Abstract
The homeobox transcription factor CDX2 plays a crucial role in intestinal cell fate specification, both during normal development and in tumorigenic processes involving intestinal reprogramming. The CDX2 regulatory network is intricate, but it has not yet been fully uncovered. Through genome-wide screening of a 3D culture system, the RNA-binding protein MEX3A was identified as putatively involved in CDX2 regulation; therefore, its biological relevance was addressed by setting up cell-based assays together with expression studies in murine intestine. We demonstrate here that MEX3A has a repressive function by controlling CDX2 levels in gastric and colorectal cellular models. This is dependent on the interaction with a specific binding determinant present in CDX2 mRNA 3′untranslated region. We have further determined that MEX3A impairs intestinal differentiation and cellular polarization, affects cell cycle progression and promotes increased expression of intestinal stem cell markers, namely LGR5, BMI1 and MSI1. Finally, we show that MEX3A is expressed in mouse intestine, supporting an in vivo context for interaction with CDX2 and modulation of stem cell properties. Therefore, we describe a novel CDX2 post-transcriptional regulatory mechanism, through the RNA-binding protein MEX3A, with a major impact in intestinal differentiation, polarity and stemness, likely contributing to intestinal homeostasis and carcinogenesis.
INTRODUCTION
The homeodomain transcription factor CDX2 is a critical determinant of intestinal homeostasis, both during development and throughout adult life. CDX2 is involved in the antero-posterior patterning of the mammalian embryo and is the key molecular mediator of intestinal differentiation (1–4). Furthermore, multiple evidences substantiate CDX2 crucial role in carcinogenesis of the digestive tract. It was shown to inhibit cell growth and migration in vitro as well as dissemination of colon tumour cells in vivo (5). CDX2 heterogeneous loss has also been observed in colorectal carcinomas (CRCs), particularly in invasive cells at the tumour edge (6). Moreover, CDX2 reduction increases the progression of chemically induced CRCs (7). Conversely, under certain pathological conditions, CDX2 becomes abnormally expressed in other organs of the digestive tract besides intestine, namely the esophagus (8) and stomach (9,10), driving a precancerous lesion known as intestinal metaplasia, a process confirmed in transgenic mouse models (11,12).
Owing to the essential function in intestinal development, differentiation and carcinogenesis, CDX2 regulation has been extensively studied. We have previously identified different mechanisms involved in the transcriptional regulation of this gene such as the Bone morphogenetic protein (BMP) pathway (13), SOX2 (14) and a CDX2 autoregulatory loop (15). Several transcription factors including HNF4α, GATA6, TCF4 and β-catenin were shown to interact with Cdx2 promoter fragments (16). However, mutations at the CDX2 locus are a rare event in CRC (17), and its expression does not depend on methylation of the proximal promoter (18). On the other hand, CDX2 protein phosphorylation has also been shown to modify its activity in intestinal cells (19,20). These and other studies support the notion that CDX2 regulation is intricate and strictly controlled.
During the past two decades, post-transcriptional regulation emerged as a fundamental mechanism guiding gene expression in higher eukaryotic cells, being at the core of normal cellular processes but also cancer initiation and development. It is now increasingly clear that RNA maturation, localization, translation and stability provide multiple layers of spatio-temporal control determining a transcript’s fate (21,22). These coupled events are generally dependent on the cooperation between cis-regulatory elements and trans-acting factors, such as non-coding RNAs and RNA-binding proteins (RBPs). RBPs have been implicated in virtually every aspect of RNA metabolism (22,23), particularly, their repressive role is critical to establish precise translational patterns that define developmental and differentiation switches in many organisms.
In Caenorhabditis elegans, MEX-3 is a translational repressor that regulates blastomere identity during early embryogenesis (24) and germline totipotency in the adult worm (25). MEX-3 has two K homology domains, which are conserved single-stranded RNA-binding motifs (26). Mutations disrupting mex-3 locus are lethal, resulting in embryos that inappropriately generate body-wall muscle from the anterior blastomere; hence, the name mex for ‘muscle excess’. This is specified, in part, by the repressive function that MEX-3 exerts over the transcription factor pal-1, the C. elegans orthologue of caudal in Drosophila and CDX in mammals (24,27). In humans, mex-3 was identified and characterized as four homologous genes, MEX3A–D (28,29), whose biological relevance is starting to be explored. Recently, the functional role of MEX3C as a RNA-binding ubiquitin E3 ligase was established, mediating the post-transcriptional decay of HLA-A allotypes (30). It was also shown to be necessary for normal postnatal growth in mutant mice by enhancing the local expression of insulin-like growth factor 1 in bone (31) and appears to be involved in metabolic regulation of energy balance (32), through yet unknown effectors. A variant form of MEX3D called TINO was shown to negatively regulate the antiapoptotic protein BCL-2 in HeLa cells (33). Finally, knockdown of MEX3A by siRNAs was shown to suppress cell proliferation and migration in human gastric cancer cells, but the molecular mechanisms behind these findings were not addressed (34). Pursuing the aim of uncovering new CDX2 regulatory mechanisms, we explored a putative translational repression by MEX3A, which was inversely correlated with CDX2 in a 3D experimental model. By studying MEX3A expression in vivo and using a cell line-based approach to modulate its levels in vitro, our study describes a novel post-transcriptional process by which CDX2 expression is impaired in the gastrointestinal setting and intestinal-like homeostasis compromised, through alterations in differentiation, polarity and stemness features. Another layer of control is thus added to the complex CDX2 regulatory network, involving MEX3A as a key regulator of intestinal homeostasis, which might have significant implications to gastrointestinal carcinogenesis.
MATERIALS AND METHODS
Cell culture and treatments
Human gastric carcinoma cell line AGS (ATCC, American Type Culture Collection) and CRC cell line Caco-2 (ATCC) were cultured under standard conditions in RPMI-1640 medium and Dulbecco’s modified Eagle’s medium, respectively, containing 10% fetal bovine serum, 100 U/ml of penicillin and 100 µg/ml of streptomycin (Life Technologies). For the AGS 3D culture, flasks were coated with 50 µl/cm2 of matrigel basement membrane (BD Biosciences) at a 1.5:1 proportion to serum-free medium. A 2 × 104 cells/cm2 suspension was seeded on top and maintained for 14 days with medium change every 2 days. For the Caco-2 3D culture, coverslips were coated with 60 µl/cm2 of matrigel. A 6 × 103 cells/cm2 suspension plus 2% matrigel was seeded on top and maintained for 8 days with medium change every 2 days. To quantify lumen formation, >100 cysts were microscopically examined. For proteasome inhibition, cells were treated with 25 µM MG132 (Calbiochem) or vehicle treated with DMSO. To inhibit transcription, cells were exposed to 10 µg/ml of Actinomycin D (Sigma).
Constructs and site-directed mutagenesis
The previously published pCMV-MEX3A expression vector (28) was used together with a pCMV-Tag3B empty vector (Agilent Technologies) in transfections. A pRLControl construct containing a humanized Renilla luciferase (Rluc) coding sequence (35) was used as a backbone to create the pRLCDX2 vector, encoding a luciferase fusion transcript to the parental CDX2 3′untranslated region (UTR). The QuickChange site-directed mutagenesis kit (Stratagene) was then used to introduce specific mutations in the previous plasmid, to generate the pRLΔCDX2 construct, with a mutated MEX-3 recognition element (MRE). Oligonucleotides containing the desired mutations were designed according to the manufacturer’s instructions (Supplementary Table S1). Taking into account the degenerate consensus sequence described for MEX-3 binding in C. elegans, a mutational background of Cytidine was used because presumably this base is not tolerated in the MRE (36).
Transfections, RNAi and luciferase assays
Transient transfections were done using Lipofectamine 2000 reagent according to the manufacturer’s guidelines (Life Technologies). A DNA (µg) to Lipofectamine 2000 reagent (µl) ratio of 1:1.5 in OPTI-MEM medium was used for routine transfection experiments of expression vectors. For stable transfections, selection was initiated 48 h post-transfection in medium supplemented with 0.6 mg/ml of G418 (Sigma). Neomycin-resistant positive clones obtained through limiting dilution were routinely maintained with 0.2 mg/ml of G418. A commercial set of three Stealth Select siRNA duplexes (HSS150674, HSS150675 and HSS150676) directed against human MEX3A (Life Technologies) and a custom set of three siRNA duplexes directed against CDX2 were used with scrambled controls. An siRNA duplexes (pmol) to Lipofectamine 2000 reagent (µl) ratio of 20:1 in OPTI-MEM medium was used for inhibition experiments. Luciferase reporter assays were performed with the Rluc Assay System (Promega), and β-galactosidase activity was used for normalization of experimental variations.
RNA isolation and quantitative real-time PCR
Total RNA was extracted using TRI Reagent (Sigma) and reverse transcribed using the Superscript II Reverse Transcriptase kit (Life Technologies). Analysis of BMI1, CDX2, GAPDH, LGR5, MEX3A and Rluc mRNA expression was performed in an ABI Prism 7500 system using SYBRgreen reagent (Life Technologies) and specific primer pairs (Supplementary Table S1). Each sample was amplified in triplicate and specificity confirmed by dissociation analysis. The 18S rRNA expression was measured for normalization of target gene abundance.
Microarrays and data processing
Experiments were performed at the National Facility for DNA Microarrays (Aveiro, Portugal). Three independent 2D and 3D AGS cell cultures were selected for total RNA extraction using TRI Reagent. RNA quantity and quality were assessed using the Nanodrop ND-1000 (Thermo Scientific) and 2100 Bioanalyzer (Agilent Technologies) systems, and only samples with a RNA integrity number above nine were considered for further study. Preparation and labelling of the RNA was performed using the One-Color Microarray-Based Gene Expression Analysis Quick Amp Labelling kit (Agilent Technologies). Briefly, 600 ng of total RNA from 2D and 3D biological replicas was used as input together with spike-in controls to generate Cyanine 3-labelled cRNA. The amplified cRNA samples were purified with RNeasy Mini kit (Qiagen) and hybridized to Human Gene expression 4 × 44K v2 Microarray slides (Agilent Technologies) at 65°C for 17 h. After washing procedures, images of the hybridizations were acquired using a G2505B Microarray Scanner (Agilent Technologies). The Feature Extraction software was used for spot identification, background subtraction and quantification of the fluorescent signal. Raw expression data were processed using BRB-ArrayTools v3.8.1 software (37). After base 2 log transformation and average of probe replicates, normalization was applied using the median intensity over the entire array to minimize systematic variance. Differentially expressed transcripts between the two culture conditions were filtered using the Class Comparison tool, performing an unpaired sample t-test with a P < 0.01 and considering a minimal 1.5-fold change. Hierarchical clustering analysis of significantly altered genes was conducted in TIGR MultiExperiment Viewer v4.8.1 software (38) using Euclidean correlation and average linkage clustering, and expression values indicated colorimetrically. Functional annotation of differentially expressed genes, identified by extending the unpaired t-test parameter to P < 0.05 and maintaining the minimal fold change was performed with DAVID program (39). Microarray data have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) and assigned the identifier E-MTAB-1234. The complete list of differentially expressed genes is detailed in Supplementary Table S2.
Protein extraction and western blot analysis
Cells were lysed for 30 min on ice in a lysis buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid and 1% IGEPAL (Sigma), supplemented with Complete protease inhibitor cocktail (Roche Applied Science), 1 mM PMSF and 1 mM Na3VO4. Lysates were centrifuged at 12 000 rpm for 20 min at 4°C and the supernatant recovered. Protein concentration was determined using the BCA Protein Assay Reagent (Thermo Scientific). Protein extracts (30–50 µg) were run on 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, blotted overnight with appropriate antibodies (Supplementary Table S1), and signals revealed with ECL detection kit (GE Healthcare Life Sciences). Actin levels were used to normalize protein expression, and quantification of western blots was performed using Fiji software (40).
Flow cytometry
Caco-2 cells were harvested 48 h after transfection at confluence and fixed with 1% paraformaldehyde for 20 min at room temperature followed by permeabilization with 0.1% Triton X-100 (Sigma) for 5 min on ice. Staining was performed with anti-MEX3A antibody and visualized with goat anti-rabbit FITC-conjugated secondary antibody. For DNA content assessment, cells were incubated with a propidium iodide (50 µg/ml) and RNAse A (200 µg/ml) solution (Sigma) for 30 min at room temperature. Samples were read in a FACSCanto II (BD Biosciences) flow cytometer, and analysis was performed using the FlowJo software.
RNA-immunoprecipitation assay
Cells were lysed 48 h after transfection as indicated for protein extraction but with the addition of 20 U/ml of RNAseOUT Ribonuclease inhibitor (Life Technologies). Before the lysis procedure, cells were washed with phosphate buffered saline (PBS) and subjected to ultraviolet crosslink (254-nm wavelength) at an energy level of 1.8 J/cm2 on ice for 20 min. Immunoprecipitation from protein extracts (750 µg) was performed overnight with anti-myc antibody (Clontech) or normal mouse immunoglobulin G (IgG) (Millipore) and RNA-immunocomplexes recovered by incubation with protein G-sepharose bead slurry (Sigma) for 2 h in the following day. After five washes in lysis buffer containing RNAseOUT, RNA was extracted from the bead slurry as indicated before and resuspended in 10 µl of dH2O. RT-quantitative real-time PCR (qPCR) analysis was directly performed on the RNA as described. Target mRNA levels immunoprecipitated in the MEX3A transfected cells were normalized to the levels recovered in the empty vector transfected cells.
Immunofluorescence and microscopy
Cells grown on coverslips were fixed using ice-cold methanol for 10 min. For Caco-2 3D assay, cysts were fixed with a 2% paraformaldehyde and 0.05% glutaraldehyde solution at room temperature for 20 min, followed by quenching with 0.1% NaBH4 and permeabilization with 0.5% Triton X-100. Paraffin-embedded wild-type murine tissue sections (4 μm) were deparaffinized with clear-rite (Thermo Scientific) and rehydrated. All samples were blocked with non-immune serum diluted in 10% BSA–PBS, followed by incubation with appropriate antibodies (Supplementary Table S1) diluted in 5% BSA–PBS. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma), and coverslips mounted on microscope slides with Vectashield (Vector). Imaging acquisition was based on optical sectioning and adjusted for brightness and contrast with Fiji software (40). Imaging was performed on an Axio Imager Z1 (Zeiss) Fluorescence microscope with an ApoTome attachment (Figures 3, 7 and 8 and Supplementary Figure S4), and on a TCS SP5 II (Leica) Laser Scanning Confocal microscope (Supplementary Figure S1). The middle focal plane of Z-stack images with similar total number of planes were used for representation of cysts polarity.
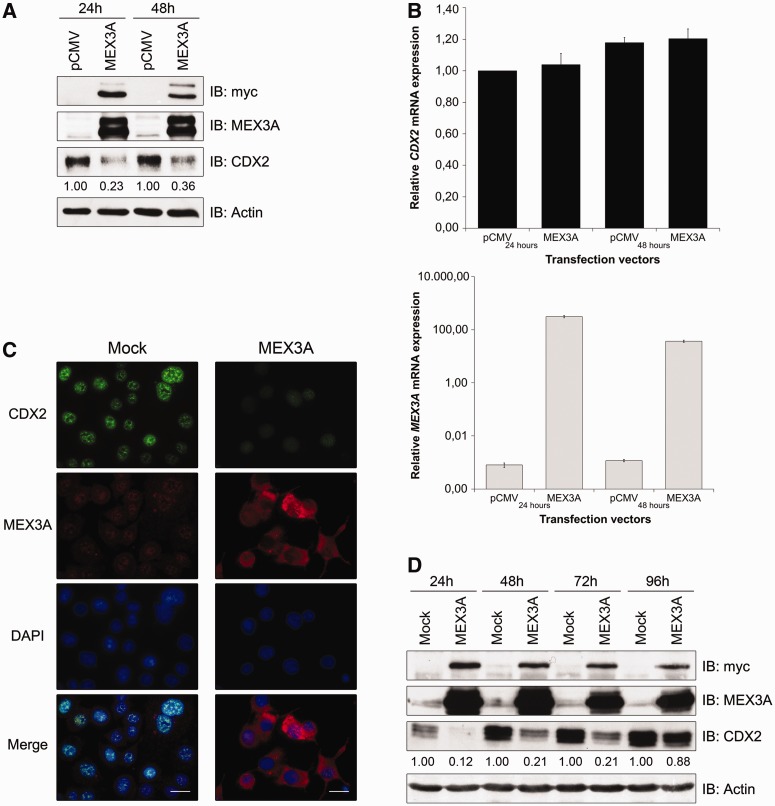
Figure 3.
CDX2 regulation by MEX3A in AGS cells. (A) Western blot of transient transfections with a myc-tagged MEX3A expression vector. (B) qPCR of CDX2 and MEX3A mRNA expression in the previous samples. The value for CDX2 mRNA expression in the empty vector transfected cells at 24 h was referred to as 1. (C) Immunofluorescence for MEX3A and CDX2 in MEX3A stably transfected cells at 48 h culture (original magnification,×400; scale bar 20 µm). (D) Western blot of MEX3A and CDX2 expression in stably transfected cells at different time points.
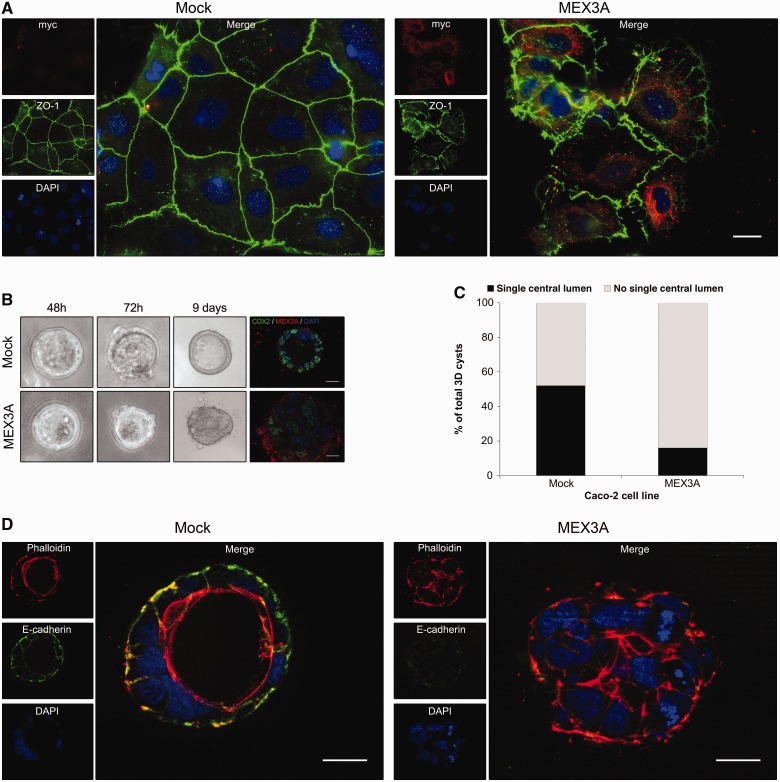
Figure 7.
Polarity alterations induced by MEX3A in Caco-2 cells. (A) Immunofluorescence showing ZO-1 expression in Caco-2 mock and MEX3A stably transfected cell lines at day −2 of culture (original magnification,×400). (B) Morphology of Caco-2 mock and MEX3A cysts in bright field microscopy during 3D culture (original magnification, ×100) and CDX2/MEX3A expression (original magnification, ×630). (C) Quantification of cysts with lumen or no lumen at culture day 8. (D) Expression of E-cadherin and Phalloidin staining in Caco-2 cysts (original magnification, ×630; all scale bars 20 µm).
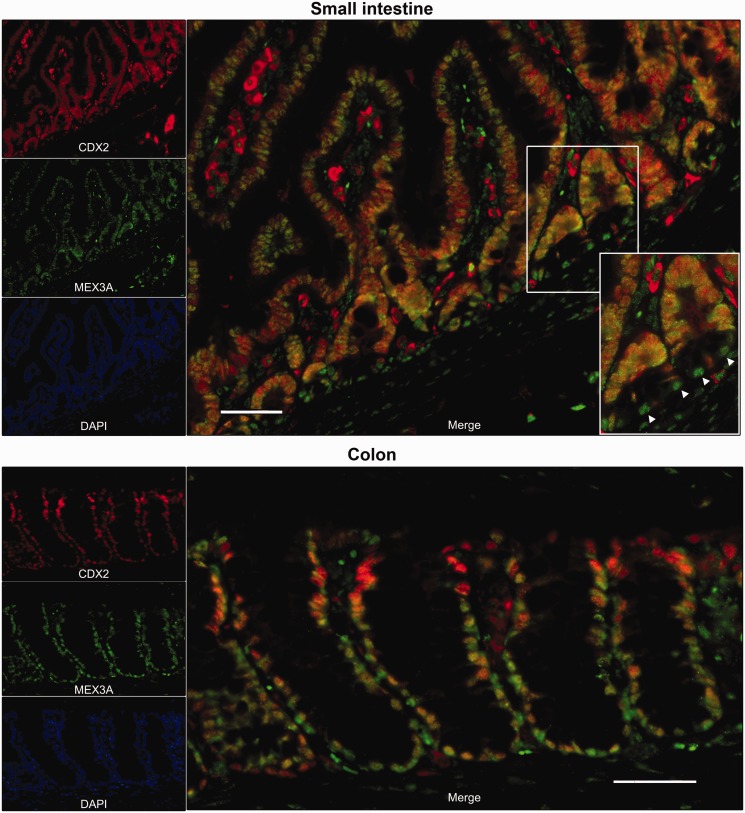
Figure 8.
Expression patterns of MEX3A and CDX2 in mouse normal intestine. Representative immunofluorescence data for MEX3A and CDX2 in small intestine and colon are shown (original magnification, ×200; scale bars 50 µm; insert original magnification, ×400).
Statistics
Each experiment was carried out in triplicates at least three times, and data were expressed as means ± SD. Statistical analysis was performed using Student’s t-test. A P < 0.05 was considered as significantly different.
RESULTS
CDX2 expression is downregulated in AGS cells cultured in matrigel
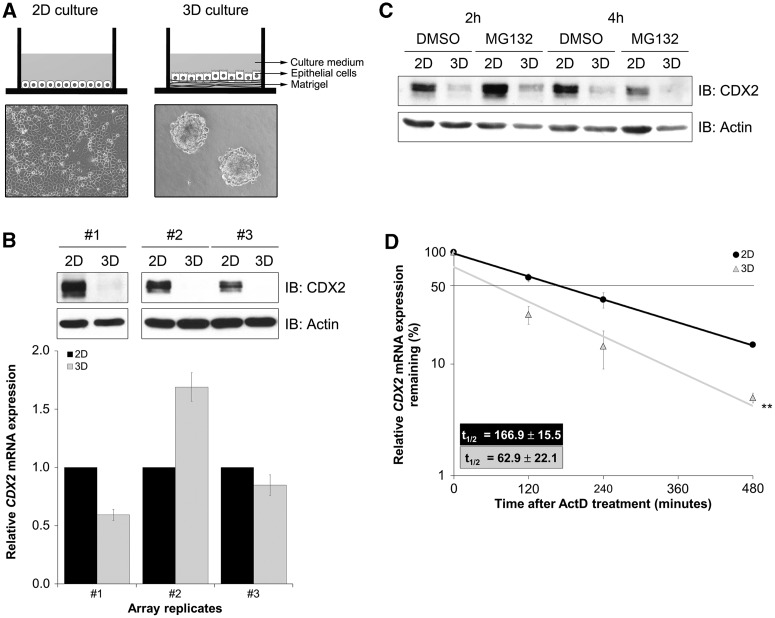
In an effort to study CDX2 regulation in a physiologically relevant microenvironment, we compared CDX2 expression in AGS cells cultured in a standard 2D condition versus a 3D condition with matrigel. Cells depicted distinct growth patterns amongst both settings, forming a compact monolayer and conspicuous multicellular aggregates, respectively (Figure 1A). Although there was a dramatic decrease in CDX2 protein levels in the 3D culture, mRNA showed heterogeneous levels without correlation with protein (Figure 1B). Treating cells with the proteasome inhibitor MG132 indicated that CDX2 downregulation was not related to an increase in proteasome-dependent degradation (Figure 1C). On the other hand, we found a significant difference in CDX2 mRNA stability after actinomycin D treatment to inhibit transcription (Figure 1D). The CDX2 mRNA half-life value was higher for the 2D culture (166.9 ± 15.5 min) than for the 3D culture (62.9 ± 22.1 min). These results suggest the existence of a post-transcriptional regulatory mechanism, probably acting over CDX2 mRNA.
Figure 1.
Establishment of AGS 3D cultures and characterization of CDX2 expression. (A) Schematic representation of the culture systems and cellular morphology at culture day 4 in bright field microscopy (original magnification, ×100). (B) Western blot and qPCR of CDX2 expression in 2D and 3D after 2 weeks culture. Cardinal numbers represent biological replicates. Values for CDX2 mRNA expression in 2D culture were referred to as 1. (C) Western blot of CDX2 protein expression on treatment with MG132. (D) qPCR of CDX2 mRNA expression on treatment with actinomycin D. Expression levels in the absence of treatment were set at 100%. Depicted half lives were calculated using exponential regression (**P = 0.003).
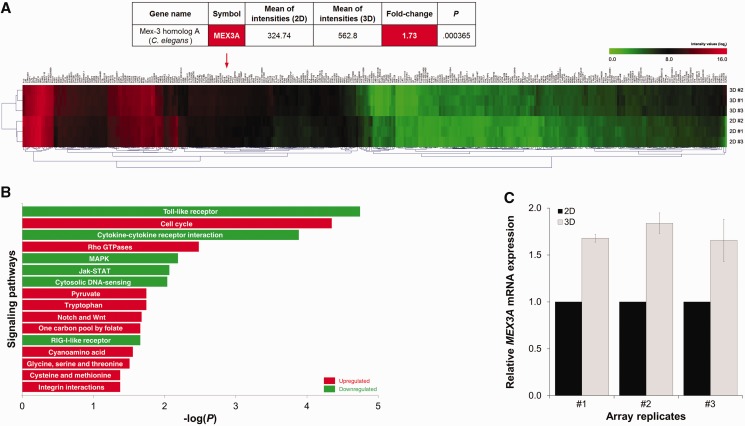
With the objective of pinpointing the mechanism underlying CDX2 downregulation, we performed a whole-genome expression array, comparing the transcriptome of AGS cells in the two culture conditions. Among the 340 differentially expressed transcripts, a 1.7-fold upregulation of MEX3A was observed in the 3D condition (Figure 2A and Supplementary Table S2). Additionally, functional annotation analysis showed alterations related to fundamental cellular processes like proliferation, differentiation and survival, namely an increase in cell cycle elements, Notch and Wnt pathways and a decrease in MAPK and Jak-STAT signalling in the 3D model (Figure 2B). An increase in RhoGTPases and integrin signalling was also registered, supporting the occurrence of increased cell-matrix interactions. Changes in metabolic patterns were evident by an increase in amino acid biosynthesis (Figure 2B). MEX3A is one of the human orthologues of MEX-3, an RBP with translational inhibitory function known to target the CDX orthologue pal-1 during C. elegans embryonic development. A novel interaction between human MEX3A and CDX2 in a gastrointestinal context could constitute a plausible explanation for the reduction of CDX2 protein levels observed in 3D culture. Accordingly, MEX3A expression was validated by qPCR (Figure 2C).
Figure 2.
Transcriptome profiling of 2D and 3D AGS cultures. (A) Microarray heat map displaying differentially expressed transcripts (P < 0.01, fold change > 1.5). Values obtained for MEX3A are highlighted. (B) Functional annotation analysis depicting the preponderance of differentially expressed genes in terms of signaling networks (P < 0.05, fold change > 1.5). (C) qPCR validation of MEX3A expression. Values for MEX3A mRNA expression in 2D culture were referred to as 1.
MEX3A overexpression leads to CDX2 downregulation in AGS cells
To define whether CDX2 expression is regulated by MEX3A, AGS cells cultured in standard conditions were transiently transfected with a myc-tagged MEX3A expression vector and collected 24 and 48 h later. We showed that the myc-tagged MEX3A fusion protein was detectable on transfection and accompanied by a 77 and 64% decrease in CDX2 protein expression, respectively (Figure 3A). In contrast, CDX2 mRNA expression did not change (Figure 3B). This result was confirmed in AGS cells stably transfected with the myc-tagged MEX3A construct up until 96 h of culture (Figure 3C and D).
It was previously published that MEX3A and MEX3B were novel components of P bodies (28,29), discrete cytosolic foci characterized in yeast and mammals as centres for translational silencing and mRNA decay (41). In our setting, on MEX3A-induced CDX2 protein decrease, we observed a specific subcellular localization of MEX3A in P bodies, as confocal microscopy analysis revealed a partial overlap between myc-tag staining and two known P body components, the human mRNA-decapping enzyme 1A (DCP1A) and the human enhancer of mRNA-decapping protein 4 (EDC4) (Supplementary Figure S1). Together, these results demonstrate that MEX3A has the ability to regulate CDX2 expression at the post-transcriptional level, probably by interplay with P bodies.
MEX3A interacts with a MRE present in CDX2 3′UTR
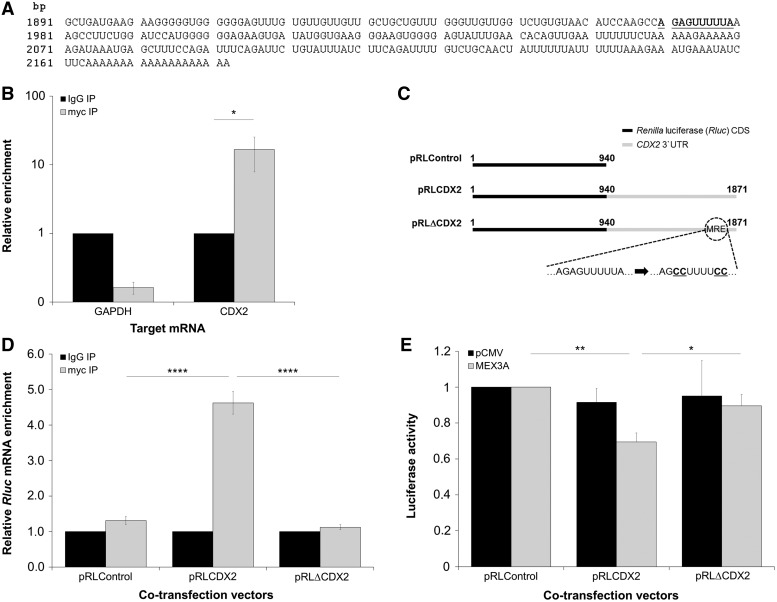
A bioinformatics search conducted throughout the CDX2 3′UTR led to the identification of the MEX-3 degenerate consensus binding sequence (36). The MRE present in CDX2 mRNA was defined as a bipartite element that consists of AGAG and UUUA motifs separated by two Uracil bases (Figure 4A). To assess whether MEX3A associates with CDX2 3′UTR, we preserved RNA–protein interactions by ultraviolet covalent linkage in AGS cells transiently transfected with the MEX3A expression vector and performed an RNA-immunoprecipitation (RIP) assay. We found a significant enrichment in CDX2 mRNA recovered with the anti-myc antibody compared with control IgG (Figure 4B). On the contrary, no enrichment for GAPDH mRNA was observed (Figure 4B). Likewise, no enrichment for TBP mRNA and 18S rRNA was detected (data not shown). Next, we asked whether MEX3A binds specifically to the MRE present within the CDX2 3′UTR. To address this question, Rluc expression vectors containing the wild-type CDX2 3′UTR (pRLCDX2) or the CDX2 3′UTR with a mutated MRE (pRLΔCDX2) were used in an RIP assay as before (Figure 4C). A complete loss in MEX3A binding capacity was detected when the MRE was mutated, as observed by comparing the levels of Rluc mRNA recovered with anti-myc antibody between the pRLCDX2 and pRLΔCDX2 transfected cells (Figure 4D). Additionally, luciferase activity assays proved that the construct with the mutated MRE was insensitive to MEX3A function, as a significant reduction in Renilla expression was only detected with the pRLCDX2 transfection (Figure 4E). We conclude that MEX3A is able to interact with CDX2 mRNA and that the major determinant for this interaction is a putative MRE located in the transcript 3′UTR.
Figure 4.
Mechanistics of MEX3A interaction with CDX2 mRNA in AGS cells. (A) Terminal sequence of CDX2 mRNA, with the predicted MRE in the 3′UTR highlighted in bold and underlined. (B) qPCR showing CDX2 and GAPDH mRNAs immunoprecipitated with myc-tag antibody in MEX3A transfected cells (*P = 0.018). The values for CDX2 and GAPDH mRNA levels in the IgG sample were referred to as 1. (C) Schematic representation of pRL constructs. (D) qPCR showing Rluc mRNA immunoprecipitated with myc-tag antibody for pRLCDX2 transfection (****P < 0.0001). Values for Rluc mRNA expression in IgG samples were referred to as 1. (E) Luciferase activity assay for the different pRL constructs (**P = 0.006 for pRLControl/pRLCDX2 and *P = 0.036 for pRLCDX2/pRLΔCDX2). The values obtained for luciferase expression in the pRLControl co-transfected cells were referred to as 1.
Intestinal phenotype of Caco-2 cells is affected by modulating MEX3A levels
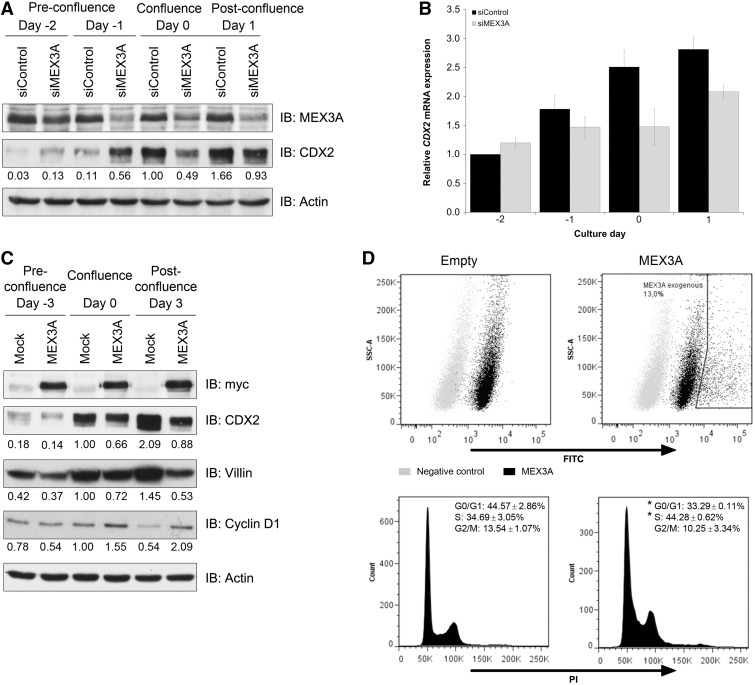
To assess the functional consequences of the previous observations and confirm that they were cell-type independent, we studied CDX2 regulation by MEX3A in another model. Caco-2 cell line was chosen for its ability to spontaneously differentiate into an enterocytic-like phenotype on reaching confluence, which has turned it into the most widely used system to study intestinal cell maturation (42). Furthermore, this process was previously shown to be accompanied by an increase in CDX2 protein expression that, in some established Caco-2 clones, is not explained by higher transcriptional levels (20). First, we evaluated the timing of CDX2 and MEX3A expression in Caco-2, harvesting cells at pre-confluence (day −2), confluence (day 0) and post-confluence (day 2–8) time points. Consistent with previous reports, CDX2 protein expression progressively increased starting from confluence and during differentiation (Supplementary Figure S2A). This trend was also verified at the mRNA level, though not to the same extent (Supplementary Figure S2B). By contrast, MEX3A mRNA expression varied in a complementary manner to that of CDX2, with a decreased expression associated to more differentiated cells (Supplementary Figure S2B), although a protein decrease was only visible at day 8 (Supplementary Figure S2A). We inhibited endogenous MEX3A using siRNA and observed that CDX2 protein started to be highly expressed at earlier time points in cells with downregulated MEX3A expression (Figure 5A). Paradoxically, this effect was reverted from confluence onwards, with lower CDX2 protein levels detected at later time points in cells with MEX3A inhibition (Figure 5A). To evaluate whether this result was dependent on MEX3A activity or due to transcriptional regulation, CDX2 mRNA expression was assessed. As expected, CDX2 mRNA levels were not altered in the pre-confluence time points but were downregulated from confluence onwards (Figure 5B), suggesting that after confluence, other MEX3A regulatory functions indirectly interfere with endogenous CDX2 levels.
Figure 5.
MEX3A modulation of intestinal phenotype in Caco-2 cells. (A) Western blot of MEX3A inhibition with transfection of siRNAs performed at day −3 and day −1. (B) qPCR of CDX2 mRNA expression in the same samples. The value for CDX2 at day −2 for the siControl sample was referred to as 1. (C) Western blot of MEX3A stably transfected cells at different confluences for differentiation and proliferation markers. (D) Flow cytometry analysis of MEX3A transiently transfected cells. Dot plots depicting negative control (secondary antibody only) and FITC-conjugated MEX3A expression levels are shown on top. DNA content histograms showing population percentages for the different cell cycle phases of the empty vector transfected cells and MEX3A-transfected cells (gate MEX3A exogenous) are shown below (*P = 0.03 for G0/G1 phase and *P = 0.05 for S phase).
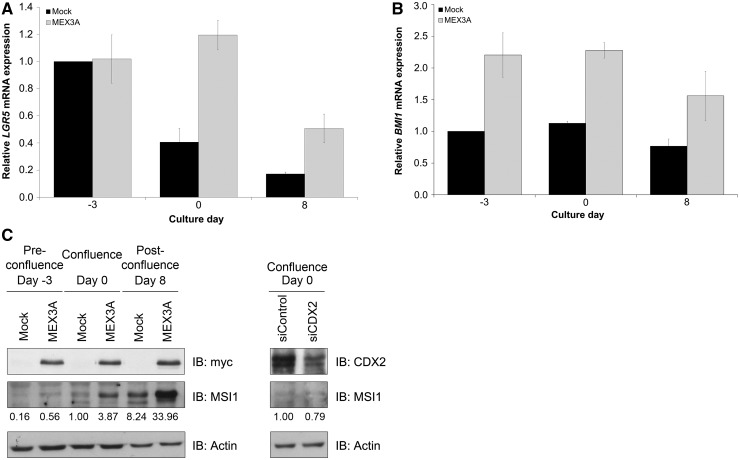
It is known that Caco-2 cells acquire expression of different intestinal columnar lineage markers on differentiation, such as Villin. To determine the effect of MEX3A overexpression in intestinal differentiation, we generated a Caco-2 cell line stably transfected with the myc-tagged MEX3A construct. Expression of the myc-tag was confirmed in different confluence states, and a concomitant reduction in CDX2 protein levels was observed (Figure 5C). Furthermore, we detected decreased Villin expression in MEX3A overexpressing cells (Figure 5C). As differentiation and growth arrest are spatially and temporally related events in the intestinal epithelium, we studied the expression of cyclin D1, a positive regulator of proliferation required for cell cycle G1/S transition. Sustained levels of cyclin D1 were detected in MEX3A overexpressing cells in latter time points, when the culture is normally already less proliferative, as observed by the striking decrease of cyclin D1 in mock cells (Figure 5C). To ascertain the cell cycle distribution related to MEX3A overexpression, we transiently transfected near-confluent Caco-2 cells with the myc-tagged MEX3A construct and performed DNA content analysis by flow cytometry. Measurements indicated that cells overexpressing MEX3A possess a different cell cycle profile (Figure 5D), with a reduced G0/G1 population (32 against 49% in the empty transfected cells) and an increased S phase population (41 against 30% in the empty transfected cells). Given the observed alterations in differentiation and proliferation parameters, we sought to determine whether MEX3A overexpressing cells possessed intestinal ‘progenitor or stem-like’ features. Assessment of expression of well-established intestinal stem cell markers (43–46) revealed that these cells show overall increased levels of LGR5, BMI1 and MSI1 (Figure 6), whereas OLFM4 basal expression was barely detectable and did not change (data not shown). Importantly, this seems to be independent of CDX2, as siRNA-mediated CDX2 downregulation did not reproduce the increase in MSI1 levels (Figure 6C).
Figure 6.
Expression profiles of intestinal stem cell markers in Caco-2 cells. (A) qPCR of LGR5 mRNA expression in mock and MEX3A overexpressing cells. The value for LGR5 at day −3 for the mock sample was referred to as 1. (B) qPCR of BMI1 mRNA expression in the same samples. The value for BMI1 at day −3 for the mock sample was referred to as 1. (C) Western blot analysis of MSI1 expression in the same samples and in Caco-2 parental cell line with CDX2 inhibition by siRNAs at the confluence time point.
Caco-2 cells retain the ability to polarize and form a transporting epithelial monolayer in culture (42). This polarization process is intimately connected with the formation of cell–cell contacts and adhesion. In standard culture conditions, Caco-2 cells overexpressing MEX3A showed alterations in the distribution of the tight junction marker ZO-1 compared with the mock cells (Figure 7A). We then used a 3D cell cyst assay (4) in matrigel to compare the phenotypic response of both cell lines. We observed that mock cells developed cyst-like structures in a few days, with ∼50% showing a well-defined central lumen (Figure 7B–D). In contrast, >80% of the MEX3A-expressing cysts, in which CDX2 reduction and MEX3A increase was confirmed, failed to elaborate such a lumen (Figure 7B–D). Additionally, we observed distinct patterns of E-cadherin and Phalloidin expression. We detected basolateral E-cadherin expression and accumulation of filamentous-actin (F-actin) around the hollow lumen in the mock cysts (Figure 7D), and a reduced E-cadherin expression in the MEX3A-expressing cysts, with F-actin appearing ubiquitously distributed (Figure 7D). Thus, we show that the establishment of proper apical-basal identity is affected on MEX3A overexpression.
MEX3A is differentially expressed in the crypt-villus unit
To address MEX3A relevance in vivo, we analysed its presence in mouse normal intestine and compared it with CDX2 expression. We observed that MEX3A is expressed in small intestine and colon, exhibiting mainly a nuclear staining pattern. It shows a stronger expression in the lower portion of the crypt-villus unit in small intestine and reduced expression in the uppermost part of the colonic crypts (Figure 8). In the small intestine, CDX2 is expressed in the same cells as MEX3A except in the base of the crypts (Figure 8, insert) that exhibit mainly MEX3A protein. In colon, CDX2 is more expressed in the surface cells (Figure 8). These results suggest that MEX3A expression might be important to define intestinal differentiation patterns.
DISCUSSION
In this study, we provide the first demonstration that MEX3A represses CDX2 expression in the gastrointestinal context, putatively as a translational repressor, with direct implications in intestinal differentiation, polarity and stemness features, key mechanisms for tissue homeostasis that are frequently altered in pathological conditions and inextricably linked with cancer.
3D cell culture systems have been shown to enable physiological and functional differentiation of several epithelial cell types (47,48), constituting a promising alternative to overcome standard cell culture limitations. Accordingly, our transcriptomic analysis performed on a 3D model of AGS cells, which have a significant level of endogenous CDX2 responsive to different molecular stimuli (13–15), allowed us to disclose MEX3A as a molecular player involved in the regulation of CDX2 translation. The array presented an increased MEX3A expression in 3D culture and no alteration in the levels of the other MEX3 family members. We further demonstrated that MEX3A overexpression leads to marked CDX2 protein decrease in two cell lines. We tried to overcome the limitation of using overexpression systems by using two different cell lines that were both transiently and stably transfected with a MEX3A plasmid, which gave concordant results. In addition, CDX2 negative regulation by MEX3A was confirmed in a more physiological context using a siRNA approach towards endogenous MEX3A in Caco-2 cells. We proved that MEX3A is able to interact with CDX2 mRNA through a canonical MRE present in the 3′UTR. This sequence, which we now show to be functionally relevant in humans, seems to be the single determinant of MEX3A binding, independently of the upstream 5′coding region. In fact, bioinformatics analysis shows that the degenerate MRE motif has been evolutionarily conserved in different CDX2 homologues (Supplementary Figure S3), namely in chimpanzee, mouse, rat, zebrafish, fruit fly and frog, suggesting that MEX3A is critical for CDX2 regulation.
Endogenously, MEX3A was mainly localized in the nuclear compartment of intestinal epithelial cells, both in vitro and in vivo. This result is crucial to show that a biological background for the regulation of CDX2 by MEX3A exists. In this regard, a variant form of MEX3D called TINO, which has been shown to negatively regulate BCL-2 expression by transcript destabilization, is also predominantly localized in the nuclei of HeLa cells (33). On the other hand, MEX3A transfectants showed predominant cytoplasmic staining. A potential problem could be antibody specificity; however, this was successfully evaluated by transfection of siRNA duplexes directed against MEX3A (Supplementary Figure S4). Therefore, the differential localization in distinct expression backgrounds might be because a certain threshold in expression levels has to be achieved for cytoplasmic translocation to occur, given that MEX3 proteins are capable of performing nucleocytoplasmic shuttling (28). On the other hand, specific subcellular distribution might be related with protein phosphorylation, in accordance with published data describing MEX3 members as phosphoproteins (28,29). Whichever the case, this does not contradict post-transcriptional regulation, as it can be elicited at multiple points of the transcript lifespan, including pre-mRNA processing in the nucleus, export from the nucleus to the cytoplasm and subsequent coordinated trafficking of the mature mRNA to the translation machinery. It also remains to be fully clarified the consequence of partial accumulation in P bodies, structures involved in processes of mRNA degradation, nonsense-mediated mRNA decay, translational repression and RNA-mediated gene silencing (49), although there are several RBPs with established roles in translational regulation known to colocalize with P bodies (50,51).
We used the Caco-2 cell line model to modulate MEX3A expression and assess its phenotype. Surprisingly, MEX3A inhibition produced distinct effects over CDX2 levels depending on cellular confluence. MEX3A might selectively regulate unique subsets of targets in different culture conditions. In agreement, it is known that transcriptomic and proteomic changes occur during the progression from the proliferative state to spontaneous differentiation of Caco-2 cells. Conversely, MEX3A might have a dual role, acting both as a repressor or enhancer contingent on the cellular microenvironment, as observed for other RBPs, like HuR (52). It is possible that intricate associations of MEX3A with other molecular effectors, namely other proteins or microRNAs, determine divergent regulatory endings. MEX3A overexpression in Caco-2 cells resulted in pronounced phenotypic alterations. CDX2 and Villin downregulation were indicative of a loss in intestinal differentiation, which was further confirmed by the flow cytometric profile, showing reduced G0/G1 population, usually associated with less-differentiated cells. Another hallmark feature of MEX3A overexpression was the altered cellular polarization in standard culture, as well as the impaired ability to form polarized structures in the presence of matrigel. These effects may be mediated by CDX2, as this transcription factor was previously shown to regulate intestinal Villin through recruitment of the Brm-type SWI/SNF complex to its promoter (53), and MEX3A-expressing cysts closely resemble the ones obtained with Caco-2 cells in which CDX2 suppression was achieved by lentiviral short-hairpin RNA particles (4). It is, therefore, important to ascertain the biological setting where this regulation might be determinant, which is suggested by the predominant expression of MEX3A in the stem, transit-amplifying and migrating post-mitotic cells of the intestine. Although CDX2 protein can be detected in most of these cells, its level is lower in the crypts compared with the uppermost differentiated cells of the villi (2,19,20). A similar increasing bottom-up gradient has been described along the colonic gland axis in the distal colon epithelium. By contrast, in situ hybridization revealed that CDX2 mRNA was homogeneously distributed along the entire crypt-villus axis (20). Given this CDX2 protein expression gradient, that we also show, and lack of correlation with mRNA, it is likely that MEX3A fine-tunes CDX2 levels in vivo as well, in a transcription-independent manner, providing swift availability of the protein to meet the physiological requirements of the continuously renewed gut epithelium. Moreover, it has been shown that intestinal stem cells cannot differentiate into any of the intestinal lineages in a background of Cdx2 ablation (54), revealing the need for tight CDX2 regulation as determinant to proper phenotype switching. Most interestingly, we observed higher expression of different intestinal stem cell markers when we overexpressed MEX3A in Caco-2, suggesting that this protein is associated with stem cell features. Our AGS 3D model also showed a significant upregulation of OLFM4 (Supplementary Table S2), previously identified as a marker for LGR5+ stem cells in human intestine (46), although this gene was not directly upregulated in Caco-2 cells, which might be due to intrinsic properties of each cell line. Strengthening the hypothesis for a role of MEX3A in stem cell potential, a recent publication showed that MEX3A is part of the molecular signature of the LGR5+ intestinal stem cells, presenting a 1.64-fold increase in relation to daughter cells, along with MSI1, LGR5 and OLFM4 (55). Furthermore, our study reinforces the increasing knowledge that other regulatory mechanisms in addition to transcriptional ones have important functions in stem cells. Although we do not know yet how MEX3A relates with the intestinal stem cell phenotype, we hypothesize that MEX3A overexpression in itself, together with the induction of a lessened differentiated phenotype and polarity defects mediated by CDX2 downregulation, might be critical to allow a permissive environment for the appearance of stemness features. Furthermore, the MEX3A–CDX2 axis might be important during early embryogenesis where CDX2 is required for correct trophectoderm differentiation, while absent from the inner cell mass, in a process that requires tight regulation (56). Interestingly, the pattern of expression of the C. elegans orthologues of both proteins was found to be mutually exclusive in early stages of embryogenesis (24,27). Still, it remains to be assessed the relevance of MEX3A in multiple pathological contexts of the gastrointestinal tract where differentiation abnormalities directed by CDX2 are key events.
In conclusion, we have identified a novel role for MEX3A protein in the regulation of intestinal differentiation, polarity and stemness features, partially mediated by the repression of CDX2. This is the first description of a CDX2 regulatory mechanism based on its mRNA control by an RBP, having a significant impact in intestinal homeostasis and likely in gastrointestinal carcinogenesis.
ACCESSION NUMBERS
The microarray data from this publication have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) and assigned the identifier E-MTAB-1234.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–4.
FUNDING
Fundação para a Ciência e a Tecnologia (FCT)—Programa Operacional Ciência e Inovação 2010 do Quadro Comunitário de Apoio III and FEDER [PTDC/SAU-OBD/64490/2006]. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partially supported by FCT. B.P. and R.B. acknowledge FCT for financial support [SFRH/BD/43168/2008 and SFRH/BPD/68276/2010, respectively]. M.B. was supported by a grant from the Fondation ARC, and N.T.C. was the recipient of a fellowship from the Fondation pour la Recherche Médicale. Funding for open access charge: FCT [SFRH/BD/43168/2008].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Professor Leonor David for fruitful discussion, valuable experimental advice throughout the project and critical reading of the manuscript. They thank Dr Catarina Leitão for technical assistance in flow cytometry and Dr Patrícia Castro for technical assistance in confocal microscopy. The overall study was coordinated by R.A. with contributions from M.B.; B.P., S.S., R.B. and L.C. performed the experiments; M.P., JP.R. and JN.F. provided essential reagents; B.P. and R.A. conceived and designed the experiments; B.P. and R.A. wrote the manuscript; all authors contributed to different aspects of data analysis and critical revision of the manuscript.
REFERENCES
- 1.Beck F, Chawengsaksophak K, Waring P, Playford RJ, Furness JB. Reprogramming of intestinal differentiation and intercalary regeneration in Cdx2 mutant mice. Proc. Natl Acad. Sci. USA. 1999;96:7318–7323. doi: 10.1073/pnas.96.13.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 3.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao N, Kaestner KH. Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev. 2010;24:1295–1305. doi: 10.1101/gad.1921510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gross I, Duluc I, Benameur T, Calon A, Martin E, Brabletz T, Kedinger M, Domon-Dell C, Freund JN. The intestine-specific homeobox gene Cdx2 decreases mobility and antagonizes dissemination of colon cancer cells. Oncogene. 2008;27:107–115. doi: 10.1038/sj.onc.1210601. [DOI] [PubMed] [Google Scholar]
- 6.Brabletz T, Spaderna S, Kolb J, Hlubek F, Faller G, Bruns CJ, Jung A, Nentwich J, Duluc I, Domon-Dell C, et al. Down-regulation of the homeodomain factor Cdx2 in colorectal cancer by collagen type I: an active role for the tumor environment in malignant tumor progression. Cancer Res. 2004;64:6973–6977. doi: 10.1158/0008-5472.CAN-04-1132. [DOI] [PubMed] [Google Scholar]
- 7.Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN, Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eda A, Osawa H, Satoh K, Yanaka I, Kihira K, Ishino Y, Mutoh H, Sugano K. Aberrant expression of CDX2 in Barrett's epithelium and inflammatory esophageal mucosa. J. Gastroenterol. 2003;38:14–22. doi: 10.1007/s005350300001. [DOI] [PubMed] [Google Scholar]
- 9.Almeida R, Silva E, Santos-Silva F, Silberg DG, Wang J, De Bolós C, David L. Expression of intestine-specific transcription factors, CDX1 and CDX2, in intestinal metaplasia and gastric carcinomas. J. Pathol. 2003;199:36–40. doi: 10.1002/path.1246. [DOI] [PubMed] [Google Scholar]
- 10.Barros R, Camilo V, Pereira B, Freund JN, David L, Almeida R. Pathophysiology of intestinal metaplasia of the stomach: emphasis on CDX2 regulation. Biochem. Soc. Trans. 2010;38:358–363. doi: 10.1042/BST0380358. [DOI] [PubMed] [Google Scholar]
- 11.Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 12.Mutoh H, Hakamata Y, Sato K, Eda A, Yanaka I, Honda S, Osawa H, Kaneko Y, Sugano K. Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem. Biophys. Res. Commun. 2002;294:470–479. doi: 10.1016/S0006-291X(02)00480-1. [DOI] [PubMed] [Google Scholar]
- 13.Barros R, Pereira B, Duluc I, Azevedo M, Mendes N, Camilo V, Jacobs RJ, Paulo P, Santos-Silva F, van Seuningen I, et al. Key elements of the BMP/SMAD pathway co-localize with CDX2 in intestinal metaplasia and regulate CDX2 expression in human gastric cell lines. J. Pathol. 2008;215:411–420. doi: 10.1002/path.2369. [DOI] [PubMed] [Google Scholar]
- 14.Camilo V, Barros R, Sousa S, Magalhães AM, Lopes T, Santos AM, Pereira T, Figueiredo C, David L, Almeida R. Helicobacter pylori and the BMP pathway regulate CDX2 and SOX2 expression in gastric cells. Carcinogenesis. 2012;10:1985–1992. doi: 10.1093/carcin/bgs233. [DOI] [PubMed] [Google Scholar]
- 15.Barros R, da Costa LT, Pinto-de-Sousa J, Duluc I, Freund JN, David L, Almeida R. CDX2 autoregulation in human intestinal metaplasia of the stomach: impact on the stability of the phenotype. Gut. 2011;60:290–298. doi: 10.1136/gut.2010.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benahmed F, Gross I, Gaunt SJ, Beck F, Jehan F, Domon-Dell C, Martin E, Kedinger M, Freund JN, Duluc I. Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology. 2008;135:1238–1247. doi: 10.1053/j.gastro.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 17.Woodford-Richens KL, Halford S, Rowan A, Bevan S, Aaltonen LA, Wasan H, Bicknell D, Bodmer WF, Houlston RS, Tomlinson IP. CDX2 mutations do not account for juvenile polyposis or Peutz-Jeghers syndrome and occur infrequently in sporadic colorectal cancers. Br. J. Cancer. 2001;84:1314–1316. doi: 10.1054/bjoc.2001.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira B, Oliveira C, David L, Almeida R. CDX2 promoter methylation is not associated with mRNA expression. Int. J. Cancer. 2009;125:1739–1742. doi: 10.1002/ijc.24544. [DOI] [PubMed] [Google Scholar]
- 19.Rings EH, Boudreau F, Taylor JK, Moffett J, Suh ER, Traber PG. Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 20.Boulanger J, Vézina A, Mongrain S, Boudreau F, Perreault N, Auclair BA, Lainé J, Asselin C, Rivard N. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J. Biol. Chem. 2005;280:18095–18107. doi: 10.1074/jbc.M502184200. [DOI] [PubMed] [Google Scholar]
- 21.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 22.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell. Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 23.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 24.Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. 1996;87:205–216. doi: 10.1016/s0092-8674(00)81339-2. [DOI] [PubMed] [Google Scholar]
- 25.Ciosk R, DePalma M, Priess JR. Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science. 2006;311:851–853. doi: 10.1126/science.1122491. [DOI] [PubMed] [Google Scholar]
- 26.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionary conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 28.Buchet-Poyau K, Courchet J, Le Hir H, Séraphin B, Scoazec JY, Duret L, Domon-Dell C, Freund JN, Billaud M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007;35:1289–1300. doi: 10.1093/nar/gkm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Courchet J, Buchet-Poyau K, Potemski A, Brès A, Jariel-Encontre I, Billaud M. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J. Biol. Chem. 2008;283:32131–32142. doi: 10.1074/jbc.M802927200. [DOI] [PubMed] [Google Scholar]
- 30.Cano F, Bye H, Duncan LM, Buchet-Poyau K, Billaud M, Wills MR, Lehner PJ. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 2012;31:3596–3606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao Y, Bishop CE, Lu B. Mex3c regulates insulin-like growth factor 1 (IGF1) expression and promotes postnatal growth. Mol. Biol. Cell. 2012;23:1404–1413. doi: 10.1091/mbc.E11-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao Y, George SK, Zhao Q, Hulver MW, Hutson SM, Bishop CE, Lu B. Mex3c mutation reduces adiposity and increases energy expenditure. Mol. Cell. Biol. 2012;32:4350–4362. doi: 10.1128/MCB.00452-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnini M, Lapucci A, Papucci L, Witort E, Jacquier A, Brewer G, Nicolin A, Capaccioli S, Schiavone N. Identification of TINO: a new evolutionary conserved BCL-2 AU-rich element RNA-binding protein. J. Biol. Chem. 2004;279:20154–20166. doi: 10.1074/jbc.M314071200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H, Zhang X, Luo J, Dong C, Xue J, Wei W, Chen J, Zhou J, Gao Y, Yang C. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol. Med. Report. 2012;6:575–580. doi: 10.3892/mmr.2012.943. [DOI] [PubMed] [Google Scholar]
- 35.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 36.Pagano JM, Farley BM, Essien KI, Ryder SP. RNA recognition by the embryonic cell fate determinant and germline totipotency factor MEX-3. Proc. Natl Acad. Sci. USA. 2009;106:20252–20257. doi: 10.1073/pnas.0907916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 38.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 39.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 40.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto M, Robine-Léon S, Appay MD, Kedinger M, Triadou N, Dussaulx E, Lacroix B, Simon-Assmann P, Haffen K, Fogh J, et al. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell. 1983;47:323–330. [Google Scholar]
- 43.Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 44.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 45.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 47.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ootani A, Toda S, Fujimoto K, Sugihara H. Foveolar differentiation of mouse gastric mucosa in vitro. Am. J. Pathol. 2003;162:1905–1912. doi: 10.1016/S0002-9440(10)64324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the cross-roads of post-transcriptional pathways. Nat. Rev. Mol. Cell. Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 50.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamichi N, Inada K, Furukawa C, Sakurai K, Tando T, Ishizaka A, Haraguchi T, Mizutani T, Fujishiro M, Shimomura R, et al. Cdx2 and the Brm-type SWI/SNF complex cooperatively regulate villin expression in gastrointestinal cells. Exp. Cell Res. 2009;315:1779–1789. doi: 10.1016/j.yexcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Stringer EJ, Duluc I, Saandi T, Davidson I, Bialecka M, Sato T, Barker N, Clevers H, Pritchard CA, Winton DJ, et al. Cdx2 determines the fate of postnatal intestinal endoderm. Development. 2012;139:465–474. doi: 10.1242/dev.070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.