Abstract
The genome-wide distribution patterns of the ‘6th base’ 5-hydroxymethylcytosine (5hmC) in many tissues and cells have recently been revealed by hydroxymethylated DNA immunoprecipitation (hMeDIP) followed by high throughput sequencing or tiling arrays. However, it has been challenging to directly compare different data sets and samples using data generated by this method. Here, we report a new comparative hMeDIP-seq method, which involves barcoding different input DNA samples at the start and then performing hMeDIP-seq for multiple samples in one hMeDIP reaction. This approach extends the barcode technology from simply multiplexing the DNA deep sequencing outcome and provides significant advantages for quantitative control of all experimental steps, from unbiased hMeDIP to deep sequencing data analysis. Using this improved method, we profiled and compared the DNA hydroxymethylomes of mouse ES cells (ESCs) and mouse ESC-derived neural progenitor cells (NPCs). We identified differentially hydroxymethylated regions (DHMRs) between ESCs and NPCs and uncovered an intricate relationship between the alteration of DNA hydroxymethylation and changes in gene expression during neural lineage commitment of ESCs. Presumably, the DHMRs between ESCs and NPCs uncovered by this approach may provide new insight into the function of 5hmC in gene regulation and neural differentiation. Thus, this newly developed comparative hMeDIP-seq method provides a cost-effective and user-friendly strategy for direct genome-wide comparison of DNA hydroxymethylation across multiple samples, lending significant biological, physiological and clinical implications.
INTRODUCTION
DNA methylation at the 5-position of cytosine (5mC) is an important epigenetic modification that plays crucial roles in mammalian development and cell differentiation (1). Recent studies show that this apparently stable modification can be removed via oxidation by the ten-eleven translocation (TET) family of proteins. TET proteins oxidize 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (2–4). Among the three 5mC oxidative derivatives, 5hmC is the most abundant form in vivo and can be detected in almost all mammalian tissues and cells (4–8). In addition to being one of the intermediate states of DNA demethylation, 5hmC is now being considered an epigenetic modification (9–11).
Genome-wide mapping of 5mC and 5hmC reveals the genomic locations of these modifications, which is important for the elucidation of their functions (10,11). Several groups have used 5hmC-specific affinity pull-down techniques [for example, hydroxymethylated DNA immunoprecipitation (hMeDIP)] followed by next-generation sequencing (NGS) or tiling arrays to profile the genome-wide 5hmC distribution in brain tissues and embryonic stem cells (12–18). However, due to a few limitations buried in the conventional hMeDIP-seq method, it remains challenging to use DNA hydroxymethylome data generated by conventional hMeDIP-seq to perform direct genome-wide comparative analysis between high- and low-5hmC samples. For example, the current standard protocol of Illumina NGS requires loading of the same amount of DNA libraries for different samples in cluster generation and sequencing, which cancels out the differences in hMeDIP products among different samples (especially for those with differential 5hmC abundance). In addition, the multiple steps of the conventional hMeDIP-seq method, including hMeDIP, library construction, cluster hybridization and sequencing, may amplify experimental variation across the samples. Many laboratories have developed elegant computational methods to normalize or adjust the DNA methylome data generated from different samples (19–22). However, the bias caused by the intrinsic limitations of the conventional MeDIP-seq or hMeDIP-seq methods has not yet been overcome. The dramatic variation of 5hmC levels among distinct tissues and cells or during cell differentiation and embryo development demands new strategies or approaches to overcome these technical difficulties (6,23).
We reasoned that if the multiple steps required for hMeDIP-seq can be performed in one reaction system with multiple samples being compared, the experimentally induced variation could be significantly reduced, therefore overcoming the aforementioned limitation of the conventional hMeDIP-seq. To address this issue, we applied barcode technology (multiple index sequencing), which has recently been developed to distinguish different DNA samples in next-generation sequencing (24,25). The sequence reads of hMeDIP, as well as input, can be sorted into several groups representing different samples based on their specific barcode, which is assigned to each sample before performing the hMeDIP. Importantly, this modified hMeDIP strategy makes the obtained hMeDIP-seq data from multiple samples with different levels of 5hmC accessible for accurate comparative analysis. This approach has been successfully used to compare the hydroxymethylomes of the benign nevus and melanoma (26). In this report, we first describe the methodology of this approach in detail and then demonstrate its broad application in understanding the functional role of 5hmC during neural differentiation. In particular, we identified the genome-wide differentially hydroxymethylated regions (DHMRs) between mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs), and analysed their correlation with changes in gene expression. Together, our studies reveal dynamic changes in DNA hydroxymethylation during neural differentiation of ESCs and suggest that this barcode-based comparative hMeDIP-seq method can be used to compare the 5hmC patterns of multiple biological or pathological samples.
MATERIALS AND METHODS
Cell culture
46C mouse ES cells (generously given by Dr Qi-Long Ying, USC) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone-ThermoScientific) supplemented with 10% fetal bovine serum (Invitrogen), 0.1 mM non-essential amino acid (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen) and 1000 U/ml leukaemia inhibitory factor (Millipore). ESC-derived NPCs were differentiated from 46C ES cells according to the monolayer neural differentiation protocol (27,28). Rosette-like Sox1-GFP+ NPCs (>95% Sox1-GFP+ NPCs) were used in our experiments at day 9 upon neural differentiation.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde and penetrated using PBS with 0.1% Triton X-100. Cellular DNA was denatured for 30 min by 2N HCl followed by neutralization for 5 min using 100 mM Tris, pH 8.0. After blocking in 5% serum for 1 h, cells were incubated with anti-5hmC antibody (1:5000) (active motif). After washing, cells were incubated with Cy3-labelled anti-rabbit IgG (beyond time), and the nuclei were labelled by DAPI. Fluorescence was observed and recorded using Leica Microscopy.
Reverse transcription-quantitative PCR (RT-qPCR)
RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. One microgram RNA was used for reverse transcription by kit (TAKARA). The 20 μl cDNA was diluted into 200 μl. The transcript levels of Tet1, Tet2 and Tet3 were measured using quantitative real-time PCR with conditions: 95°C for 2 min followed by 40 cycles at 95°C for 15 s and 60°C for 31 s. Quantitative real-time PCR was carried in an iQ5 real-time PCR cycler (Bio-Rad) using the premixed 2× real-time SYBR Green reagent (TAKARA). Sequences of primers are described in Supplementary Table S1.
Dot blot analysis
Genomic DNA was extracted from cells using DNeasy Blood and Tissue kit (Qiagen). Denatured DNA was spotted on a nitrocellulose membrane (Waterman) and cross-linked by UVC (ultraviolet C) irradiation (Hoefer) for 5 min. The membrane was then blocked with 5% milk in TBS-Tween 20 for 1 h and incubated with anti-5hmC antibody (1:10 000) (active motif) at 4°C overnight. After incubation with a horse radish peroxidase (HRP)-conjugated anti-rabbit IgG (beyond time) for 1 h at room temperature, the membrane was washed with TBS-Tween 20 three times and then DNA was detected by Western blotting analysis system (Kodak) using enhanced chemiluminescence (GE-Healthcare). The dot blot intensity was quantified by Image-J software (NIH).
Comparative hMeDIP-seq
Library construction (adding adaptor containing barcode sequence)
Genomic DNA was sonicated to <500 bp by Bioruptor sonicator (Diagenode) and quantified using the Quant-iTdsDNA HS assay kit (Invitrogen) according to the manufacturer’s manual. The sonicated DNA fragments (4 μg of each sample) were end-repaired using the End-It DNA End Repair Kit (EPICENTRE Biotechnologies) according to the manufacturer’s instructions, followed by treatment with Klenow fragment 3′–5′exo (NEB) and dATP to generate a protruding 3′A for ligation with the adaptor containing a specific barcode sequence. The barcode sequence (four-base index: CCAG and TAGC) within the adaptor was designed as described previously (24) with some modifications.
hMeDIP
Equal amounts (∼4 μg) of barcode-tagged gDNA from ESCs and NPCs were pooled together in one tube. The mixed DNA was denatured and diluted by IP buffer (10% was taken off as input at this step). The denatured DNA was incubated with 4 μl anti-5hmC antibody (active motif) at 4°C overnight. Antibody–DNA complexes were captured by protein A/G beads, and the enriched 5hmC-containing DNA fragments (hMeDIP product) were purified.
Library amplification
The hMeDIP products, as well as input DNA, were amplified for 10–12 cycles, and the PCR products were purified by Qiagen Mini Gel Purification Kit.
Next-generation sequencing
The amplified libraries were submitted for cluster growth and sequencing by the Illumina Genome Analyzer II (GAII).
hMeDIP-seq data processing
The image analysis and base calling were performed with the Illumina package OLB (v1.8). We separated the raw sequence reads of hMeDIP and Input into different files, according to the specific barcode sequences (4-bp) at 5′-end of each sequence read.
Sequence reads were mapped onto the reference mouse genome (NCBI Build UCSC mm9) using the Bowtie (v0.12.7) algorithm (29). Unique and monoclonal reads were used for further analysis. Since the average DNA fragment length used in hMeDIP-seq was 350 bp, each sequence was extended to 350 bp. Summary of hMeDIP-seq data is shown in Supplementary Table S2. The sequencing data have been deposited in the Gene Expression Omnibus database under accession number GSE40810.
The distribution of 5hmC reads at promoters or in gene body regions was analysed by house-made software (13). The 5hmC-enriched regions (5hmC peaks) were identified using MACS (v1.4) at P < 1e-5 and FDR < 0.01 (30). The 5hmC peaks were annotated to mouse Refseq genes and the genes with 5hmC peaks at promoters or in gene body regions were chosen for further analysis.
Analysis of the relationship between DNA hydroxymethylation and gene expression
Gene expression data of ESCs (31) and NPCs (32) were downloaded from GEO dataset [Accession number of GEO: ESCs (ES_rep1: GSM198062, ES_rep2: GSM198063, ES_rep3: GSM198064), NPCs (NPC_rep1: GSM618399, NPC_rep2: GSM618400)]. The raw CEL files were processed by robust multi-array average (RMA), and log2 intensities were used to represent the expression levels after normalization between samples. Genes were separated into five groups with respect to their average signals of probes, and the 5hmC distribution at promoters and in gene body regions of these five groups of genes were plotted for ESCs and NPCs, respectively.
The genes were also classified into ‘5hmC+’ and ‘5hmC−’ groups according to the presence or absence of 5hmC peaks at either promoters [TSS (transcription start site ) ± 1 kb] or gene body regions [from ‘TSS + 1 kb’ to ‘TES (transcription end site)’]. The differences in average gene expression levels between these groups of genes were analysed by ANOVA.
Identification of DHMRs
Pooled 5hmC peaks were called from the un-separated hMeDIP-seq data by MACS (30), and the densities of 5hmC peaks were compared between ESCs and NPCs. For the quantitative analysis, the densities of 5hmC peaks between ESCs and NPCs were normalized by their input respectively. The threshold was set at 1-fold (NPCs versus ESCs, log2 5hmC density ratio ≥ 1 or ≤ −1) for identification of ‘DNA de-hydroxymethylation (loss of 5hmC)’ regions and ‘de novo DNA hydroxymethylation (gain of 5hmC)’ regions during neural differentiation.
hMeDIP-qPCR
Two microgram sonicated gDNA was denatured and incubated with 1 μl anti-5hmC antiserum (active motif) for hMeDIP as described previously (13,33). Normal IgG antibodies were used as control. Input and hMeDIP products were used as templates for quantitative real-time PCR. Relative 5hmC enrichment was calculated by 2dCt (dCt = CtInput − CthMeDIP). Primers used in hMeDIP-qPCR are described in Supplementary Table S3.
Glucosylation of genomic DNA followed by methylation-sensitive qPCR (glucMS-qPCR)
The ‘5hmC’ and ‘5hmC + 5mC’ levels in DHMRs containing MspI/HpaII sites were measured by a restriction enzyme-based assay (EpiMark kit, NEB) (34). Genomic DNA was treated with or without T4 Phage β-glucosyltransferase and then digested by MspI, HpaII or no enzyme (mock digestion). The MspI- and HpaII-resistant fraction was quantified by qPCR using primers designed around at least one HpaII/MspI site, and normalizing to the mock digestion. Primers used in glucMS-qPCR are described in Supplementary Table S4.
Gene ontology (GO) analysis
GO analysis for genes with ‘gain of 5hmC’ DHMRs was performed by the database for annotation, visualization and integrated discovery (DAVID) website (35,36). GO analysis for the ‘gain of 5hmC’ DHMRs was also performed by the Genomic Regions Enrichment of Annotations Tool (GREAT) (37).
Analysis of the relationship between the changes in DNA hydroxymethylation and gene expression during neural differentiation
The genes were sorted according to the changes in gene expression level: genes that were down-regulated and genes that were up-regulated during neural differentiation (log2 mRNA value ≤−1 or ≥1). The changes in tag density of 5hmC peaks at promoters (TSS ± 1 kb) or in gene body regions (from TSS + 1 kb to TES) were compared between the up- and down- regulated genes by ANOVA.
All 5hmC peaks located at promoters (TSS ± 1 kb) or gene body regions (from TSS + 1 kb to TES) were selected for dot plotting. The alteration in 5hmC density for each 5hmC peak was plotted on the y-axis and the change in corresponding gene expression was plotted on the x-axis. Correlation between the changes in 5hmC and gene expression was analysed by linear regression analysis.
RESULTS
Strategy for comparative hMeDIP-seq method
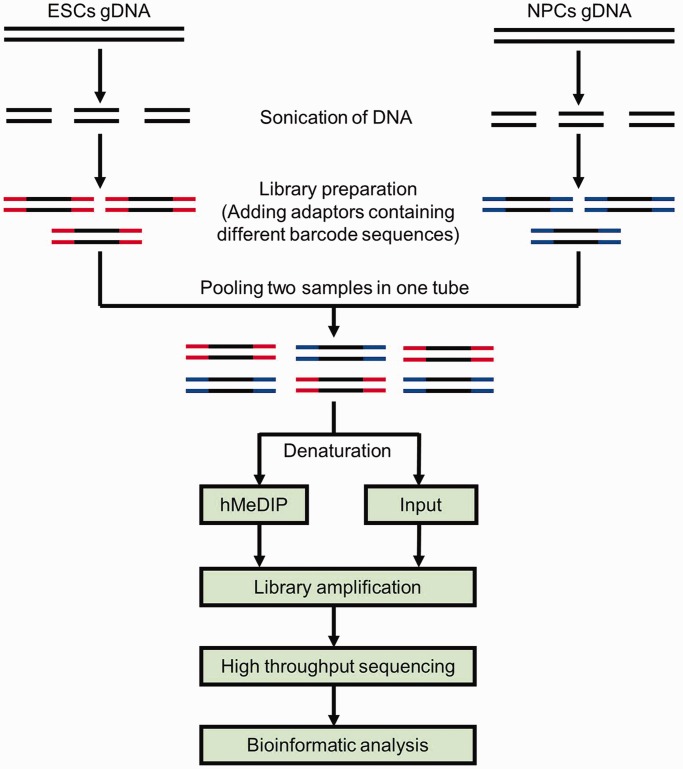
To compare the DNA hydroxymethylomes of diverse cell types, we optimized the conventional hMeDIP-seq method in multiple aspects (as illustrated in Figure 1). For instance, after sonication, the genomic DNA fragments from different samples were labelled with adaptors containing specific barcode sequences. Equal amounts of barcode-tagged DNA fragments from different samples were then pooled together in one tube for hMeDIP (the input was taken out before IP). The hMeDIP and input libraries were amplified and sequenced using Illumina GAII. The raw reads from different samples were sorted according to their specific barcode sequences, and mapped to the genome for further bioinformatic analysis. Immunoprecipitation and sequencing of multiple samples in one reaction system markedly reduce the experimental variation among samples and enable direct comparison of the DNA hydroxymethylation data across samples. Thus, we refer to this new approach as the ‘comparative hMeDIP-seq method’.
Figure 1.
Schematic diagram of the comparative hMeDIP-seq method.
Reduced global 5hmC level in NPCs as compared with ESCs
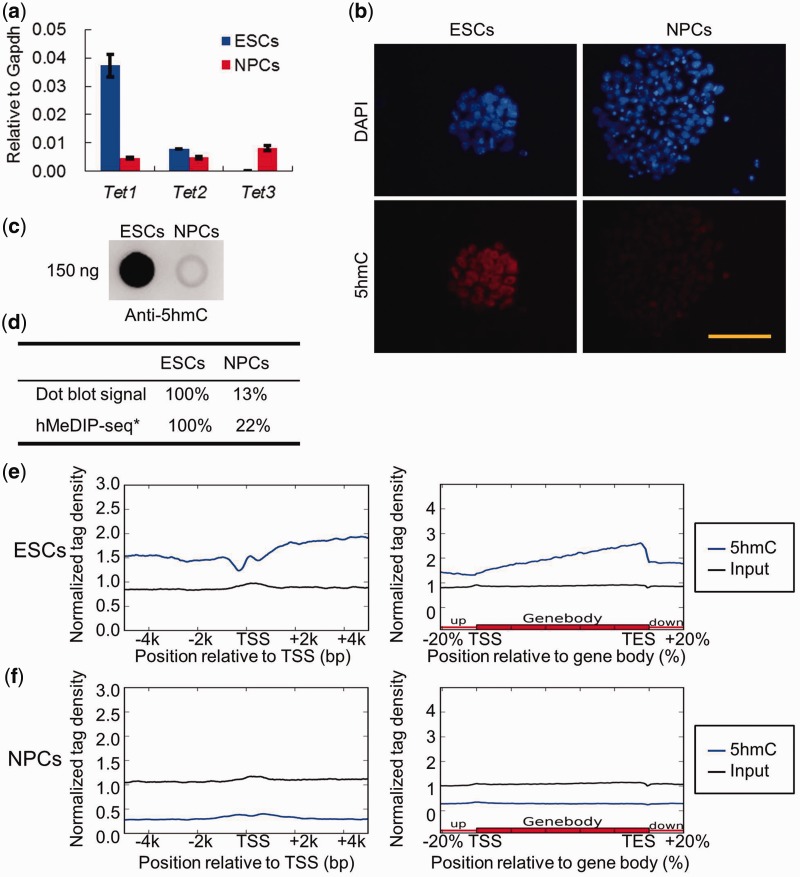
Epigenetic modifications undergo dynamic changes during cell differentiation and development. Directed differentiation of pluripotent mouse ESCs into NPCs in vitro is a well-established model for the study of molecular mechanisms controlling neural induction. Therefore, we used the newly developed comparative hMeDIP-seq method to examine the genome-wide differences in DNA hydroxymethylation in ESCs and ESC-derived NPCs. A sufficient number of NPCs were obtained with high purity from mouse ESCs via N2B27 serum-free monolayer differentiation in vitro (Supplementary Figure S1a–c). We compared the expression levels of Tet family genes as well as the global 5hmC levels in ESCs versus NPCs. RT-qPCR analysis revealed that Tet1 mRNA level, which was high in ESCs, decreased dramatically in NPCs, while Tet3 was up-regulated in NPCs but only marginally expressed in ESCs (Figure 2a). However, Tet2 mRNA levels were comparable between ESCs and NPCs (Figure 2a). Both immunofluorescence (Figure 2b) and dot blot analysis (Figure 2c) showed that the global 5hmC levels in NPCs were much lower than those in ESCs, suggesting that pluripotent ESCs and neural lineage-committed NPCs are in two different developmental stages with high and low 5hmC abundance, respectively. As expected, comparative hMeDIP-seq data showed that the 5hmC reads of ESCs and NPCs (after normalized to the corresponding input) were in complete agreement with changes of the global 5hmC levels detected between ESCs and NPCs by dot blot (Figure 2d). These data indicate that the new comparative hMeDIP-seq method preserves the relative difference in 5hmC abundance across different samples.
Figure 2.
Differential Tet1/2/3 gene expression and global 5hmC levels in ESCs and NPCs. (a) RT-qPCR analysis of Tet1, Tet2 and Tet3 mRNA levels in ESCs and NPCs (mean values ± SD, n = 3). (b) Immunofluorescence analysis of 5hmC levels in ESCs and NPCs. Bar: 50 μm. (c) Dot blot analysis of the global 5hmC levels in the gDNA of ESCs and ESC-derived NPCs. One hundred fifty nanogram gDNA for each dot. (d) The relative 5hmC signal of dot blot and comparative hMeDIP-seq in ESCs and NPCs. *The ratio of hMeDIP/Input reads numbers in ESCs was set as 100%. (e) Distribution of 5hmC in TSS ± 5 kb and gene body regions in ESCs. (f) Distribution of 5hmC in TSS ± 5 kb and gene body regions in NPCs.
The average distribution of 5hmC was mapped onto an average gene model. ESCs exhibited a hypo-hydroxymethylated ‘valley’ with a small ‘bumper’ around TSS regions, but significant 5hmC enrichment was found across gene body regions (Figure 2e), consistent with the previously reported 5hmC pattern in ESCs (13–15). However, neither a 5hmC dip with a small ‘bumper’ around TSS regions nor significant 5hmC enrichment in gene body regions was observed in NPCs (Figure 2f). This suggests that both the total level of 5hmC and the genomic distribution of 5hmC are dramatically different between ESCs and NPCs (38).
Recent reports have suggested significant 5hmC enrichment at enhancers (defined by the enrichment of both H3K4me1 and H3K27ac histone modifications) in ESCs (17,39–42). Therefore, we next examined the association of 5hmC with the annotated enhancers and DNase I hypersensitive sites (DHSs), as well as the loss of 5hmC at these newly defined genomic elements during ESC to NPC differentiation. Consistent with previous findings, we observed significant 5hmC enrichment at the enhancers in ESCs (Supplementary Figure S2a). Interestingly, this 5hmC enrichment at ESC enhancers is lost in NPCs (Supplementary Figure S2a). We also analysed the relationship of 5hmC and DHSs. We separated DHSs into three classes: (i) those lacking the enhancer histone modifications H3K4me1 and H3K27ac; (ii) putative poised enhancers bearing only H3K4me1; and (iii) putative active enhancers with both modifications. DHSs with poised or active enhancer chromatin signatures exhibit 5hmC enrichment, and poised enhancers have even higher 5hmC levels than active enhancers (Supplementary Figure S2b–d). These observations indicate that DNA hydroxymethylation may be involved in regulating the function of enhancers.
It is noteworthy that the comparative hMeDIP-seq approach (two samples in one lane) generated fewer 5hmC reads and peaks for individual samples (for example ESCs) than the conventional hMeDIP-seq approach (one sample in one lane). However, the 5hmC distribution patterns were consistent between the comparative and conventional hMeDIP-seq methods (Supplementary Figure S3a–d), suggesting that the new comparative hMeDIP-seq method yields similar results, albeit at the expense of throughput. If higher throughput data is required, it would be necessary to sequence the library in more lanes and combine the data together for analysis.
Correlation between DNA hydroxymethylation and gene expression status
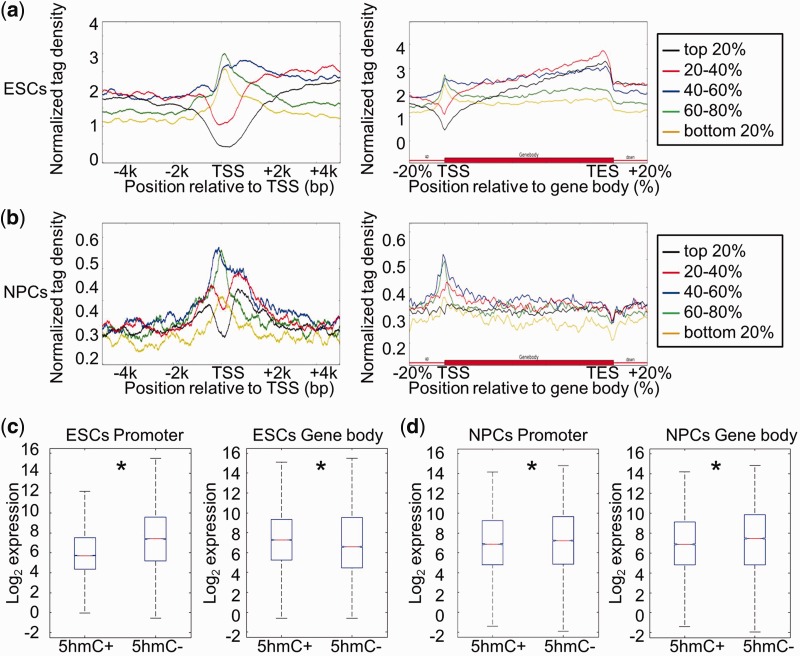
To determine whether there is a relationship between DNA hydroxymethylation and gene expression status in the same cell type, we classified genes into five groups based on their transcriptional levels and plotted their 5hmC distribution patterns at promoters and gene body regions. As shown in Figure 3a, genes with high expression levels in ESCs exhibited 5hmC depletion around TSS regions, whereas genes with low expression levels displayed 5hmC enrichment at promoters. A similar negative correlation between 5hmC density around TSS regions and gene expression was also observed in NPCs (Figure 3b). Consistent with the previous findings, all five groups of genes have abundant 5hmC enrichment in gene body regions in ESCs, and genes with high expression levels displayed higher 5hmC enrichment than genes at low expression levels (Figure 3a). However, in NPCs, the 5hmC curves of all five groups of genes could not be easily separated from each other because 5hmC was not particularly enriched in gene body regions (Figure 3b).
Figure 3.
Correlation between DNA hydroxymethylation and gene expression in ESCs and NPCs. (a) Genes were separated into five groups (from high to low) according to their expression levels in ESCs: top 20%; 20–40%; 40–60%; 60–80%; bottom 20%. The average 5hmC densities of the five groups of genes were plotted across the promoter or gene body regions. Left: TSS ± 5 kb regions. Right: gene body regions. (b) Genes were also separated into five groups according to their expression levels in NPCs. The average 5hmC densities of the five groups of genes were plotted across the promoter or gene body regions. Left: TSS ± 5 kb regions. Right: gene body regions. (c) Genes were classified into two groups with respect to the presence (5hmC+) or absence (5hmC−) of 5hmC peak(s) at their promoters (TSS ± 1 kb) or in gene body regions (from TSS + 1 kb to TES) in ESCs. Gene expression levels were compared between ‘5hmC+’ and ‘5hmC−’ groups. Left: promoter, P = 0; Right: gene body region, P = 1.08358e-13. *P < 0.01. (d) Genes were classified into two groups with respect to the presence (5hmC+) or absence (5hmC−) of 5hmC peak(s) at their promoters (TSS ± 1 kb) or in gene body regions (from TSS + 1 kb to TES) in NPCs. Gene expression levels were compared between ‘5hmC+’ and ‘5hmC−’ groups. Left: promoter, P = 0.0001; Right: gene body region, P = 0. *P < 0.01.
We also separated genes into ‘5hmC+’ and ‘5hmC−’ groups according to the presence or absence of 5hmC peak (s) at promoters (TSS ± 1 kb) or gene body regions (from TSS + 1 kb to TES) and compared their average gene expression levels. In ESCs, genes with 5hmC peaks at promoters (TSS ± 1 kb) had lower average expression levels than those without 5hmC peaks, while genes with 5hmC peaks in the gene body regions (from TSS + 1 kb to TES) had higher expression levels than those without 5hmC peaks (Figure 3c). In NPCs, genes with 5hmC peaks at promoters (TSS ± 1 kb) also had lower expression levels than those without 5hmC peaks (Figure 3d). However, unlike the relationship observed in ESCs, genes with 5hmC peaks in gene body regions (from TSS + 1 kb to TES) in NPCs had lower expression levels than the genes without 5hmC peaks (Figure 3d). Thus, these data suggest that DNA hydroxymethylation in gene body regions correlates differently with gene expression depending on the cell type.
Identification of DHMRs between ESCs and NPCs
Since the numbers and widths of 5hmC peaks were not consistent between two different samples, it was difficult to compare the peaks of ESCs and NPCs directly using the conventional hMeDIP-seq method. It is now possible to identify peaks from the un-separated (pooled) hMeDIP-seq data of ESCs and NPCs as the DNA hydroxymethylation-sensitive regions in both cells because the comparative hMeDIP-seq reactions for two different samples are performed in one reaction system. The density of pooled 5hmC peaks in ESCs and NPCs can then be directly compared after normalization by the ratio of corresponding input throughput.
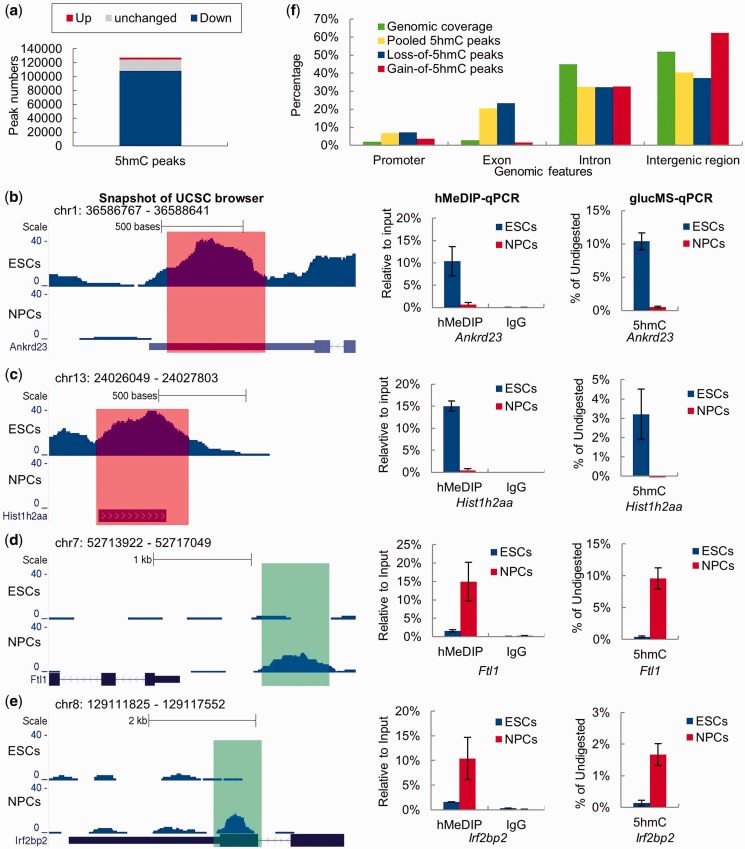
Overall, we identified 127 256 pooled 5hmC peaks from the un-separated hMeDIP-seq data (Figure 4a). Among them, 107 834 regions lose 5hmC (NPCs versus ESCs, log2 5hmC density ratio ≤ −1) during neural differentiation (Figure 4a). However, we also detected 2748 regions that underwent ‘de novo DNA hydroxymethylation (gain-of-5hmC)’ (NPCs versus ESCs, log2 5hmC density ratio ≥ 1) (Figure 4a). Importantly, the DHMRs between ESCs and NPCs can be validated by hMeDIP-qPCR and glucMS-qPCR analysis using a representative set of DHMRs. As shown in Figure 4b–e, the 5hmC peaks at Ankrd23 and Hist1h2aa exhibited ‘loss of 5hmC’ upon neural differentiation, while the 5hmC peaks at Ftl1 and Irf2bp2 genes underwent ‘de novo hydroxymethylation (gain of 5hmC)’.
Figure 4.
Identification of DHMRs between ESCs and NPCs. (a) All 5hmC peaks called from the pooled hMeDIP-seq data were separated into three groups: down-regulated (log2 5hmC density ratio ≤ −1), up-regulated (log2 5hmC density ratio ≥ 1) and no significant change (−1 < log2 5hmC density ratio < 1) 5hmC peaks. (b–e) UCSC genome (mmp9) browser screen shots, hMeDIP-qPCR and glucMS-qPCR validation (mean values ± SD, n = 3) of two representative DHMRs with de-hydroxymethylation (Ankrd23 and Hist1h2aa) and two representative DHMRs with de novo hydroxymethylation (Ftl1 and Irf2bp2). (f) Percentages of pooled 5hmC peaks, ‘gain of 5hmC’ peaks and ‘loss of 5hmC’ peaks located at different genomic features.
The distributions of pooled 5hmC peaks, ‘loss of 5hmC’ peaks and ‘gain of 5hmC’ peaks in different genomic features were compared with the sequence coverage of these genomic features in the mouse genome. As previously reported (13,16), the pooled 5hmC peaks are associated with exons and promoters with a much higher frequency than with introns and intergenic regions (relative to the percentage of these genome elements in proportion to the genome size) (Figure 4f). Strikingly, although the ‘loss of 5hmC’ is a genome-wide feature of the neural lineage commitment, it is most dramatic in exons and promoters and less evident in introns and intergenic regions (Figure 4f), suggesting that the dynamic change of 5hmC likely has a more profound effect on regulation of the promoters and exons during ESC differentiation. The ‘gain of 5hmC’ peaks show no apparent preference for the enrichment of these peaks in promoters, exons, introns or intergenic regions. Nonetheless, Gene Ontogeny (GO) analysis of the genes with de novo DNA hydroxymethylation showed an enrichment of genes involved in ‘dendrite morphogenesis’ and other neural system functions (Supplementary Figure S4 and Supplementary Table S5). Thus, the functional potential and mechanistic insight of the ‘gain of 5hmC’ peaks during neural differentiation may warrant future investigation.
Relationship between the dynamic changes of DNA hydroxymethylation and gene expression during neural lineage commitment of ESCs
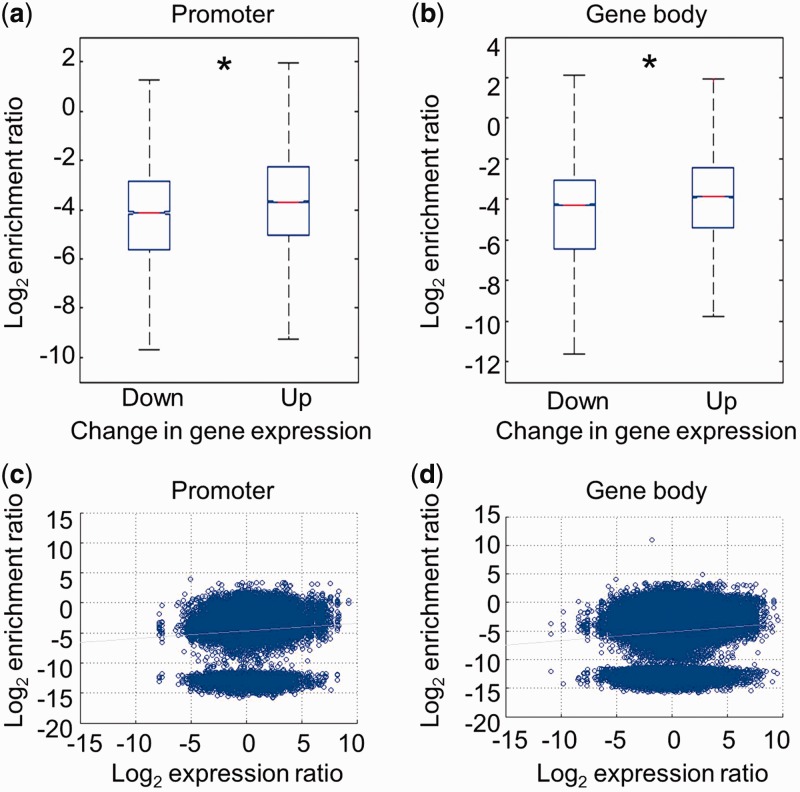
We next asked whether the alteration in DNA hydroxymethylation at promoters (TSS ± 1 kb) or gene body regions (from TSS + 1 kb to TES) is associated with the changes in gene expression during neural lineage commitment of ESCs. We stratified genes into two groups: down-regulated (log2 gene expression ratio ≤ −1) and up-regulated (log2 gene expression ratio ≥ 1) genes during neural differentiation (NPCs versus ESCs) and compared their changes in 5hmC density. Although the 5hmC peaks at both groups of genes displayed a dramatic decrease during neural lineage commitment of ESCs, the 5hmC peaks at down-regulated genes decreased more than those at up-regulated genes (Figure 5a and b). Pooled 5hmC peaks at promoters or gene body regions were plotted according to their changes in 5hmC density and gene expression during neural lineage commitment of ESCs. The majority of 5hmC peaks exhibited a decrease in 5hmC density upon neural lineage commitment of ESCs while a small fraction of 5hmC peaks showed an increase (Figure 5c and d). As a whole, there is a weak positive correlation between the changes in 5hmC peak density and corresponding gene expression during neural lineage commitment of the ESCs (Figure 5c and d).
Figure 5.
Relationship between the changes in DNA hydroxymethylation and gene expression during neural differentiation. (a) Down- and up-regulated genes during neural differentiation were sorted (NPC versus ESC, log2 mRNA expression ratio ≤ −1 or ≥1). The changes in 5hmC peak density at promoters (TSS ± 1 kb) in down- and up-regulated genes were compared during neural differentiation (P = 0). *P < 0.01. (b) The changes in 5hmC peak density at gene body regions (from TSS + 1 kb to TES) in down- and up-regulated genes were compared during neural differentiation (P = 0). *P < 0.01. (c) Dot plotting of the alteration in 5hmC peak density at promoters and the gene expression change during neural differentiation. The changes in 5hmC peak density at promoters (TSS ± 1 kb) were plotted on the y-axis and the corresponding gene expression changes were plotted on the x-axis. R2 = 0.0048. (d) Dot plotting of the alteration in 5hmC peak density in gene body regions and the gene expression change during neural differentiation. The changes in 5hmC peak density in gene body regions (from TSS + 1 kb to TES) were plotted on the y-axis and the corresponding gene expression changes were plotted on the x-axis. R2 = 0.0059.
DISCUSSION
The barcode technology that distinguishes DNA fragments of different samples allows performance of hMeDIP of multiple samples in one reaction system and sequencing the products in one lane. Although barcode technology has been widely used in high throughput sequencing of multiple samples and reduces cost in sequencing (24,25), this is the first time (to our knowledge) it has been applied as the first step of hMeDIP-seq. This new application achieves sequencing of multiple samples in one lane and preserves the 5hmC differences among the samples throughout the whole process of hMeDIP-seq (the latter is the most important innovation in this method). We could eliminate most of the systematic and technical errors that arise with conventional hMeDIP-seq method, since the experiment is processed in identical conditions and is internally controlled. The new method has several advantages: (i) Performing hMeDIP in one tube for several samples (especially samples with differential 5hmC abundance) allows the equal IP efficiency across compared samples; (ii) One reaction system with multiple samples for all steps after barcode tagging, including hMeDIP, library amplification, cluster formation and sequencing, can reduce accidental and systematic errors of the experiments; (iii) Sequencing multiple samples in one lane can reduce the cost; (iv) For the comparison across different samples by this method, it is not necessary to reach saturated sequencing coverage in any samples, although more hydroxymethylated loci could be identified with more sequencing coverage; and (v) Finally and most importantly (also see below), the comparative hMeDIP-seq data generated by this new approach is ready for direct comparative analysis across multiple samples after normalization of hMeDIP data by the corresponding input data of each sample.
The new strategy used in the comparative hMeDIP-seq method greatly facilitates later data processing. The computational analysis for all the reads obtained from deep sequencing is processed as a whole for the different samples (ESCs versus NPCs) within the data collection. Thus, it is simplified, straightforward and unbiased for all reads alignment and all 5hmC peak calling. Indeed, we have made significant improvements in peak calling and DHMR identification. Pooled 5hmC peaks can be called from the un-separated hMeDIP-seq data of multiple samples as the sensitive DNA hydroxymethylated regions. Then, we can compare the 5hmC density at these sensitive DNA hydroxymethylated regions across multiple samples. With this new strategy, we have bypassed the problem of asymmetric peak widths and numbers between samples and significantly reduced the work load for the data processing.
Recently, two new techniques (oxBS-seq and TAB-seq) have been reported to distinguish 5hmC from 5mC at single-base resolution (40,43). These two methods also provided means to detect the absolute quantitative value (percentage) of 5hmC in the genome. The single-base resolution 5hmC sequencing approach revealed that the absolute percentage of 5hmC in gene body regions appears to be low, contrary to previous data from the conventional hMeDIP-seq approach, which suggested that gene body regions of ESCs tend to have a large amount of 5hmC. Therefore, the conclusions drawn from the two different approaches seem contradictory. We believe that this discrepancy is likely caused by the different measurement techniques of the two 5hmC-detecting approaches. Gene body DNA (especially in exons) has a relatively high frequency of CpG content but low percentage of 5hmC at each CpG site. It is true that the TAB-Seq method represents a more accurate methodology to measure the absolute abundance of 5hmC at base pair resolution. However, as pointed out by Yu et al. (40) in their Discussion, the TAB-seq method likely underestimates relatively low abundances of 5hmC sites within gene body, as these sites might have escaped detection because of insufficient coverage of sequencing depth. Accordingly, an unexpectedly low (underestimated) percentage of 5hmC within the gene bodies was observed in this study (40). This shortcoming could most likely be compensated by increasing the sequencing depth, although this would increase the cost even more, another limitation for routine usage of these methods in an average laboratory at the current stage. Conversely, conventional affinity-based 5hmC mapping (hMeDIP) is not only a cost-effective way to quickly project the overall landscape of 5hmC along the genome, but it also allows amplification of high frequency but weak 5hmC signals within gene bodies. The downside of this measurement is that it determines 5hmC distribution at low resolution (counting 200–400 bp as one unit). It may also potentially over-amplify the weak signals of those clustering individual 5hmC sites with low abundances, adding up multiple weak 5hmC signals to make it appear like one strong signal within 200–400 bp window. Therefore, it may introduce a potential overestimation of the abundance of 5hmC in the gene bodies, outweighing the low frequency, but high abundance, of 5hmC at distal-regulatory elements such as the enhancers. Clearly, both single-base resolution 5hmC-seq and hMeDIP-seq approaches have their advantages and disadvantages.
The comparative hMeDIP method we describe in this article retained all of the advantages from the conventional hMeDIP-seq, while minimizing the aforementioned shortcomings. The unique features of the comparative hMeDIP-seq method, particularly the newly developed algorithm for DHMR identification, systematically eliminate the conventional hMeDIP-seq-induced bias or potential overestimation of 5hmC abundance for the low-5hmC sample when the 5hmC abundance was compared among multiple biological samples. If the research aim is to identify DHMRs among samples, the optimized comparative hMeDIP-seq method can fulfill this aim with much lower cost and less experimental time. With no doubt, if the research goal is to map the absolute 5hmC distribution at base-pair resolution, oxBS- or TAB-seq is the method to choose. However, to precisely locate 5hmC sites with low abundance, oxBS- or TAB-seq would require much larger sequencing coverage than hMeDIP-seq, thus increasing the cost and amount of labour required. In the future, it would be a good strategy to combine the advantages of the comparative hMeDIP-seq with the oxBS- or TAB-seq approaches for studying the dynamic changes and precise role of 5hmC in genome regulation during cell differentiation and development. For example, we may use the comparative hMeDIP-seq to locate the genes or genomic regions with most dramatic changes of 5hmC (DHMRs). Then we can use targeted oxBS- or TAB-seq approaches to sequence these targeted genomic fragments (located by the comparative hMeDIP-seq approach) to map the precise changes of 5hmC within the region at single base resolution. By combining these two approaches, we envision that we could quantitatively compare the localized 5hmC changes between the different biological samples in a cost-effective user-friendly manner while yielding better resolution.
Upon neural differentiation, ESCs lose pluripotency and gain the characteristics of neural lineage cells (44,45). Cell differentiation involves drastic epigenomic reprogramming, including the establishment of cell-specific DNA methylome and hydroxymethylome (11,46). The DNA methylomes of ESCs and NPCs have been well established by MeDIP-seq and Bisulfite-seq in the past few years (47,48). Although the DNA hydroxymethylome of ESCs has been established recently by affinity-based approach or new sequencing methods at single base resolution, the DNA hydroxymethylome of NPCs remained unknown. In this study, using the newly developed comparative hMeDIP-seq method, we revealed a dramatic loss of 5hmC during neural differentiation of mouse ESCs and provided the first genome-wide comparative 5hmC map of NPCs in conjunction with that of the ESCs. We found that the 5hmC distribution pattern of NPCs is vastly different from that of ESCs: the majority of 5hmC peaks that exist in ESCs are lost in NPCs, whereas a small fraction of gene loci undergo de novo DNA hydroxymethylation. Many regions that undergo de novo DNA hydroxymethylation are located at genes associated with mature neuronal functions. We speculate that these genes may be repressed by 5hmC or its associated cellular complex (es) in NPCs, but are poised for DNA demethylation, and as a result can undergo gene activation upon later stages of neural differentiation and maturation.
In agreement with the previous findings, the comparative hMeDIP-seq method revealed a complex correlation between 5hmC distribution and gene activity in both ESC and NPC. We found that the 5hmC levels at promoters (TSS ± 1 kb) had a negative correlation with gene expression in both ESCs and NPCs, whereas the correlation between 5hmC levels in gene body regions and gene expression varied depending on the cell type. ‘Loss of 5hmC’ happens at down-regulated, up-regulated and unchanged genes during neural differentiation; however, there was a weak, but significant, positive correlation between the changes in 5hmC and gene expression. These results suggest that 5hmC patterns are likely another layer of largely uncharacterized epigenetic regulation within the larger landscape of cell-specific chromatin structure. It is possible that 5hmC in the gene body may not be an essential and driving force for gene transcription, but rather, gene transcriptional activity at a steady state may affect 5hmC generation by regulating the access of TET proteins towards their DNA substrates. Finally, it has been proposed that DNA methylation in gene body regions may inhibit cryptic transcription and correlate with gene expression (49). DNA methylation in gene body regions has also been reported to regulate alternative promoters and splicing (50,51). In the future, it will be of great interest to investigate whether DNA hydroxymethylation in gene body regions also affects alternative promoters and mRNA splicing.
In summary, future investigation is required to dissect the intrinsic and intricate relationship between dynamic changes of 5hmC and specific gene expression within the same cell type or during cell differentiation. Nonetheless, our data demonstrate that the comparative hMeDIP-seq method is a powerful approach for genome-wide comparisons of DNA hydroxymethylation across multiple samples. Using this method, we have uncovered the difference in DNA hydroxymethylation between ESCs and NPCs. Notably, the strategies described here can also be applied to the genome-wide quantitative comparison of other DNA modifications (such as 5mC, 5caC and 5fC) across different biological or pathological samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–4.
FUNDING
National Natural Science Foundation of China [31200966]; ‘985’ Project of Ministry of Education of China; ‘973’ National Basic Research Program of China [2009CB825602, 2009CB825603]; NIH [GM078458, DK077036 to Y.G.S.]. Funding for open access charge: ‘973’ National Basic Research Program of China [2009CB825602].
Conflict of interest statement. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Y.G.S. was a Pew Scholar.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Miss Julia Pian (Harvard University) and Dr Xiaodong Zhao (Shanghai Jiaotong University) for their critical reading of this manuscript.
REFERENCES
- 1.Latham T, Gilbert N, Ramsahoye B. DNA methylation in mouse embryonic stem cells and development. Cell Tissue Res. 2008;331:31–55. doi: 10.1007/s00441-007-0537-9. [DOI] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruzov A, Tsenkina Y, Serio A, Dudnakova T, Fletcher J, Bai Y, Chebotareva T, Pells S, Hannoun Z, Sullivan G, et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Ed. Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 9.Munzel M, Globisch D, Carell T. 5-Hydroxymethylcytosine, the sixth base of the genome. Angew. Chem. Int. Ed. Engl. 2011;50:6460–6468. doi: 10.1002/anie.201101547. [DOI] [PubMed] [Google Scholar]
- 10.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 11.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 17.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39:5015–5024. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, Su J, Li X, Wu Q, Wang F, et al. QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res. 2011;39:e58. doi: 10.1093/nar/gkr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R, Adjaye J. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20:1441–1450. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 28.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gennet N, Gale E, Nan X, Farley E, Takacs K, Oberwallner B, Chambers D, Li M. Doublesex and mab-3-related transcription factor 5 promotes midbrain dopaminergic identity in pluripotent stem cells by enforcing a ventral-medial progenitor fate. Proc. Natl Acad. Sci. USA. 2011;108:9131–9136. doi: 10.1073/pnas.1016679108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 34.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, Feng S, Stroud H, Jacobsen SE, Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J. Biol. Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Peron CL, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–8265. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 44.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du ZW, Zhang SC. Neural differentiation from embryonic stem cells: which way? Stem Cells Dev. 2004;13:372–381. doi: 10.1089/scd.2004.13.372. [DOI] [PubMed] [Google Scholar]
- 46.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 47.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin. Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 50.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.