX is the different in chemical shift between comparable resonances in different 2D spectra (39).
+ (ΔδN/6.5)2]½, where ΔδX is the different in chemical shift between comparable resonances in different 2D spectra (39).
Multi-angle laser light scattering
Fifty-microlitre samples of TRBP-D1 (1.8 mg/ml), TRBP-D2 (1.7 mg/ml), TRBP-D12 (2 mg/ml) and pre-miR-155 (4 mg/ml) were loaded onto an analytical Superdex S200 size exclusion chromatography (SEC) column (GE Healthcare) pre-equilibrated with NMR buffer. SEC was performed at 0.5 ml/min with an in-line multi-angle laser light scattering (MALLS) spectrometer (DAWN EOS, Wyatt Instruments) with 18 angles measuring at 690 nm. An in-line refractive index detector (RI 2000, Spares) was used to follow the differential refractive index relative to the solvent (1.34). Masses were estimated with the Debye model using ASTRA software version 5.3.4.20 (Wyatt Instruments) using a theoretical dn/dc value of 0.185 ml/g.
Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity experiments were performed with a Beckmann Coulter XLI analytical ultracentrifuge at 50 000 rpm (185 000g), 25°C with an An-50Ti rotor. Samples or reference (NMR buffer) were loaded into sapphire-windowed cells with 12-mm optical paths, which had previously been treated with RNase ZAP (Sigma-Aldrich). Acquisition was performed at 260 nm and 280 nm, and using interference optics. The molar masses for the polymers were calculated from their composition. A partial specific volume (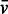 ) of 0.74 ml/g was estimated for TRBP-D12 using the software SEDNTERP version 2 (40). The
) of 0.74 ml/g was estimated for TRBP-D12 using the software SEDNTERP version 2 (40). The 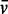 used for pre-miR-155 (0.508 ml/g) was taken from a study of a viral RNA in dilute aqueous KCl solutions (41). The data were analysed in SEDFIT [Version 12.52; (42)] using the c(s) analysis and non-interacting species models. The latter was used to estimate buoyant molar masses, Mb, using the equation, Mb = M × [1 − (ρ ×
used for pre-miR-155 (0.508 ml/g) was taken from a study of a viral RNA in dilute aqueous KCl solutions (41). The data were analysed in SEDFIT [Version 12.52; (42)] using the c(s) analysis and non-interacting species models. The latter was used to estimate buoyant molar masses, Mb, using the equation, Mb = M × [1 − (ρ × 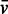 )], where M is the molar mass and ρ the solvent density. Hybrid models and multi-wavelength analysis were performed in the program SEDPHAT (43). Results in terms of buoyant mass and multi-wavelength analysis are given for the more concentrated sample, which yielded higher signal-to-noise ratios. The mass density (1.005 g/ml) and viscosity (0.898 cP) of the buffer were calculated using SEDNTERP (v20120328 beta, www.jphilo.mailway.com).
)], where M is the molar mass and ρ the solvent density. Hybrid models and multi-wavelength analysis were performed in the program SEDPHAT (43). Results in terms of buoyant mass and multi-wavelength analysis are given for the more concentrated sample, which yielded higher signal-to-noise ratios. The mass density (1.005 g/ml) and viscosity (0.898 cP) of the buffer were calculated using SEDNTERP (v20120328 beta, www.jphilo.mailway.com).
The 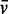 for possible protein/RNA complexes was estimated by summing the individual
for possible protein/RNA complexes was estimated by summing the individual 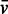 values of each component weighted by their fractional mass in the complex (Supplementary Table S1). The sedimentation coefficient, s, of each complex estimated from the distribution of sedimentation coefficients, c(s), was not affected by the choice of the three possible values. c(s) profiles reported here were fitted with a value of 0.64 ml/g. The Svedberg and Stokes equations were used to analyse the value of the experimental sedimentation coefficients.
values of each component weighted by their fractional mass in the complex (Supplementary Table S1). The sedimentation coefficient, s, of each complex estimated from the distribution of sedimentation coefficients, c(s), was not affected by the choice of the three possible values. c(s) profiles reported here were fitted with a value of 0.64 ml/g. The Svedberg and Stokes equations were used to analyse the value of the experimental sedimentation coefficients.
Isothermal titration calorimetry
All isothermal titration calorimetry (ITC) experiments were conducted using a MicroCal ITC-200 at 25°C. ITC experiments were performed in two ways: either with the ‘ligand’, in this case the protein, in the cell, or with ‘ligand’ in the syringe. Protein concentrations varied between 30 and 60 µM in the sample cell and 250 and 500 µM in the syringe. RNA concentrations varied between 25 and 40 µM in the sample cell and 150 and 300 µM in the syringe. The heat evolved after each injection of protein or RNA was obtained from the integral of the baseline-corrected calorimetric signal. The equations used to analyse ITC data of TRBP-D12 were derived from the classical sequential binding equation using activity coefficients of 1. Previous assumptions of independency or co-operativity were not made. The resulting binding isotherms were fitted to maximize the likelihood of the data assuming a Gaussian noise distribution. The titration data were globally fitted for each pair of experiments using previously described protocols (44). The standard deviation of the estimation of each binding parameter was determined by computing 100 Monte Carlo simulations of each experiment performed assuming a Gaussian noise of 70 nanocals (estimated from our data) followed by a global fitting of the data. The stoichiometry for each protein/RNA pair was estimated by comparing the fits of sequential models for 1:1 and 1:2 protein/RNA complexes (see Supplementary Material).
RESULTS
Two independent dsRBDs comprise the RNA-binding region of TRBP
Three constructs derived from the RNA-binding region of TRBP were produced (Figure 1a) and characterized by SEC. Analysis of the eluted protein by SEC-MALLS reported values of 8.5 ± 0.5 kDa, 8.4 ± 0.5 kDa and 23.2 ± 0.9 kDa for TRBP-D1, TRBP-D2 and TRBP-D12, respectively (Figure 1b). These values correspond well with the respective monomeric masses of each protein construct.
Figure 1.
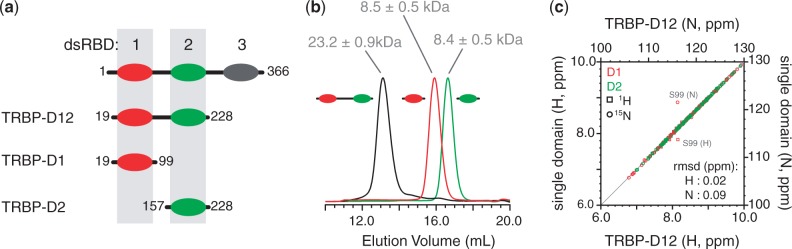
Analysis of the solution behaviour of dsRBDs from TRBP. (a) Diagram of the three constructs of TRBP used in this study. The same colour scheme for TRBP-D1 (red) and TRBP-D2 (green) is used throughout; (b) analytical SEC profiles showing elution volume vs. absorbance at 280 nm, A280, for TRBP-D1 (red), TRBP-D2 (green) and TRBP-D12 (black). The molecular weight of each species determined by MALLS is given; (c) site-by-site comparison of the (1H,15N) chemical shift of a residue in the single domain and the corresponding residue in TRBP-D12. Circles denote (15N) chemical shifts and squares (1H) chemical shifts. Amide groups from TRBP-D1 are coloured in red and those from TRBP-D2 in green. The points corresponding to the large outliers are annotated in grey. RMSD values for 1H and 15N chemical shifts are provided.
Two dimensional (1H,15N) HSQC spectra of each of the three constructs displayed good 1H resonance dispersion, which indicated that the dsRBDs adopt folded 3D structures. The spectrum of the double-domain construct, TRBP-D12, can be reconstituted by overlaying the spectra of TRBP-D1 and TRBP-D2 (Supplementary Figure S1a–d). The additional peaks visible in the spectrum of TRBP-D12 correspond to residues 96–151, which are located in the linker region. The (1H,15N) resonance frequencies of a given residue in a single-domain construct compare well with their counterparts in the double-domain construct, with root mean squared deviation (RMSD) values of 0.02 and 0.09 ppm for 1H and 15N frequencies, respectively (Figure 1c). The largest deviations in compound chemical shift differences between comparable sites are predictably found at the C-terminus of TRBP-D1 (Supplementary Figure S1e). These differences can be easily attributed to the presence of the inter-domain linker in the double-domain construct. The similarity in corresponding NMR signals strongly suggests that the two domains do not interact in the tandem dsRBD construct. This observation is consistent with previously reported (1H,15N) heteronuclear NOE data, which demonstrated the flexibility of the inter-domain linker (35). Taken together, these data suggest that the RNA-binding region of TRBP is composed of two independent dsRBDs connected by a flexible linker and, therefore, that it is similar to the tandem dsRBDs that form the dsRNA-interacting region of PKR (45).
Both dsRBDs of the RNA-binding region of TRBP interact with pre-miR-155
Large, site-specific changes in (1H,15N) chemical shifts are observed when unlabelled pre-miR-155 is added to constructs of TRBP (Figure 2a). Backbone resonance assignments were obtained of TRBP-D12 in the presence of an excess of pre-miR-155. Using these sequence-specific assignments, it was possible to evaluate which regions of each dsRBD were involved in the formation of protein/RNA complexes. A comparison of the amide chemical shifts of apo- and RNA-bound TRBP-D12 revealed large RMSD values between the two data sets (0.11 ppm for 1H and 0.54 ppm for 15N frequencies; Supplementary Figure S2). The largest chemical shift changes observed on complex formation are restricted to amino acids located in the two dsRBDs. Only negligible chemical shift changes occur in the linker region (Figure 2a–c). The profiles of residue-by-residue chemical shift changes of the two dsRBDs in TRBP-D12 are highly comparable (Figure 2a–c).
Figure 2.
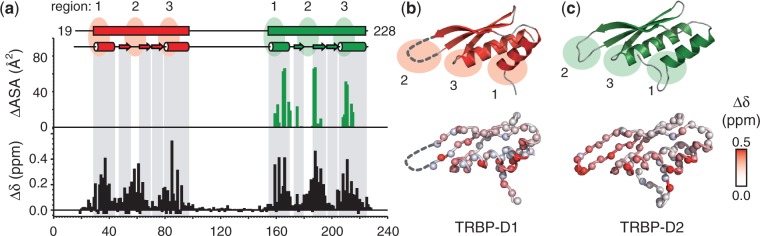
NMR analysis of the interaction between TRBP-D12 and pre-miR-155. (a) Compound chemical shift values (Δδ) measured when TRBP-D12 interacts with pre-miR-155 plotted as a function of residue number (black). Δδ = [ + (ΔδN/6.5)2]½, where ΔδX is the difference in chemical shift between apo- and RNA-bound spectra (39). Prolines and unassigned residues have been given values of −0.05 and −0.1 ppm, respectively. For comparison, the change in accessible surface area (ΔASA) when TRBP-D2 forms a complex with dsRNA is shown (green). ΔASA were calculated using the POPS* server (46) from 3ADL (18). The boundaries of secondary structure elements were taken from the 3D structures of TRBP-D1 [3LLH; (31)] and TRBP-D2 [3ADL; (18)]; (b,c) TOP: cartoon representation of the 3D structures of TRBP-D1 and TRBP-D2 showing the three regions in canonical dsRBDs that are implicated in dsRNA binding; (b,c) BOTTOM: Δδ values plotted on the respective 3D structures. Each amide nitrogen is represented by a sphere and coloured according to the Δδ scale provided. The β2/3 loop was not resolved in the 3D structure of TRBP-D1 and is shown by a broken line.
+ (ΔδN/6.5)2]½, where ΔδX is the difference in chemical shift between apo- and RNA-bound spectra (39). Prolines and unassigned residues have been given values of −0.05 and −0.1 ppm, respectively. For comparison, the change in accessible surface area (ΔASA) when TRBP-D2 forms a complex with dsRNA is shown (green). ΔASA were calculated using the POPS* server (46) from 3ADL (18). The boundaries of secondary structure elements were taken from the 3D structures of TRBP-D1 [3LLH; (31)] and TRBP-D2 [3ADL; (18)]; (b,c) TOP: cartoon representation of the 3D structures of TRBP-D1 and TRBP-D2 showing the three regions in canonical dsRBDs that are implicated in dsRNA binding; (b,c) BOTTOM: Δδ values plotted on the respective 3D structures. Each amide nitrogen is represented by a sphere and coloured according to the Δδ scale provided. The β2/3 loop was not resolved in the 3D structure of TRBP-D1 and is shown by a broken line.
Three critical regions in canonical dsRBDs mediate the interaction with dsRNA (47): region 1, α-helix 1 (α1); region 2, the loop between strands β2 and β3 (β2/3); and region 3, the N-terminus of α2 (Figure 2b and c). The NMR chemical shift perturbation profiles showed that both TRBP-D1 and TRBP-D2 interact with pre-miR-155 through the classical dsRBD interaction site. Residues located in regions 1, 2 and 3 exhibit the largest changes in chemical shift or disappear entirely from the spectrum, even after the addition of 2.3-fold excess of pre-miR-155 (Figure 2). These chemical shift changes are consistent with previously published structural and biochemical analyses of other dsRBD/dsRNA complexes (31, 47–50). The 3D structure of TRBP-D2 in complex with a short siRNA was reported in 2010 (18). The residue-by-residue changes in solvent accessible surface area, ΔASA, that occur on the formation of this complex highlight the location of the three critical RNA interaction regions in TRBP-D2 (Figure 2a). The profile of ΔASA for the TRBP-D2/siRNA complex and Δδ observed for dsRBD-2 in the TRBP-D12/pre-miR-155 complex correspond well. These observations suggest that the interaction between TRBP-D2 and the imperfect stem-loop of pre-miR-155 in solution is similar to that reported for a perfect A-form dsRNA duplex and, furthermore, that highly similar protein/RNA contacts are formed in the context of the tandem dsRBD construct.
Titration of [15N]-labelled TRBP-D1 or TRBP-D2 with unlabelled pre-miR-155 revealed that both of the isolated dsRBDs of TRBP can interact with pre-miR-155. Two-dimensional (1H,15N) HSQC spectra of either TRBP-D1 or TRBP-D2 showed clear and significant chemical shift perturbations after the addition of RNA. NMR data from sub-equimolar titration points were characterized by severe line-broadening. Many cross-peaks broaden beyond detection after the addition of 0.1 equivalents of RNA and reappear when the RNA/protein ratio exceeds 0.3–0.5 (Supplementary Figure S3). Two-dimensional (1H,15N) HSQC spectra of TRBP-D12 in complex with pre-miR-155 superpose nicely with spectra of either of the single dsRBDs in complex with the same RNA (Figure 3a–d). As was the case for the apo-protein (Supplementary Figure S1), the spectrum of the double-domain construct in complex with pre-miR-155 is essentially a superposition of the spectra of the individual dsRBDs in complex with the same RNA (Figure 3a–d). Site-by-site differences in (1H,15N) resonance frequencies between single-domain and double-domain data sets are very small, with RMSD values of 0.02 and 0.11 ppm for 1H and 15N frequencies, respectively (Figure 3e). That is, a cross-peak for an amide group in the TRBP-D1/pre-miR-155 complex, for example, has almost exactly the same (1H,15N) resonance frequencies, and therefore experiences a near-identical local chemical environment, as the comparable amide group in the tandem domain/pre-miR-155 complex. This similarity of resonance frequencies between single- and double-domain constructs suggests that each dsRBD forms essentially the same interaction regardless of whether it is a component of the tandem domain construct. If the interaction with pre-miR-155 led to (or resulted from) the formation of contacts between the two dsRBDs, this would result in chemical shift differences between NMR spectra of the single- and double-domain constructs. However, the small RMSD between these data sets suggests that each dsRBD in TRBP-D12 interacts independently with pre-miR-155.
Figure 3.
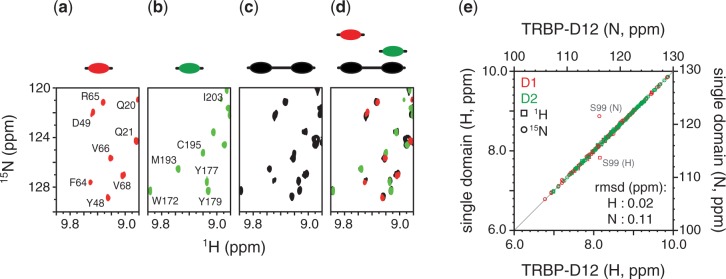
Comparison of NMR data collected of single- and double-domain constructs in complex with pre-miR-155. Two dimensional (1H,15N) spectra of (a) TRBP-D1/pre-miR-155 in red, (b) TRBP-D2/pre-miR-155 in green, and (c) TRBP-D12/pre-miR-155 in black. A coloured graphical representation of each construct is provided above the spectrum. (d) Superposition of the spectra shown in (a-c) retaining the same colour scheme and construct graphics. (e) Comparison of the (1H,15N) resonance frequencies of a residue in the single domain and the corresponding residue in TRBP-D12. Circles denote (15N) chemical shifts and squares (1H) chemical shifts. Amide groups from TRBP-D1 are coloured in red and those from TRBP-D2 in green. The points corresponding to the large outliers are annotated. RMSD values for 1H and 15N chemical shifts are provided.
The RNA-binding region of TRBP can form a 2:1 complex with pre-miR-155
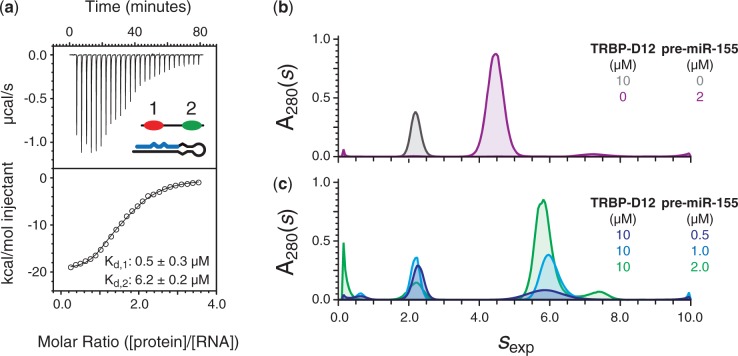
ITC experiments were conducted to further characterize the complex formed between the dsRBDs of the RNA-binding region of TRBP and pre-miR-155. The resulting binding isotherms were globally fitted and best described by a two-site sequential binding model in which the first molecule of TRBP-D12 binds pre-miR-155 approximately 10 times more strongly than the second molecule (Kd,1 = 0.5 ± 0.03 µM; Kd,2 = 6.2 ± 0.2 µM; Figure 4a and Supplementary Table S2). The difference in affinity measured for the two molecules of TRBP-D12 is most likely due to a decrease in the number of sites available for the second binding event with respect to the first. The interaction between TRBP-D12 and pre-miR-155 exhibits an exothermic character, which suggests that polar interactions are the primary driving force of the binding event and which is consistent with the electrostatic nature of interactions between dsRNAs and dsRBDs (47).
Figure 4.
Characterization of the interaction between TRBP-D12 and pre-miR-155. (a) An example of ITC data recorded of the titration of pre-miR-155 with TRBP-D12. The top panel shows power vs. time together with a schematic of the components of the sample. The bottom panel shows the integrated injection enthalpy per mole of injectant plotted as a function of the ratio of protein and RNA concentrations. The data were fitted to a two-site sequential binding model shown by a solid black line. The two dissociation constants are given. The thermodynamic parameters calculated are provided in Supplementary Table S2. (b) Comparison of c(s) distributions of TRBP-D12 (grey) and pre-miR-155 (purple); (c) c(s) distributions of a sample prepared with 10 µM TRBP-D12 and different concentrations of pre-miR-155. The protein and/or RNA concentration corresponding to each curve is provided.
A comparison between the binding modes of single- and double-domain constructs was also performed. ITC data obtained from the titration of TRBP-D1, TRBP-D2 or TRBP-D12 with pre-miR-155 were analysed using the standard single-site model. The resulting macroscopic dissociation constant, within the context of a identical independent sites model for the complex formed between TRBP-D12 and pre-miR-155, is 3.4 ± 0.8 µM (Supplementary Figure S4b and c and Supplementary Table S2). ITC analyses of the interactions between single-domain constructs and pre-miR-155 revealed that both single-domain constructs, TRBP-D1 and TRBP-D2, interact with pre-miR-155 to form 4:1 complexes (Supplementary Figure S4b and c and Supplementary Table S2). All macroscopic binding constants measured were in the mid-to-low micromolar range, consistent with results of TRBP-D12. TRBP-D2 (Kd = 3.7 ± 0.5 µM) interacts 4 times more strongly than TRBP-D1 (15.4 ±0.8 µM). Like TRBP-D12, both TRBP-D1 and TRBP-D2 exhibit an exothermic binding enthalpy, again suggesting that polar interactions mediate binding. Furthermore, the macroscopic binding enthalpy obtained for TRBP-D12 is approximately equal to the sum of that of TRBP-D1 and TRBP-D2, which indicates that the polar interactions formed by each single domain and pre-miR-155 are unaffected in the context of the full-length protein. Thus, these ITC data support the conclusions derived from NMR analysis that each dsRBD interacts independently with pre-miR-155.
The formation of a complex between TRBP-D12 and pre-miR-155 in solution was evaluated by sedimentation velocity analytical ultracentrifugation (svAUC). A series of samples were prepared to characterize the behaviour of the free protein, free RNA and the protein/RNA complex under different conditions. SEC-MALLS (Figure 1b) strongly suggests that TRBP-D12 is monomeric in solution. This observation was supported by svAUC data at 10 and 20 µM, which revealed an experimental sedimentation coefficient, sexp, of 2.19 S and which was independent of the protein concentration used (Figure 4b). A frictional ratio of 1.43 for TRBP-D12 was calculated by combining sexp and the molecular mass (22.3 kDa) of the monomeric protein. This value is in excellent agreement with one previously reported for a protein composed of two dsRBDs connected by a flexible linker (51). When a single sedimenting species is considered, a molar mass of 22.4 kDa is calculated from the sedimentation profile of TRBP-D12, without assumptions about the shape of the sedimenting species, which is in excellent agreement with the expected molecular mass. Similar svAUC experiments indicated that pre-miR-155 is largely monomeric in solution at sample concentrations of 1 and 2 µM. A sexp of 4.4 S is obtained for pre-miR-155 at both 1 µM and 2 µM, which corresponds to a frictional ratio of 1.35. Non-interacting species analysis gives a molecular mass of 18.5 kDa, which is again close to the theoretical value of 19.4 kDa. An additional minor contribution at 7.2 S (6% of the main peak) was also detected. This peak is likely to result from a small fraction of dimerized pre-miR-155. This species was also observed by SEC-MALLS (data not shown).
SvAUC analysis of the TRBP-D12/pre-miR-155 complex was performed using three samples, each with a different excess of protein. In the resulting sedimentation profiles, the RNA contribution at 4.4 S has disappeared and a new faster contribution is visible, which sediments at 5.9 S (Figure 4c). The composition of the 5.9 S peak was estimated using the data obtained from the three different optical signals. The ratios of these signals as well as theoretical values calculated for TRBP-D12/pre-miR-155 complexes with different stoichiometries are given in Supplementary Table S1. The experimental ratios measured in the c(s) analysis for the fast boundary (Supplementary Table S1) reflect an enrichment of protein in the complex and strongly suggest that a protein/RNA complex with a stoichiometry of 2:1 can be formed in solution.
The combination of ITC and AUC analyses demonstrates that two molecules of TRBP-D12 can interact with a single molecule of pre-miR-155. These data are consistent with ITC analysis of the two single-domain constructs, which demonstrated that four molecules of either TRBP-D1 or TRBP-D2 can interact with a single pre-miR-155. It is therefore likely that pre-miR-155 has a minimum of four dsRBD-binding sites.
The RNA-binding region of TRBP interacts with the substrates and products of Dicer
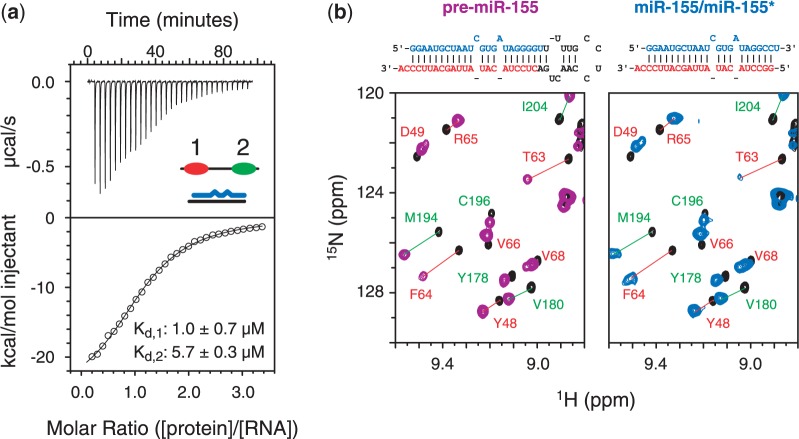
Dicer excises the apical loop of pre-miR-155 (nucleotides 24–39) to generate a largely double-stranded molecule containing the 23-nucleotide miR-155 and its complement, miR-155* (Supplementary Figure S3a). ITC experiments were conducted to characterize the interaction between miR-155/miR-155* and the RNA-binding region of TRBP (Figure 5a). These data revealed many similarities to those measured for the interaction between TRBP-D12 and pre-miR-155. The interaction between TRBP-D12 and miR-155/miR-155* was also exothermic in character (Figure 4a). In addition, the binding isotherms were also optimally described by a two-step sequential binding model (Kd,1 = 1.0 ± 0.7 µM; Kd,2 = 5.7 ± 0.3 µM; Supplementary Table S2). Finally, as was seen for the TRBP-D12/pre-miR-155 complex, the first molecule of TRBP-D12 binds miR-155/miR-155* more strongly than the second molecule. The difference in dissociation constants is again likely due to the reduced probability of the second molecule of TBRP-D12 binding.
Figure 5.
Interaction of the RNA-binding region of TRBP with the substrate and product of Dicer. (a) An example of an ITC analysis of the titration of miR-155/miR-155* with TRBP-D12 showing power vs. time data (top, with a schematic of the components of the sample) and the integrated injection enthalpy per mole of injectant plotted as a function of the protein/RNA ratio (bottom). The data were fitted to a two-step sequential binding model shown by a solid black line. The two dissociation constants and their standard deviations are given. The complete thermodynamic parameters derived are provided in Supplementary Table S2. (b) Comparison of the changes in (1H,15N) NMR spectra of TRBP-D12 resulting from the addition of either pre-miR-155 (left) or miR-155/miR-155* (right). The nucleotide sequence and predicted secondary structure of each RNA are provided. (1H,15N) assignments of cross-peaks in each spectrum are annotated and coloured according to the dsRBD in which they reside (red, dsRBD-1; green, dsRBD-2).
NMR spectroscopy was used to assess which regions of TRBP-D12 interact with the miR-155/miR-155* duplex. Large chemical shift changes are observed in 2D (1H,15N) HSQC spectra of [15N]-labelled TRBP-D12 on the addition of unlabelled miR-155/miR-155* (Figure 5b and Supplementary Figure S5b and c). Comparison of the spectra of the two TRBP-D12/RNA complexes shows little difference between the magnitude or sequence location of chemical shift perturbations. The changes in chemical shift for TRBP-D12 on binding miR-155/miR-155* are largest in the three RNA interaction regions of each dsRBD (Supplementary Figure S3b). Again, residues in the linker are barely perturbed. NMR spectra of TRBP-D12 in complex with either pre-miR-155 or miR-155/miR-155* are nearly identical (Figure 5b and Supplementary Figure S5b and c). Site-by-site differences in (1H,15N) resonance frequencies between single-domain and double-domain data sets are very small, with RMSD values of 0.02 and 0.11 ppm for 1H and 15N frequencies, respectively (Supplementary Figure S5c). These data suggest that the complexes formed between TRBP-D12 and either pre-miR-155 or miR-155/miR-155* are highly similar.
ITC and NMR data demonstrate that TRBP-D12 interacts with two different precursors of miR-155. These data support the formation of two highly similar complexes, which are dependent on the same protein/RNA interaction surfaces. These data also suggest that the apical loop of pre-miR-155 is neither directly recognized by TRBP nor is it required for the interaction. The similarity in the measured affinities also suggests that the RNA-binding region of TRBP alone cannot differentiate between the two precursor molecules.
DISCUSSION
dsRBDs often appear in modular RNA-binding proteins, either in multiple copies or in combination with other functional domains such as RNase III, RNA helicases and PAZ domains (47). Structural studies of modular proteins containing single or multiple copies of dsRBDs have frequently revealed that these domains do not interact with other structured regions in the same protein. An NMR spectroscopic study of the dsRNA-binding region of PKR revealed no interaction between the two dsRBDs (52). Likewise, the two dsRBDs of ADAR2 (50) and HYL1 (19) were also shown to behave independently. In PKR, ADAR2 and HYL1, a long, unstructured linker connects the two dsRBDs. We have shown here that the first two dsRBDs of TRBP are also independent. Solution NMR analysis of different constructs of the RNA-binding region of TRBP revealed no contact between the two dsRBDs (Figure 1c and Supplementary Figure S1), while SEC-MALLS indicated that each isolated dsRBD is monomeric in solution.
NMR (Figure 3) and ITC analysis (Supplementary Figure S4) of TRBP-D1 and TRBP-D2 revealed that each isolated domain interacts with pre-miR-155, which is consistent with previous conclusions that these two dsRBDs constitute the main interaction site for immature miRNAs in TRBP (18,31). However, little biophysical or structural analysis of how multiple-domain constructs of TRBP interact with immature miRNAs has been previously reported. The NMR analysis of the interaction between TRBP-D12 and pre-miR-155 presented here reveals several interesting features: first, comparison of NMR spectra of single- and double-domain constructs in complex with pre-miR-155 shows no evidence of an interaction between the two dsRBDs (Figure 3). NMR spectra of each complex studied revealed no examples of peak doubling that would typically indicate the formation of multiple different protein/RNA or protein/protein interfaces. Furthermore, the very small perturbations of the (1H,15N) chemical shifts of the inter-domain linker indicate that this region does not interact with pre-miR-155 nor does RNA binding induce contacts with either dsRBD. The elevated signal intensity of residues in the linker with respect to the two dsRBDs is consistent with this region remaining flexible in the TRBP-D12/pre-miR-155 complex. Taken together, these data strongly suggest that each dsRBD interacts with pre-miR-155 independently. This mode of interaction is similar to that used by ADAR2. A recently elucidated 3D structure of the complex formed by ADAR2 and a 71-nt stem-loop RNA revealed that the two dsRBDs bind to distinct regions of the ds stem without any inter-domain contact or contribution from the inter-domain linker (50,53).
dsRBDs in multiple arrays do not always interact with dsRNA independently. An interesting example that contrasts TRBP is DGCR8, a component of the Microprocessor complex, which performs the first processing step in miRNA biogenesis. The two dsRBDs of DGCR8 interact with each other through a large interface, which fixes their relative orientation and has important implications for pri-miRNA recognition. DGCR8 interacts with pri-miRNAs in preference to pre-miRNAs (54,55), whereas our data show that TRBP can interact with both pre-miRNAs and miRNA/miRNA* duplexes with similar affinities. These different patterns of RNA selectivity displayed by DGCR8 and TRBP could result from the contrasting ways in which the tandem dsRBDs interact with immature miRNAs. The compact structure formed by the DGCR8 core means that the two dsRBDs are not independent and that a pri-miRNA must bend to interact with both RNA-binding surfaces (56). A sufficient conformational change may not be feasible for the shorter pre-miRNA, leading to the preference for pri-miRNAs. Conversely, given that TRBP has been implicated in both the Dicer cleavage reaction and the transfer of the miRNA/miRNA* duplex to RISC, the independence of its two dsRBDs demonstrated here may reflect a broader range of binding targets. Taken together, these results suggest that there are considerable differences in how RNA is recognized by the different accessory proteins involved in the two miRNA biogenesis steps.
A second important feature of the TRBP-D12/pre-miR-155 complex is the stoichiometry. Both ITC (Figure 4a and Supplementary Figure S4) and svAUC (Figure 4b) experiments indicate that a complex containing four dsRBDs can be formed; that is, a single pre-miR-155 interacts with four single dsRBD constructs or two double-domain constructs. ITC data of the interactions between TRBP-D12 and pre-miR-155, and between TRBP-D12 and miR155/miR155*, revealed no evidence of positive co-operativity. This result is consistent with earlier studies (34).
The data presented here indicate that one pre-miR-155 can associate with four dsRBDs to form a complex in which the dsRBDs interact with similar sites on the RNA but do not form inter-domain contacts. Is there sufficient space on pre-miR-155 to permit this? The theoretical secondary structure of pre-miR-155 is typical of pre-miRNAs and comprises an imperfect ds stem containing two GU base-pairs, two unpaired nucleotides and a four-base apical loop (Figure 5b). The majority of pre-miRNAs in the miRbase (23) are predicted to adopt 3D structures similar to A-form dsRNA (57). Three-dimensional structures of large stem-loop RNAs have revealed that features such as non-Watson–Crick base-pairs or nucleotide bulges often cause minimal perturbation to the overall ds character (53,58). Canonical dsRBDs typically interact with a 12–16-bp region of dsRNA (47). However, the RNA-binding region of PKR, which also contains two dsRBDs, can interact with 20–40-bp RNA duplexes with stoichiometries that are incompatible with a simple 1D lattice model and which would require binding to multiple surfaces of the dsRNA with a binding site overlap of 4 bp (51). Several 3D structures of protein/dsRNA complexes have also demonstrated that multiple dsRBDs can bind in proximity without forming inter-dsRBD interactions (53,59).
A structural model of the interaction between pre-miR-155 and dsRBDs was generated to evaluate the feasibility of a protein/RNA complex containing the four dsRBDs per pre-miR-155 suggested by ITC and AUC analysis (Supplementary Figure S6). The model demonstrates that it is theoretically possible to arrange four non-contacting dsRBDs on a single molecule of pre-miR-155. The dsRBDs tessellate optimally when they interact with the RNA in alternating directions. The inter-dsRBD linker in TRBP is more than 60 residues and therefore places little limitation on the respective orientation of the two dsRBDs in the TRBP-D12 construct. In addition, NMR data suggest that the linker remains flexible in both of the TRBP-D12/RNA complexes analysed here. The arrangement shown in Supplementary Figure S6 is also compatible with NMR and ITC data of the interaction between TRBP-D12 and the Dicer product miR-155/miR-155*, as none of the four proposed dsRBD-binding sites in the model involve nucleotides from the excised apical loop.
The functional stoichiometry of the Dicer/TRBP complex has not been determined (32). However, the report that a tertiary complex containing Dicer, TRBP and PACT can be formed implies that two TRBP-like proteins can interact simultaneously with Dicer in the presence of pre-miRNAs (21). In addition, the plant DCL1 accessory protein HYL1 has been shown to form 2:1 protein/RNA complexes with dsRNA sequences of suitable length (18). Thus, high occupancy of dsRBDs on a relatively short sequence of dsRNA, as seen here with TRBP and two different precursors of miR-155, is potentially functionally relevant. Whether the 2:1 ratio observed here is a standard feature of the complexes formed between TRBP and immature miRNAs or whether the stoichiometry is determined by features in the target immature miRNA, such as the length or percentage of canonical dsRNA, remains to be determined.
Much of the information concerning the role of TRBP in small regulatory RNA synthesis has been obtained from studies into siRNAs. The data presented here represent the first in-depth biophysical analysis of the interaction between TRBP and immature miRNAs that have an imperfect dsRNA stem. Analysis of ITC data yielded macroscopic dissociation constants for the TRBP/pre-miR-155 complex in the low µM range, which is also consistent with other investigations of dsRBD/dsRNA interactions (18,19,56,60,61). The affinities measured for the isolated dsRBDs of TRBP indicate that TRBP-D2 interacts with pre-miR-155 approximately 4 times more strongly than TRBP-D1. This observation is consistent with earlier interaction studies of TRBP and siRNAs (31). Differences between the binding affinities of dsRBDs in tandem or multi-dsRBD proteins are relatively common—e.g. HYL1 (18,19) or PKR (45)—and may play a role in target selection by fine-tuning the overall affinity of a protein for dsRNA molecules that possess many potential dsRBD-binding sites.
In addition to its function in the Dicer complex, TRBP is a component of the RLC (4,11,12). The precise role of TRBP in the formation and/or activity of RISC is less clear. We have shown here that the RNA-binding region of TRBP interacts with both pre-miR-155 and miR-155/miR-155*—i.e. both the substrate and product of Dicer—with a comparable affinity (Figure 5a). Furthermore, NMR spectra of TRBP-D12 in complex with either pre-miR-155 or miR-155/miR-155* are highly similar (Figure 5b), which indicates that TRBP interacts with pre-miR-155 and miR-155/miR-155* through essentially identical sites. These results suggest that TRBP could participate in the recognition of Dicer substrates as well as the transfer of the cleavage product to RISC through the RLC.
CONCLUSIONS
Despite several structural studies, many of the mechanistic details that underpin human miRNA biogenesis remain unclear. One area in which the link between 3D structure and function remains particularly obscure concerns the role of the non-catalytic proteins of that associate with Drosha or Dicer. We have demonstrated here that the region of TRBP responsible for interacting with immature miRNAs is composed of two dsRBDs that recognize RNA independently. The linker connecting the two dsRBDs in the RNA-binding region of TRBP remains flexible and does not interact with the bound RNA. Therefore, the formation of a complex between TRBP and immature miRNAs is unlikely to require bending of the ds stem of the RNA, as has been reported for DGCR8. Finally, the observation that RNA-binding region of TRBP can interact with both the substrates and products of Dicer suggests that TRBP may function at several stages in the miRNA biogenesis pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–6, Supplementary Methods and Supplementary References [62–64].
FUNDING
This work was supported by grants from the European Marie Curie Programme (International Reintegration Grant 231082 to M.J.P.) and l’Agence Nationale de la Recherche (06-JCJC-0034 to J.B.). M.J.P. acknowledges a post-doctoral fellowship from l’Association pour la Recherche sur le Cancer, France, and M.P.M.H.B. acknowledges a Ph.D. fellowship from le Commissariat à l'Énergie Atomique et aux Energies Alternatives (CEA), France. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. A. Favier and E. Condamine for help with the acquisition of NMR data; Drs. R. Rasia, B. Bersch, C. Petosa and A. Leech for critical reading of the manuscript; and the Grenoble Partnership for Structural Biology (PSB) for access to integrated structural biology platforms, including: A. le Roy from the IBS/PSB AUC platform for help in the acquisition and analysis of AUC data; Dr. M. Noirclerc-Savoye, from the IBS/PSB RoBioMol platform, for the generation of the constructs used in this study; and the IBS/PSB high-field NMR spectroscopy platform; and IBS/PSB cell free expression platform.
REFERENCES
- 1.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 3.Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 6.Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl Acad. Sci. U.S.A. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosentino GP, Venkatesan S, Serluca FC, Green SR, Mathews MB, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 12.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc. Natl Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 14.Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 17.Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 18.Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasia RM, Mateos J, Bologna NG, Burdisso P, Imbert L, Palatnik JF, Boisbouvier J. Structure and RNA interactions of the plant MicroRNA processing-associated protein HYL1. Biochemistry. 2010;49:8237–8239. doi: 10.1021/bi100672x. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok KH, Ng MHJ, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 22.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 25.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr. Opin. Cell Biol. 2009;21:470–479. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 30.Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Güntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011;20:118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau PW, Potter CS, Carragher B, MacRae IJ. Structure of the human Dicer-TRBP complex by electron microscopy. Structure. 2009;17:1326–1332. doi: 10.1016/j.str.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J. Mol. Biol. 2008;384:967–979. doi: 10.1016/j.jmb.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benoit MP, Plevin MJ. Backbone resonance assignments of the micro-RNA precursor binding region of human TRBP. Biomol. NMR Assign. 2013 doi: 10.1007/s12104-012-9416-8. August 9 (doi:10.1007/s12104-012-9416-8; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 36.Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J. Magn. Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 39.Mulder FA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 40.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Horton J, Rowe A, Harding SE, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge: Royal Society of Chemistry; pp. 90–125. [Google Scholar]
- 41.Bonifacio GF, Brown T, Conn GL, Lane AN. Comparison of the electrophoretic and hydrodynamic properties of DNA and RNA oligonucleotide duplexes. Biophys. J. 1997;73:1532–1538. doi: 10.1016/S0006-3495(97)78185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc. Natl Acad. Sci. USA. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavallo L, Kleinjung J, Fraternali F. POPS: a fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 2003;31:3364–3366. doi: 10.1093/nar/gkg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Welch JL, Arnold JJ, Boehr DD. Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry. 2010;49:9361–9371. doi: 10.1021/bi100833r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Hartman E, Roy K, Chanfreau G, Feigon J. Structure of a yeast RNase III dsRBD complex with a noncanonical RNA substrate provides new insights into binding specificity of dsRBDs. Structure. 2011;19:999–1010. doi: 10.1016/j.str.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FHT. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14:345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 51.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 52.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 55.Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA. 2010;16:1570–1583. doi: 10.1261/rna.2111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 57.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 58.Nomura Y, Kajikawa M, Baba S, Nakazato S, Imai T, Sakamoto T, Okada N, Kawai G. Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2. Nucleic Acids Res. 2006;34:5184–5193. doi: 10.1093/nar/gkl664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 60.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl Acad. Sci. USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wostenberg C, Quarles KA, Showalter SA. Dynamic origins of differential RNA binding function in two dsRBDs from the miRNA “microprocessor” complex. Biochemistry. 2010;49:10728–10736. doi: 10.1021/bi1015716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akaike H. A new look at statistical model identification. IEEE Trans. Automat. Contr. 1973;19:716–723. [Google Scholar]
- 63.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2nd edn. Springer; 2004. [Google Scholar]
- 64.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.