Abstract
miRNAs play important roles in many biological processes, including erythropoiesis. Although several miRNAs regulate erythroid differentiation, how the key erythroid regulator, GATA-1, directly orchestrates differentiation through miRNA pathways remains unclear. In this study, we identified miR-23a as a key regulator of erythropoiesis, which was upregulated both during erythroid differentiation and in GATA-1 gain-of-function experiments, as determined by miRNA expression profile analysis. In primary human CD34+ hematopoietic progenitor cells, miR-23a increased in a GATA-1-dependent manner during erythroid differentiation. Gain- or loss-of-function analysis of miR-23a in mice or zebrafish demonstrated that it was essential for normal morphology in terminally differentiated erythroid cells. Furthermore, a protein tyrosine phosphatase, SHP2, was identified as a downstream target of miR-23a that mediated its regulation of erythropoiesis. Taken together, our data identify a key GATA-1–miRNA axis in erythroid differentiation.
INTRODUCTION
Transcription factors and miRNAs are key regulators of gene expression in higher eukaryotes (1). Gene expression is thought to be primarily regulated by transcription factors. Moreover, the ultimate fate of a gene is carefully controlled at the post-translational level by miRNAs (2). miRNAs have been discovered in multiple organisms, and many are evolutionarily conserved. They regulate various developmental and physiological processes and are often implicated in human disease (3).
Hematopoiesis is highly orchestrated by the interaction of lineage-specific transcription factors driving pluripotent precursors to differentiate toward mature blood cells (4). Increasing evidence suggests that this differentiation, along the various hematopoietic lineages, including erythropoiesis, is, in part, regulated by miRNAs. For example, miR-223 enhances retinoic acid-induced granulocytic differentiation by targeting nuclear factor I/A (5); miR-21 acts as a monopoietic promoter, while miR-196b functions as an antagonist of granulopoiesis (6); miR-221 and miR-222 inhibit normal erythropoiesis and erythroleukemic cell growth by downregulating the Kit protein (7); miR-451 is required for erythrocyte maturation in both zebrafish and mouse development (8–11); and miR-210 increases the expression of the γ-globin gene in differentiating erythroid cells (12). Our previous work demonstrated that miR-223, miR-103 and miR-376a inhibited erythroid differentiation by targeting different protein-coding genes (13–16).
These hematopoiesis-associated miRNAs, whose expression is strictly controlled during lineage differentiation, are extensively regulated by lineage-specific transcription factors. For example, the activation of miR-223 by C/EBPα can trigger neutrophil differentiation and is necessary for normal myelopoiesis (17); the downregulation of miR-21 and miR-196b during myelopoiesis is largely dependent on the inhibition by transcriptional repressor Gfi1 (6); and GATA-1 activates miR-451 and comprises a regulatory circuit that modulates erythroid maturation (11). Thus, the combined involvement of miRNAs and transcription factors in these processes makes the study of miRNA regulation complex and requires a change of perspective. While past studies seeking to identify functionally significant miRNAs have begun with miRNA profiling, it has become clear that the incorporation of their upstream regulation, especially in response to essential transcription factors, is of critical importance for the screening of candidate miRNA genes.
In this respect, we adopted a strategy combining global miRNA profiles during human erythropoiesis with changes in miRNA expression derived from GATA-1, which is a specific and critical erythroid transcription factor that regulates genes implicated in nearly all facets of erythroid cell maturation (18,19). We generated miRNA expression profiles using an illumina microarray platform in K562 cells that underwent either erythroid differentiation or GATA-1 manipulation. An integrated analysis of these data revealed several miRNA genes that were not only functional during erythropoiesis but also regulated by GATA-1. The initial expression and functional screening of candidate miRNAs in K562 cells highlights the significance of miR-23a in human erythroid differentiation. Remarkably, ectopic expression of miR-23a promoted the accumulation of mature erythroid cells and the formation of erythroid clones in primary cultured CD34+ hematopoietic progenitor cells (HPCs). Alternatively, inhibition of miR-23a delayed erythroid maturation. Furthermore, zebrafish lacking miR-23a displayed an impaired erythroid phenotype during the primitive wave of hematopoiesis. Overall, our strategy successfully identified an erythroid miR-23a that plays important regulatory roles in hematopoiesis.
MATERIALS AND METHODS
Cell culture
Human erythroleukemia cell line K562 was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone). Erythroid differentiation of K562 cells was obtained using 30 µM hemin (Sigma-Aldrich, Deisenhofen, Germany) over 24, 48 and 72 h. The different degrees of differentiation were determined by benzidine staining for hemoglobin expression. 293T cells were obtained from American Type Culture Collection and grown in DMEM media with 10% FBS.
miRNA microarray and data analysis
Two groups of miRNA microarrays were carried out with illumina microRNA Expression Beadchip (Human V2). For erythroid differentiation array (group 1), K562 cells were induced by hemin at 0, 48 and 72 h. For GATA-1 over-expression array (group 2), K562 cells were transfected with either pcDNA3.1 vector with full-length cDNA of GATA-1 or empty vector. Forty-eight hours after transfection, RNA was purified from cells using the RNeasy kit with on-column DNase digestion. Over-expression efficiency was confirmed by western blot. Two biological replicates for each group were performed. miRNA gene expression levels were calculated using the PLIER algorithm after a quantile normalization. For group 1, we considered a change in expression significant if fold change was >2, P-value was <0.05, and continuous increase from 0 to 72 h. For group 2, we just considered fold change (>2) and P-value (<0.05). To find miRNA genes potentially activated by GATA-1, we compare up-regulated miRNAs from group 1 with up-regulated miRNAs from group 2 based on the initial screening. MiRNA microarray data has been deposited in the Gene Expression Omnibus database under accession number GSE30380.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from the cell harvest using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instruction. The RNA was quantified by absorbance at 260 nm. cDNA was synthesized by M-MLV reverse transcriptase (Invitrogen) from 2 µg of total RNA or 20 ng of small RNA. Oligo (dT) 18 were used as the RT primers for reverse transcription of mRNAs. Stem-loop RT primers were used for reverse transcription of miRNAs. For mRNAs, quantitative real-time PCR was carried out in BIORAD IQ5 real-time PCR System (Biorad, Foster City, CA, USA) using SYBR Premix Ex Taq kit (Takara, Dalian, China) according to manufacturer’s instruction. For measurement of miR-23a expression in CD34+ HPCs, qPCR was performed using Taqman probes (Applied Biosystems, Foster City, CA, USA): miR-23a (TM399), RNU6B (TM1093) according to manufacturer’s instruction. For mRNAs, the data were normalized using the endogenous GAPDH control. For miRNAs, U6 snRNA was used as the endogenous control. The oligonucleotides used for PCR are listed in Supplementary Table S4.
Oligonucleotides and transfection
miRNA-23a mimics, miRNA-23a inhibitors and negative control molecules (scramble control mimic and inhibitor) were obtained from Dharmacon (Austin, TX, USA) and transfected with DharmFECT1 (Dharmacon) in K562 cells at a final concentration of 60 nM. K562 cells were washed the next day with PBS and plated for hemin induction. siRNAs smart pools (specifically for GATA-1 or SHP2) and control siRNA pools were synthesized by Dharmacon and transfected into K562s (100 nM) using DharmFECT1. Medium was changed after 6 h, cells were cultured for 48 h and harvested for western blot analyses as described below.
Constructs and lentivirus
The reverse complementary sequence of miR-23a was inserted into pGL3 downstream of the firefly luciferase gene (Promega, WI, USA) to generate a reporter system (pGL3-miR-23a) to detect mature miRNA expression in 293T cells. The 3′ untranslated region (UTR) of human SHP2 mRNA was PCR amplified and cloned into pGL3 to generate the corresponding reporter. A mutation in this mRNA sequence was created using the QuickChange SiteDirected Mutagenesis kit (Stratagene, CA, USA). For GATA-1 overexpression, full-length cDNA of GATA-1 was cloned into pcDNA3.1 vector. The primers were listed in Supplementary Table S4. The self-inactivating transfer vector plasmid containing miR-23a (pMIR-lenti-23a), complementary sequence of miR-23a (Lenti-Zip-23a) and the packaging kit were purchased from System Biosciences (SBI, CA, USA) and operated according to the manufacturer’s instructions. The harvested viral particles (Lenti-23a or Lenti-Zip-23a) were added to the CD34+ cultured cells. Cells were washed the next day with PBS and plated for colony-forming experiments and liquid cultures. The shRNA lentivirus plasmids specific to GATA-1 (sc-29330-SH, lenti_si_GATA-1) and SHP2 (sc-36488-SH, lenti_si_SHP2) were purchased from Santa Cruz Biotechnology and operated according to the manufacturer’s instructions.
Isolation and culturing of CD34+ hematopoietic progenitor cells
The isolation system yielded ∼90% CD34-positive cells. Human umbilical cord blood (UCB) was obtained from normal full-term deliveries after informed consent as approved by the Research Ethics Committee of Peking Union Hospital (Beijing, China). Mononuclear cell (MNC) fractions were isolated from UCB by Percoll density gradient (d = 1.077; Amersham Biotech, Germany). CD34+ cells were enriched from MNCs through positive immunomagnetic selection (CD34 MultiSort kit, Miltenyi Biotec). The isolated CD34+ cells were cultured in IMDM supplemented with 30% fetal bovine serum (Hyclone), 1% BSA, 100 µM 2-ME, 2 ng/ml recombinant human IL-3, 100 ng/ml recombinant human stem cell factor (SCF) (Stem Cell Technologies, Vancouver, BC, Canada), 2 U/ml recombinant human erythropoietin (EPO) (R&D Systems, Minneapolis, MN, USA), 60 mg/ml penicillin and 100 mg/ml streptomycin. Cells were harvested every 3–5 days.
Northern and western blot analysis
Northern blot analysis of miRNAs was done as described (15). The oligonucleotide probe sequences are listed in Supplementary Table S2. Whole-cell lysate or nuclear extract was subjected to western blot analysis as detailed elsewhere (20). The following antibodies were used for western blot. GAPDH was purchased from Santa Cruz Biotechnology. SHP2 (BS1322) was purchased from Bioworld Company. GATA-1 (ab11963) was purchased from Abcam.
Luciferase reporter assay
For miRNA target analysis, the 293T cells were co-transfected with 0.4 µg of the reporter construct, 0.02 µg of pRL-TK control vector and 5 pmol of miRNA mimic or scramble controls. Cells were harvested 48 h post-transfection and assayed with Dual Luciferase Assay (Promega) according to manufacturer’s instructions. All transfection assays were carried out in triplicates.
Colony forming assay and Giemsa staining
The colony forming cell assay was performed in triplicate using human methylcellulose media (R&D Systems) according to the manufacturer’s instructions. Human CD34+ cells were cultured in 35 mm plates with medium containing 1.3% methylcellulose, 25% FBS, 2% bovine serum albumin, 2 mmol/l l-glutamine, 0.05 mmol/l 2-mercaptoethanol, 50 ng/ml SCF, 10 ng/ml interleukin-3 (IL-3) and 2 U/ml EPO. To quantify the number of colony-forming unit-erythroid (CFU-E) colonies, the dishes were examined for hemoglobinized and colonies with eight or more cells on day 7 of culture. Burst-forming unit-erythroid (BFU-Es) were scored on day 15 and identified as large aggregates of 64 or more hemoglobinized cells, or as clusters of three or more subcolonies with eight or more hemoglobinized cells per subcolony. For morphological analysis, cells were smeared on glass slides by cytopin centrifugation, stained with Giemsa and analyzed at ×400 magnification under a microscope (Nikon TE2000) equipped with a digital camera.
Rescue assay of miRNA targets
For the detection of effects of miRNAs and target genes on phenotype changes, K562 cells in six-well plates were first transfected with scramble control miRNA inhibitor or miRNA-23a inhibitors (100 nM). After 24 h in culture, these cells were then co-transfected with combination of scramble control miRNA inhibitor (50 nM) and control siRNA (50 nM), miRNA inhibitors and control siRNA, or miRNA inhibitors and siRNA to SHP2 24 h before hemin induction. Cells were harvested at indicated time points after hemin addition and assayed as required.
In vivo functional analysis of miRNAs in zebrafish
Zebrafish was raised and maintained by standard methods (21). Total RNA was isolated from the fertilized eggs at different stages using Trizol reagent (Invitrogen) according to manufacturer’s instruction. Morpholinos (MOs) (Gene Tools, LLC, Philomath, OR, USA) were designed to target the mature miR-23a (miR-23a MO). The standard control MO from Gene Tools was used as a control (ctrl MO). MOs were injected into the yolk of one-cell stage embryos at 8 ng. Whole-mount in situ hybridizations of MO-injected embryos were carried out as described previously by the use of digoxigenin-labeled riboprobes for hbbe3, scl and gata-1 at indicated times (22). Staining of hemoglobin by o-dianisidine was performed on MO-injected embryos at 48 h post-fertilization (hpf), dechorionated and fixed with 4% paraformaldehyde overnight. Fixed embryos were washed in Phosphate-Buffered Saline/Tween (PBST) for three times and then incubated in the staining buffer [0.6 mg/ml o-dianisidine, 10 mM sodium acetate (pH 5.2), 0.65% hydrogen peroxide and 40% ethanol] for 15 min in the dark. For morphological analysis, stained embryos were analyzed at ×400 magnification under a microscope (Nikon TE2000) equipped with a digital camera.
Mice and transplantation assays
All animal experiments were performed with the approval of the Research Ethics Committee of Peking Union Hospital. Bone marrow cells were obtained from 6- to 8-week-old male C57Bl/6 mice injected intravenously with 5 mg 5-fluorouracil. Briefly, bone marrow was flushed from femurs and tibias, and red blood cells were lysed (RBCL buffer, Sigma, CA, USA). Cells were cultured overnight with 2 ng/ml IL-3, 10 ng/ml IL-6 and 100 ng/ml SCF (Stem Cell Technologies) in IMDM supplemented with 30% FBS. Cells were spin-infected for 2 h at a concentration of 1 × 106 cells/ml with the scramble control (Lenti-control) or miR-23a (Lenti-23a) lentiviral supernatant (purchased from GeneChem Company, Shanghai, China) plus 5 μg/ml of polybrene. After a second round of spin infection with lentiviral supernatant, cells were washed and resuspended in PBS containing 2% FBS and injected (1 × 106 cells/0.1 ml) into the lateral tail vein of lethally irradiated (2 × 450 cGy) male NOD/SCID recipient mice. Our results are from three mice per group. Mice were killed after 8 weeks post-transplantation. Blood, bone marrow and spleen were harvested and processed into single-cell suspensions. Samples of spleen were fixed in 10% neutral buffered formalin for paraffin sectioning.
Flow cytometry
K562 cells and CD34+ HPCs were harvested at indicated times and washed twice at 4°C in PBS/0.5% BSA to block Fc receptors. Cells were incubated with PE-conjugated anti-CD71 and FITC-conjugated anti-CD235a antibodies for 30 min (eBioscience; 1 μg/ml). Mice cells were stained with PE-conjugated anti-Ter119 and FITC-conjugated anti-CD71 antibodies (eBioscience). Flow cytometry was carried out on a C6 Flow Cytometer® Instrument (BD Biosciences, Franklin Lakes, NJ, USA). The average value of each experiment was shown in Supplementary Table S3.
Statistics
Student’s t-test (two-tailed) was performed to analyze the data. P values <0.05 were considered significantly as indicated by asterisk (*P values <0.05; **P values <0.01).
RESULTS
miRNA expression profiling in K562 cells
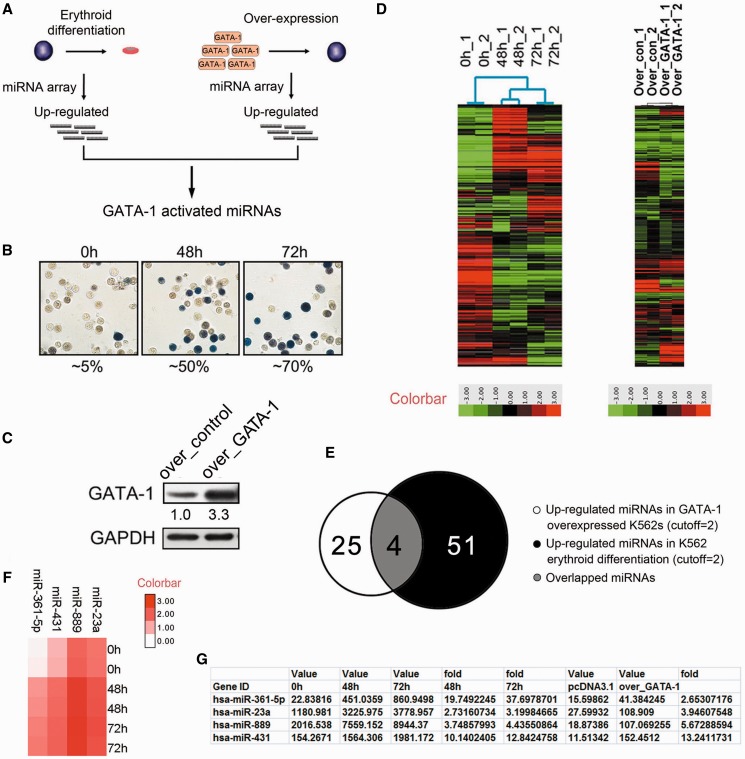
We planned to identify critical miRNA genes that modulate erythroid differentiation and are regulated by the erythroid transcription factor GATA-1. To this end, our strategy was to perform miRNA expression profiling in erythrocytes undergoing either erythroid differentiation or GATA-1 overexpression (Figure 1A). Therefore, we constructed an erythroid differentiation model by using the hemin-induced K562 erythroleukemia cell line. Benzidine staining, which was performed to identify hemoglobin-containing cells and was representative of differentiated erythrocytes, showed that hemin treatment dramatically increased the proportion (from 5 to 50% after 48 h and to 70% after 72 h) of benzidine-positive K562 cells (Figure 1B). We then profiled miRNA gene expression with the illumina hybridization system in undifferentiated and hemin-induced differentiated K562 cells for 48 and 72 h (Figure 1D). Sixty-five miRNA genes showed differential expression during erythroid differentiation (cutoff = 2; P < 0.05; continuous increase from 0 to 72 h). Of the identified miRNA genes, 51 genes were upregulated, and 14 were downregulated (Supplementary Table S1). To define GATA-1-regulated miRNA genes, a comprehensive analysis of GATA-1-induced miRNA gene expression changes was performed in K526 cells overexpressing GATA-1 using the illumina miRNA profiling Beadchip system. Immunoblots were used to identify changes in the GATA-1 protein level (Figure 1C), and miRNA expression was profiled in the identified K562 cells (Figure 1D). Seventy-nine miRNA genes were differentially expressed upon GATA-1 overexpression (cutoff = 2; P < 0.05), of which 25 were upregulated, and 54 were downregulated (Supplementary Table S2).
Figure 1.
Identification of GATA-1-activated miRNAs in K562 cells. (A) The strategy to screen for GATA-1-activated miRNA genes in erythroid cells. (B) Benzidine staining of K562 cells after hemin treatment for 0, 48 or 72 h. Hemoglobinized cells were stained dark blue/brown. The percentage of benzidine-positive cells was indicated below the panel. (C) Western blot analysis of K562 cells transfected with a pcDNA construct overexpressing GATA-1 (over_GATA-1) for 48 h. Signals are normalized to GAPDH, and the values, expressed as fractions with respect to the empty vector (over_control), are indicated below. (D) miRNA microarray of hemin-treated and GATA-1 overexpressing K562 cells at 0, 48 and 72 h. Each sample is biologically duplicated. (E) Venn diagram showing the number of upregulated miRNA genes after hemin treatment (black), the number of upregulated miRNA genes after GATA-1 overexpression (white) and the overlap (gray) between the two. (F) Heatmap showing the expression profiles of miRNA genes that are potentially activated by GATA-1 during K562 erythroid differentiation. (G) Detailed array information of potential GATA-1-activated miRNA genes.
Analysis of GATA-1 activated miRNA genes
Because GATA-1 mainly transactivated its targets during erythroid differentiation, we first focused on identifying GATA-1-activated miRNA genes. To find the most significant miRNAs activated by GATA-1, we analyzed upregulated miRNA genes based on the fold change and P-value of each miRNA from both the erythroid differentiation and GATA-1 overexpression profiles. Our analysis earlier revealed 51 miRNA genes that were upregulated during K562 erythroid differentiation and 25 that were upregulated in GATA-1-overexpressing K562 cells (Figure 1E). Because GATA-1 expression increased during erythropoiesis, miRNA genes with enhanced expression after hemin treatment were likely to be activated by GATA-1. Therefore, we compared the two sets of upregulated miRNA genes and identified four miRNA genes that were potentially activated by GATA-1 (Figure 1F): miR-23a, miR-889, miR-431 and miR-361-5p. None of these miRNAs have been previously implicated in GATA-1 control or erythropoiesis (Figure 1G), and they had the highest potential to be directly or indirectly regulated by GATA-1 on the basis of their absolute expression values (Figure 1G).
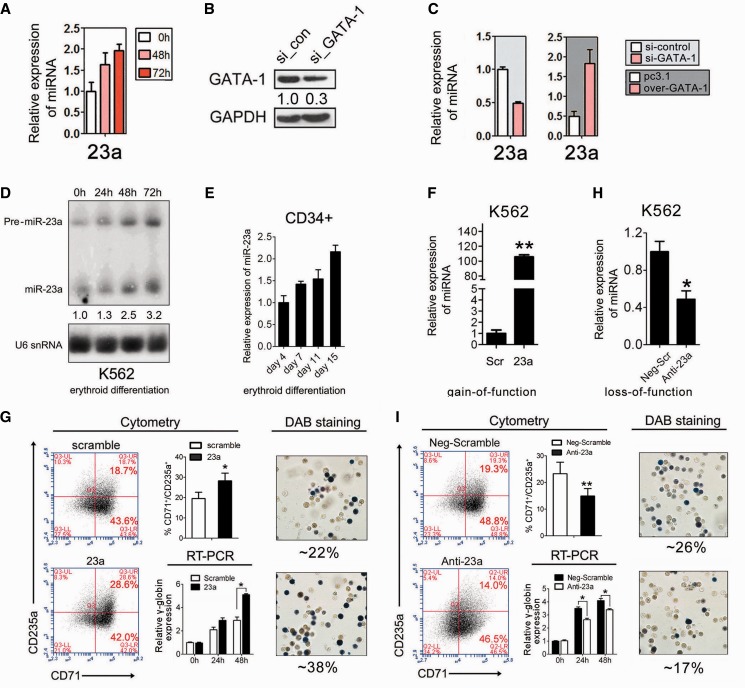
GATA-1 activates miR-23a in K562 cells
Quantitative PCR (qPCR) was conducted to validate the expression of the four miRNA genes that were identified from expression profiling. Of these genes, miR-23a was continuously upregulated at 48 and 72 h in hemin-treated K562 cells (Figure 2A), indicating its importance in erythroid differentiation. To further test the direct relevance of miRNA upregulation in response to GATA-1 activation along with erythroid differentiation, we conducted qPCR to analyze the expression of miR-23a in K562 cells after GATA-1 overexpression or knockdown. To knock-down GATA-1, siRNAs specifically targeting GATA-1 were transfected into K562 cells, and immunoblotting was conducted to assess the efficiency of GATA-1 inhibition (Figure 2B). Indeed, miR-23a showed a 3-fold increase in GATA-1 overexpressing K562 cells. Similarly, miR-23a was downregulated after GATA-1 knockdown in K562 cells (Figure 2C). To further validate our findings, northern blot analysis was used to measure the level of miR-23a in hemin-induced erythroid differentiation of K562 cells. As expected, mature miR-23a expression was similar to that observed above in the miRNA expression array (Figure 2D). In addition, northern blot analysis indicated that the level of the miR-23a precursor (Pre-miR-23a) progressively increased during erythroid differentiation (Figure 2D). Furthermore, qPCR using specific TaqMan probes also showed that mature miR-23a increased during erythroid differentiation of human CD34+ HPCs (Figure 2E). These results raise the possibility that miR-23a is a critical erythroid miRNA gene that is not only a positive regulator of erythroid differentiation but also activated by GATA-1.
Figure 2.
Validation of miRNA genes activated by GATA-1. (A) qPCR showing that miR-23 a is significantly upregulated in hemin-treated K562 cells at 0, 48 and 72 h. Error bars represent the standard deviation obtained from three independent experiments. (B) Western blot analysis of K562 cells after GATA-1 knockdown for 48 h. Signals are normalized to GAPDH, and the values, expressed as fractions with respect to scramble oligonucleotide (si_con), are indicated below. (C) qPCR analysis of miR-23 a in K562 cells with GATA-1 overexpression or knockdown. (D) Northern blot analysis of mature and precursor miR-23 a in hemin-induced K562 cells at 0, 24, 48 and 72 h. U6 snRNA was used as a loading control. (E) qPCR analysis of mature miR-23 a in CD34+ HPCs induced into erythroid differentiation for 4, 7, 11 or 15 days. (F) qPCR analysis of miR-23 a in miRNA mimic-transfected K562 cells after 24 h of hemin treatment. (G) FACS analysis of K562 cells following transfection with scrambled control or miR-23 a mimic and hemin induction for 24 h (left). Summary of data obtained in the left, showing the percentage of CD71+/CD235a+ population was shown in the middle top. qPCR analysis of γ-globin expression in K562 cells transfected with scrambled control or miR-23 a mimic, following hemin induction for 24 or 48 h (middle bottom). Representative benzidine staining of K562 cells treated as described above (right). Mean ± SD were obtained from three independent experiments. (*) γ-globin significantly increased in miR-23 a mimic-transfected K562 cells compared with controls (*P < 0.05). (H) qPCR analysis of miR-23 a in miRNA inhibitor-transfected K562 cells after 24 h of hemin treatment. (I) FACS analysis of K562 cells transfected with scrambled control or miR-23 a inhibitor transfection following hemin induction for 24 h (left). Summary of data obtained in the left, showing the percentage of CD71+/CD235a+ population was shown in the middle top. qPCR analysis of γ-globin expression in K562 cells transfected with scrambled control or miR-23 a inhibitor following hemin induction for 24 or 48 h (middle bottom). Representative benzidine staining of K562 cells treated as described above (right). Mean ± SD were obtained from three independent experiments. (*) γ-globin decreased in miR-23a inhibitor-transfected K562 cells compared with controls (*P < 0.05).
miR-23a promotes erythroid differentiation of K562 cells
To validate our findings, a miR-23a mimic was transfected into K562 cells, and qPCR was performed to assess transfection efficiency (Figure 2F). To assess the influence of miR-23a mimic on erythroid differentiation, we first evaluated hemin-driven differentiation in miRNA mimic transfected K562 cells by staining hemoglobin with benzidine (DAB), fluorescence activated cell sorting (FACS) and qPCR analysis. As expected, DAB staining demonstrated that miR-23a overexpression increased the proportion of benzidine-positive cells after 24 h of hemin treatment in K562 cells (Figure 2G). In addition, a 1.5-fold elevation of γ-globin expression was observed at 48 h in K562 cells transfected with the miR-23a mimic compared with scrambled control (Figure 2G). To further validate our findings, K562 cells were analyzed by FACS using two major erythroid cell surface markers (CD71/CD235a). Overexpression of miR-23a raised the percentage of CD71+/CD235a+ cells (∼9%) compared with the scrambled control (Figure 2G; Supplementary Figure S3 and Table S3). Conversely, a miRNA inhibitor of miR-23a (Anti-23a) (Figure 2H) inhibited the erythroid phenotype in K562 cells, as indicated by the reduced percentage of CD71+/CD235a+ cells (∼6%), decreased benzidine-positive cells and repressed γ-globin accumulation compared with the scrambled control (Figure 2I; Supplementary Figure S3 and Table S3). These results indicate that miR-23a promotes erythroid differentiation in K562 cells.
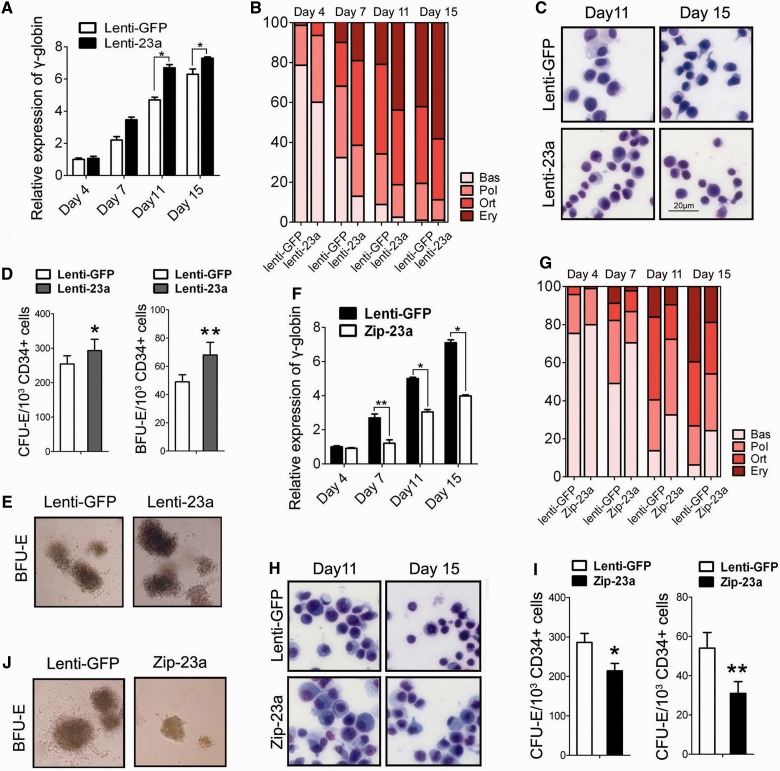
miR-23a promotes erythroid differentiation of human CD34+ HPCs
To assess the influence of miR-23a on primary erythroid differentiation, we transduced miR-23a into HPCs using a recombinant lentivirus harboring miR-23a (Lenti-23a) and subsequently evaluated the expression of γ-globin and CD235a in liquid E culture at 4, 7, 11 and 15 days after miRNA transduction. Before this evaluation, qPCR was conducted to measure the expression of miR-23a, and a 2- to 3-fold increase was observed at the above times (Supplementary Figure S1A). The expression of γ-globin and CD235a was consistently higher in the miR-23a-transduced cells compared with the Lenti-GFP-infected cells (Figure 3A; Supplementary Figure S1C). Cell counting analysis showed a significant increase in the proportion of mature erythroblasts (orthochromatic and erythrocyte) in the miR-23a-transduced HPCs and a concomitant decrease in immature erythroblasts (basophilic and polychromatic erythroblasts), as evaluated at different stages of differentiation (Figure 3B). Likewise, these cells showed morphologic evidence of erythroid differentiation after miRNA transduction. As erythroid differentiation proceeds, erythroblasts display a gradual decrease in cell size and increase in chromatin condensation. Indeed, the miR-23a-transduced HPCs appeared smaller and contained more condensed chromatin than the control cells (Figure 3C). Furthermore, we performed colony formation assays on methylcellulose medium to assess the erythroid clonogenic capacity of transduced HPCs. The numbers of CFU-E and BFU-E colonies were scored after 7 and 15 days of culture. The miR-23a-transduced cells produced more and larger colonies than the Lenti-GFP-infected HPCs (Figure 3D). A representative micrograph of BFU-Es from Lenti-23a or Lenti-GFP transduced HPCs after 15 days E culturing was shown in Figure 3E.
Figure 3.
Functional analysis of miR-23a during erythroid differentiation of human CD34+ HPCs. (A) qPCR analysis of γ-globin mRNA in CD34+ HPCs infected with lentivirus expressing mature miR-23a (Lenti-23a) or a control virus (Lenti-GFP) and cultured in erythroid induction medium for the indicated times. Mean ± SD were obtained from three independent experiments. (*) γ-globin significantly increased in Lenti-23a-transduced CD34+ cells compared with Lenti-GFP-transduced cells (*P < 0.05). (B) Detection of the degree of erythroid differentiation of CD34+ HPCs, as described above. Percentages of basophilic (Bas), polychromatophilic (Pol) and orthochromatic (Ort) erythroblasts and erythrocytes (Ery) were determined by May-Grunwald Giemsa staining of cytospin preparations. (C) Representative May-Grunwald Giemsa staining of CD34+ HPCs infected with Lenti-GFP or Lenti-23a at days 11 and 15 of erythroid induction. (D) A comparison of the erythroid colony-forming capacity of CD34+ HPCs transduced with each virus. Mean ± SD were obtained from three independent experiments. (*) Significantly more clones formed in Lenti-23a transduced CD34+ cells compared with the Lenti-GFP-transduced cells (*P < 0.05; **P < 0.01). (E) Representative erythroid-colony forming assays at day 15 of erythroid induction in CD34+ HPCs transduced with Lenti-GFP or Lenti-23a in semisolid media. (F) qPCR analysis of γ-globin mRNA in CD34+ HPCs infected with lentivirus expressing the miR-23a inhibitor (Zip-23a) or Lenti-GFP and cultured in erythroid induction medium for the indicated times. Mean ± SD were obtained from three independent experiments. (*) γ-globin decreased in the Zip-23a-transduced CD34+ cells compared with the Lenti-GFP-transduced cells (*P < 0.05; **P < 0.01) (G) Detection of the degree of erythroid differentiation of CD34+ HPCs transduced with Lenti-GFP or Zip-23a. (H) Representative May-Grunwald Giemsa staining of CD34+ HPCs infected with Lenti-GFP or Zip-23a at days 11 and 15 of erythroid induction. (I) A comparison of the erythroid colony-forming capacity of CD34+ HPCs transduced with Lenti-GFP or Zip-23a. Mean ± SD were obtained from three independent experiments. (*) Significantly more clones formed in Lenti-23a transduced CD34+ cells than Lenti-GFP-transduced cells (*P < 0.05; **P < 0.01). (J) Representative erythroid colony-forming assays at day 15 of erythroid induction in CD34+ HPCs transduced with Lenti-GFP or Zip-23a in semisolid media.
Next, we performed miR-23a loss-of-function experiments in CD34+ HPCs using recombinant lentivirus carrying antisense RNAs specific for miR-23a (Zip-23a) (the efficiency of miRNA inhibition is shown in Supplementary Figure S1B). In agreement with our gain-of-function data, miR-23a loss of function impaired erythroid maturation, as revealed by expression analysis of the erythroid markers γ-globin and CD235a (Figure 3F; Supplementary Figure S1D), morphological analysis (Figure 3G and H) and colony formation assays (Figure 3I and J). Taken together, these results demonstrate that miR-23a enhances human primary erythroid differentiation.
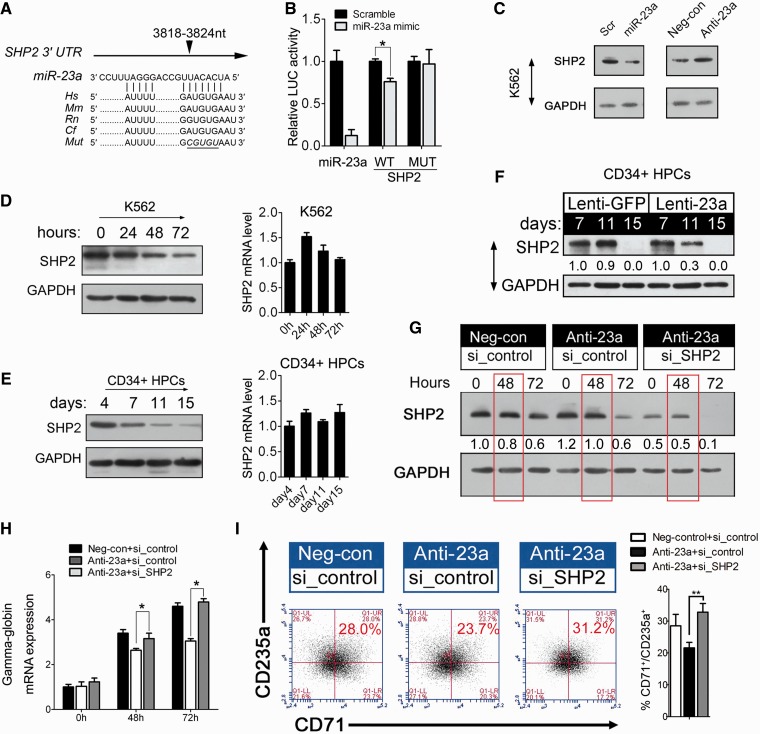
SHP2 is a direct target of miR-23a in erythroid cells
Because miRNAs function by negatively regulating their targets, we decided to search for targets of miR-23a. To this end, we used TargetScan algorithms to predict potential mRNAs with miR-23a-binding sites. Indeed, SHP2 mRNA was predicted as a potential target of miR-23a on the basis of the presence of miRNA-binding sites in its 3′ UTR (Figure 4A). We cloned the 3′ UTR of SHP2 into a luciferase reporter construct (pGL3). Reporter assays in 293T cells revealed miRNA-dependent repression of this 3′ UTR, and mutation of the miRNA-binding site abrogated this reduction in luciferase activity (Figure 4B). Consistent with the reporter assay, we observed a 2-fold decrease of SHP2 expression in the presence of miR-23a mimic in K562 cells compared with the scrambled control, which had no effect (Figure 4C). Conversely, SHP2 was increased after endogenous miR-23a was blocked with the miRNA inhibitor (Figure 4C).
Figure 4.
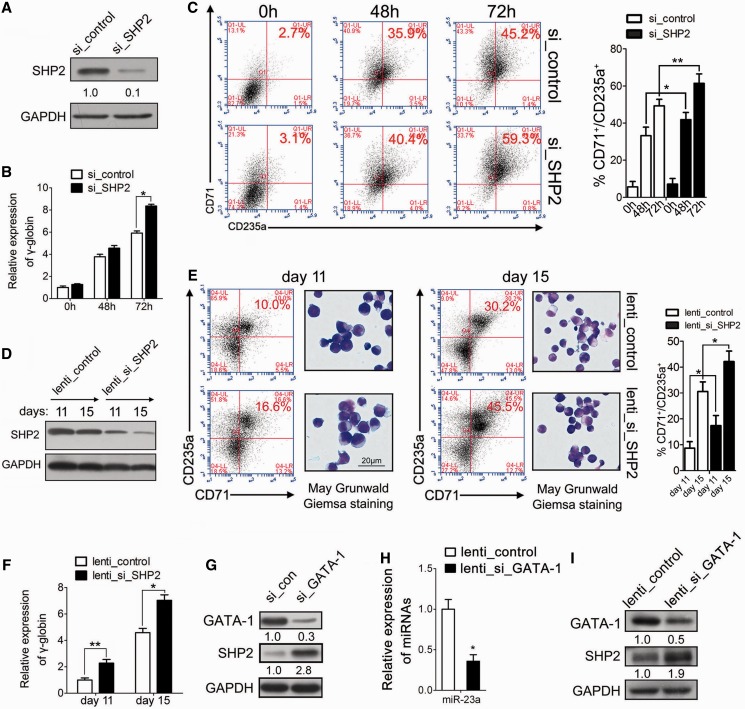
miR-23a directly targets SHP2 to promote erythroid differentiation. (A) Computer prediction of miRNA-binding site in the 3′ UTR of SHP2. (B) Relative luciferase activity of the indicated reporter constructs. Firefly luciferase activity was normalized to the activity of coexpressed Renilla luciferase. Results represent three independent experiments. Mean ± SD were obtained from three independent experiments. (*) Luciferase activity was significantly reduced in miR-23a mimic-transfected 293T cells compared with the scrambled control (*P < 0.05). miR-23a represents a positive construct containing the perfect complementary sequence to mature miR-23a. (C) Western blot analysis of SHP2 expression in K562 cells transfected with scrambled oligonucleotide or miRNA mimic/miRNA inhibitor. (D) Western blot analysis of SHP2 in K562 cells undergoing erythroid differentiation (left). qPCR analysis of SHP2 in K562 cells undergoing erythroid differentiation (right). (E) Western blot analysis of SHP2 in CD34+ HPCs during erythroid induction (left). qPCR analysis of SHP2 in CD34+ HPCs in erythroid induction culture (Right). (F) Western blot analysis of SHP2 in the CD34+ HPCs that were infected with Lenti-23a or Lenti-GFP and cultured in erythroid induction medium for the indicated times. (G) Western blot analysis of SHP2 in K562 cells 24 h after treatment with scrambled or miRNA inhibitor. The cells were subsequently treated for another 24 h with control siRNAs or siRNAs specific to SHP2 and treated with hemin for 0, 48 or 72 h. (H) qPCR analysis of γ-globin mRNA in K562 cells, as described in (G). Error bars represent the standard deviation obtained from three independent experiments. (*) Significant changes of globin expression in the indicated group compared with the control (*P < 0.05). (I) K562 cells were hemin-treated for 48 h as in (G) and stained for CD71 and CD235a to determine the degree of erythroid differentiation. Summary of data obtained in FACS, showing the percentage of CD71+/CD235a+ population was shown in the right. Signals from western blots are normalized to GAPDH, and the values, expressed as fractions with respect to the control, are indicated below.
To further establish the regulatory interaction between miR-23a and SHP2, we evaluated the expression of SHP2 during K562 and CD34+ HPC erythroid differentiation. A continuous decrease in the protein level of SHP2 was observed in both erythroid models (Figure 4D and E), whereas SHP2 mRNA remained unchanged (Figure 4D and E). In addition, we observed enhanced accumulation of mature miR-23a in erythroid cultured HPCs (Figure 2E), and the reciprocal expression of miR-23a and SHP2 verified the functional significance of their interaction during erythroid differentiation. Next, we transduced CD34+ HPCs withLenti-23a to evaluate its regulation of SHP2 in primary cultured erythroid cells. After 11 days, miR-23a transduction decreased SHP2 expression to 30% of the control value (Figure 4F). Thus, our attempt to investigate the molecular mechanism of miR-23a activity led to the identification of SHP2 as a direct target of miR-23a during erythropoiesis.
miR-23a-mediated inhibition of SHP2 expression is required for erythroid differentiation
To clarify the biological link between miR-23a, SHP2 and the erythroid phenotype, we designed a rescue experiment to assess their functional relevance in differentiating K562 cells. First, an increase in SHP2 was observed after Anti-miR-23a treatment, confirming the effect of miR-23a on target expression in this process (Figure 4G, Anti-23a+si_control versus Neg-con+si_control). Furthermore, the addition of specific SHP2 siRNAs led to a further inhibition of SHP2 protein, in addition to the pre-existing increase (Figure 4G, Anti-23a+si_control versus Anti-23a+si_target). For quantification, protein levels of SHP2 were normalized to GAPDH at 48 and 72 h after hemin induction in K562 cells (Figure 4G). In agreement with the rescue of SHP2 protein expression, restored γ-globin accumulation (Figure 4H) was observed at 48 and 72 h after hemin induction in K562 cells transfected with miR-23a inhibitor followed by siRNA treatment. Similarly, the percentage of differentiated erythroid cells (CD71+/CD235a+ cells) that were rescued was calculated at 48 h after hemin induction in K562 cells, as described earlier (Figure 4I; Supplementary Figure S4 and Table S3). These data confirm the biological relevance of miR-23a-mediated regulation of SHP2 in erythropoiesis.
SHP2 acts as a negative regulator of erythroid differentiation
To date, the function of SHP2 in erythropoiesis is not well known. Because SHP2 was downregulated in both the K562 and CD34+ erythroid differentiation models, it was reasonable to hypothesize that SHP2 is a negative regulator of erythroid differentiation. To investigate its biological role in erythroid differentiation, we performed a loss-of-function experiment using SHP2 siRNAs in K562 cells (Figure 5A). As expected, SHP2 silencing increased the expression of γ-globin in K562 cells at 48 and 72 h (Figure 5B). Similarly, the percentage of CD71+/CD235a+ cells (∼5% at 48 h and ∼14% at 72 h) was increased in SHP2 siRNA-transfected K562 cells (Figure 5C; Supplementary Table S3). To further validate our findings in human primary CD34+ HPCs, lentivirus was used to express specific siRNAs that inhibited endogenous SHP2. Immunoblots were performed to test the efficiency of lentivirus infection and showed 2- to 4-fold decreases of SHP2 protein expression following Lenti_si_SHP2 treatment at the indicated times (Figure 5D). In agreement with the data from K562 cells, the loss-of-function study showed impaired erythroid maturation, as revealed by the increased percentage of CD235a+/CD71+ cells, morphological analysis (Figure 5E; Supplementary Table S3) and expression of γ-globin (Figure 5F).
Figure 5.
SHP2 is a negative regulator of late erythroid differentiation. (A) Western blot analysis of SHP2 expression in K562 cells transfected with control siRNAs or SHP2 siRNAs for 48 h. (B). qPCR analysis of γ-globin expression in K562 cells treated as described in (A) at 0, 24, 48 and 72 h. (C) K562 cells were hemin-treated for 48 or 72 h in (B) and were stained for CD71 and CD235a to determine the degree of erythroid differentiation. Summary of data obtained in FACS, showing the percentage of CD71+/CD235a+ population was shown in the right. (D) Western blot analysis of SHP2 in CD34+ HPCs infected with lentivirus expressing SHP2 siRNAs (Lenti_si_SHP2) or a control virus (Lenti_control) and cultured in erythroid induction medium for 11 or 15 days. (E) FACS analysis by CD71/CD235a double staining at days 11 and 15. CD34+ HPCs were treated as described in (D) (left). Representative May-Grunwald Giemsa staining of CD34+ HPCs infected with Lenti_control or Lenti_si_SHP2 at days 11 and 15 of erythroid induction (right). Summary of data obtained in FACS, showing the percentage of CD71+/CD235a+ population was shown in the far right. (F) qPCR analysis of γ-globin expression in CD34+ HPCs, treated as described in (D). (G) Western blot analysis of SHP2 in K562 cells transfected with GATA-1 siRNAs for 48 h. (H) qPCR analysis of miR-23a in the CD34+ HPCs that were infected with lentivirus expressing GATA-1 shRNA (Lenti_si_GATA-1) or a control virus (Lenti_control) and cultured in erythroid induction medium for the indicated times. (*) Significant reduction of miR-23a expression in Lenti_GATA-1-transduced HPCs compared with the control (*P < 0.05). (I) Western blot analysis of SHP2 in the CD34+ HPCs, treated as described in (H). Signals from western blots are normalized to GAPDH, and the values, expressed as fractions with respect to the control, are indicated below.
According to our data, miR-23a directly suppresses SHP2 expression and is activated by the accumulation of GATA-1 during erythropoiesis. A genetic cascade mediated by miRNAs might occur between GATA-1 and SHP2 in erythroid cells. To determine whether GATA-1 could regulate SHP2 through the activation of miR-23a, we first treated K562 cells with a siRNA specific to GATA-1 to discern the responses of miRNA target genes. miRNA repression upon GATA-1 silencing resulted in an increase in the total protein level of SHP2 (Figure 5G), confirming that the production of SHP2 was affected by GATA-1. Then, qPCR was performed to measure miR-23a in lenti_si_GATA-1-infected human CD34+ HPCs. Mature miR-23a was downregulated in HPCs in which GATA-1 was silenced (Figure 5H). Immunoblots showed increased SHP2 in the Lenti_si_GATA-1-transduced HPCs compared with the control cells (Figure 5I).
Suppression of miR-23a blocks erythroid differentiation in zebrafish
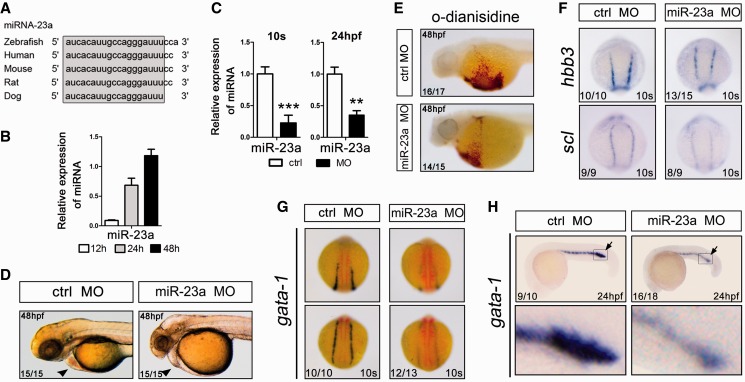
Because miR-23a is highly conserved among multiple species (Figure 6A) and is upregulated during early zebrafish development, we decided to analyze its function in vivo (Figure 6B). To this end, we used miRNA MOs to test whether knocking down the endogenous miR-23a would affect zebrafish erythropoiesis. MOs complementary to mature miR-23a (miR-23a MO) were injected into zebrafish embryos at the one-cell stage. qPCR analysis showed a 3- to 4-fold reduction in the expression of mature miRNAs in MO-injected embryos (Figure 6C). At 24 hpf, miRNA MO-injected embryos showed a reduced number of blood cells in circulation in the presence of a beating heart. Interestingly, there were negligible or fewer blood cells inside the heart and blood vessels at 48 hpf compared with the control embryos (Figure 6D), suggesting perturbed hematopoiesis during early zebrafish development.
Figure 6.
Suppression of miR-23a blocks erythroid differentiation in zebrafish. (A) Manual alignment of mature miR-23a genomic sequences in five vertebrate species. Sequences derived from the miRBase database (Release 18). (B) qPCR analysis of miR-23a at 12, 24 and 48 h of zebrafish embryo development. Error bars represent standard deviation obtained from three independent experiments. (C) qPCR analysis of miR-23a in 10 somites and 24 hpf embryos after the indicated treatments. (*) Significant changes in embryos injected with the morpholino antagonist of miR-23a (miR-23a MO) compared with control morpholino (Ctrl MO) (*P < 0.05; **P < 0.01). (D) Lateral view of the heart and yolk sac of Ctrl MO- or miR-23a MO-injected embryos. (E) o-Dianisidine staining of hemoglobin in randomly selected 48 hpf embryos injected with Ctrl MO or miR-23a MO. (F) Expression of the hematopoietic lineage markers hbbe3 and scl in 10 somites embryos by whole-mount in situ hybridization. Randomly selected 10 somites embryos, injected with Ctrl MO or miR-23a MO, showing a normal degree of hbbe3 and scl staining in both groups. (G and H) Expression of gata-1 in 10 somites (G) and 24 hpf (H) embryos by whole-mount in situ hybridization. Randomly selected 10 somites and 24 hpf embryos, injected with Ctrl MO or miR-23a MO, showing a normal degree of gata-1 staining in both groups.
To further examine the requirement of miR-23a in erythropoiesis, we used o-dianisidine staining to detect the number of circulating erythrocytes. In control embryos, hemoglobin-positive erythrocytes were robustly stained with o-dianisidine, whereas miRNA MO-injected embryos relatively lacked staining (Figure 6E). Furthermore, the expression of two erythroid markers hbbe3 and scl in miRNA MO-injected embryos notably decreased compared with control embryos at 10 somites (Figure 6F). In agreement with this, a reduction in gata-1 staining was also observed in miR-23a MO-injected embryos at 10 somites (Figure 6G) and 24 hpf (Figure 6H), suggesting that miRNA MO treatment impaired erythroid differentiation. These results are compatible with the notion that, in zebrafish, miR-23a is required for erythroid differentiation during primitive hematopoiesis.
Forced expression of miR-23a enhances mature erythroid populations in mice
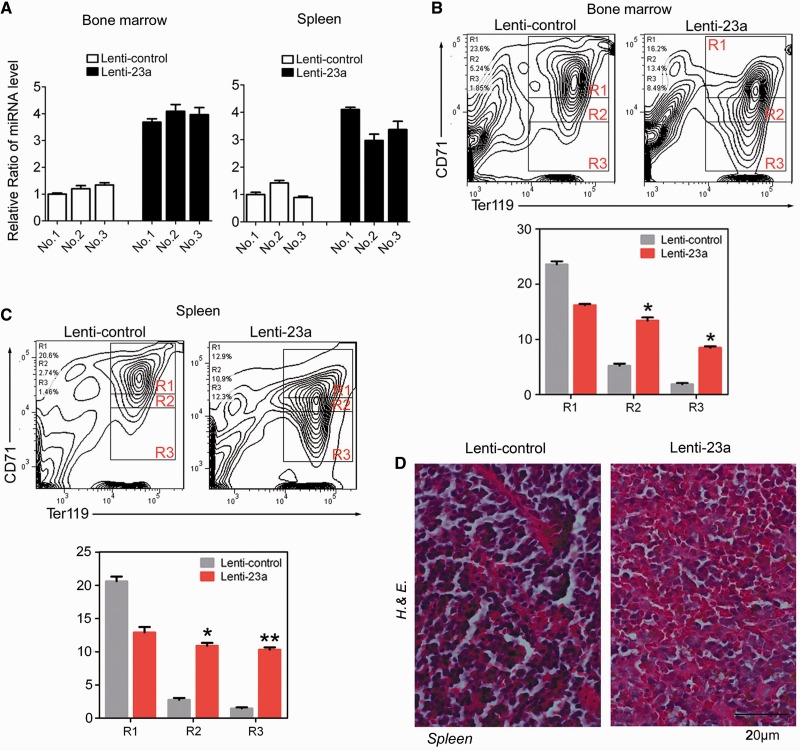
To further test the role of miR-23a in vivo, we performed transplantation experiments in mice. To this end, empty control or miR-23a-transduced bone marrow cells were transplanted into lethally irradiated NOD/SCID mice. Engraftment into recipient mice was confirmed by analyzing miRNA expression in bone marrow and spleen after 8 weeks (Figure 7A). To determine the effect of miR-23a on erythroid differentiation in adult hematopoietic tissues, flow cytometry analysis was performed 8 weeks post-transplantation. As expected, animals overexpressing miR-23a displayed an increase in region 3 of CD71low/TER119high erythrocytes and a concomitant decrease in region 1 of CD71high/TER119high erythroblasts from bone marrow and spleen (Figure 7B and C). This change likely contributed to the accelerated hematocrit of the miRNA-transduced animals compared with the controls (Supplementary Figure S2). In addition, histologic analysis of spleens revealed fewer erythroid cell clusters in the miR-23a-transduced mice, indicating increased terminal erythroid maturation (Figure 7D). Thus, forced expression of miR-23a enhanced the mature erythroid populations in these transplanted mouse models.
Figure 7.
miR-23a overexpression enhances erythropoiesis in mice. (A) qPCR analysis of miR-23a in bone marrow and spleen of mice, 8 weeks post-transplantation with empty control- or miRNA-transduced bone marrow cells. (B and C) FACS analysis of bone marrow (BM) (B) and spleen (SP) (C) harvested from mice at 8 weeks post-transplantation, stained for CD71 and TER119. Representative FACS plots from one mouse are shown. The percentage of cells gated within each region is quantified below. Data from control- (Lenti-control, n = 3) or miR-23a-transduced (Lenti-23a, n = 3) animals are shown as means ± SD. *P < 0.05; **P < 0.01. (D) Hematoxylin and eosin-stained sections of spleens harvested from mice at 8 weeks post-transplantation. A ×400 magnification of a representative field is shown.
DISCUSSION
In the last few years, our growing understanding of miRNAs has uncovered intricate gene-regulatory networks modulated by these post-transcriptional repressors (23,24). As the complement to classic transcription factor-governed gene regulation, miRNAs exert their functions as fine regulators to achieve efficient and elaborate control of gene expression. Most miRNA genes are differentially expressed at different locations, at different times during development or in response to environmental signals (25–28). Differential miRNA expression is largely controlled by various transcription factors (29–31). Moreover, the regulatory effects of miRNAs are often highly coordinated with such transcription factors, which control miRNA expression either directly or indirectly (32). Although numerous methods exist for elucidating miRNA- or transcription factor-related regulatory networks, the comparable information to explicitly connect them is still lacking. In this work, we investigated erythroid-associated miRNAs from coordinated miRNA- and GATA-1-regulated miRNA expression profiles to identify significant erythroid miRNAs. After a stringent filtering process, our study identified four previously uncharacterized miRNAs that were evaluated in the context of erythrocyte maturation and GATA-1 activation. We further characterized miR-23a because it showed a considerable ability to promote erythroid differentiation. A known GATA-1-regulated miRNA, mmu-miR-451, did not emerge from our analysis, possibly as a result of species diversity between human and mouse or our stringent filter settings.
miR-23a has been studied in various biological systems, including osteoblast differentiation, angiogenesis, cardiac remodeling, skeletal muscle atrophy and B-cell development (29,33,34). In different tissue types, miR-23a displays distinct, differential activities, which are coordinated to specific transcription factors (35–42). miR-23a is involved in antagonizing lymphoid cell fate acquisition, as a downstream target of PU.1 (28). Runx2, a transcription factor essential for osteoblastogenesis, negatively regulates miR-23a to induce osteoblast differentiation (29). miR-23a also functions downstream of the prohypertrophic transcription factor NFATc3 to regulate cardiac hypertrophy (34). Although there have been a few reports concerning the upregulation of miR-23a in hematopoiesis (43,44), the role for miR-23a in erythroid differentiation is not fully understood. Here, we demonstrated that miR-23a promoted the erythroid development of K562 cells and CD34+ primary erythroid progenitor cells by both gain- and loss-of-function experiments. Furthermore, we confirmed the requirement of miR-23a for zebrafish and mouse erythropoiesis through in vivo studies. Recently, the miR-23a/27a/24-2 tricistron has been reported to highly express in normal megakaryocytes and blocks maturation and platelet formation (45). Megakaryocytes and erythrocytes are derived from a process by which megakaryocyte-erythrocyte progenitors commit to becoming either erythroid precursors, yielding mature red blood cells, or megakaryocytes, yielding mature platelets. Thus, miR-23a might be an important regulator of hematopoietic lineage specification.
We also identified the tyrosine phosphatase SHP2, which functions in cell survival, proliferation and differentiation in many tissues, as a direct target of miR-23a. SHP2 is highly expressed in hematopoietic cells and participates in hematopoietic growth factor signal transduction through IL-3, EPO and SCF (46,47). However, compared with the SHP1 phosphatase, our understanding of the physiological and biochemical functions of SHP2 in hematopoietic cells is incomplete. Germ line and somatic gain-of-function mutations in SHP2 are associated with juvenile myelomonocytic leukemia (JMML), a myeloproliferative disease of early childhood (48,49). Expression of the leukemia-associated mutant SHP2 (D61Y) in hematopoietic cells causes elevated ERK and STAT5 activation in response to growth factor stimulation and produces an increased number of immature progenitor cells (47). This observation is consistent with the essential role for SHP2 in survival and maintenance of the hematopoietic stem and progenitor cells (47,50). SHP2-deficient human and mouse hematopoietic stem cells show reduced proliferation and survival, in addition to defective ERK and AKT activation in response to SCF (43). However, there are few reports concerning the role of SHP2 in the late stage of erythropoiesis. SHP2 (D61Y) hematopoietic stem cells yield increased numbers of erythrocyte-committed progenitors but fewer erythrocyte colony-forming units than wild-type SHP2, indicating defective terminal erythroid differentiation upon SHP2 activation (47). We found that SHP2 continuously decreased during terminal erythroid differentiation of both K562 cells and primary CD34+ HPCs. Knockdown of SHP2 in K562 cells and HPCs efficiently increased the proportion of differentiated cells, as reduced SHP2 is required for cells to withdraw from the hyperproliferative progenitor state during the terminal stage of differentiation. Our finding that miR-23a regulates SHP2 suggests that miR-23a may have inhibitory roles in the SHP2-mediated cell proliferation and survival pathways and may serve as a therapeutic target for JMML.
In conclusion, our investigation of GATA-1-regulated miRNAs identified a critical regulatory link comprised GATA-1, miR-23a and SHP2 during erythroid differentiation. In differentiated erythroid cells, accelerated GATA-1 expression increases the accumulation of miR-23a, and miR-23a directly represses the negative modulator SHP2 to promote terminal erythropoiesis. Thus, with rapidly increasing information concerning miRNA profiles and transcription factor regulation, the integration of these data may improve our understanding of the complex and combinatorial nature of the eukaryotic genome, representing a prominent area in the investigation of miRNA function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–4.
FUNDING
The National Natural Science Foundation of China [2010, 31040021 to J.Y.; 2011, 31150002 to F.W.; 2010, 60905014 to T.T.L.; 2012, 81271643 to J.G.Z.]; the National Key Basic Research Program of China [2011CBA01100 to J.Y.]; the IBMS, CAMS [2009RC03 to J.Y., 2010PYB06 to J.Y.]; the Beijing Municipal Science & Technology Commission [2010B071 to J.Y.]. Funding for open access charge: National Key Basic Research Program of China [2011CBA01100 to J.Y.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Bin Zhu from Peking Union Hospital for assistance in umbilical cord blood preparation; H.-L. Zhao for technical assistance.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Lawrie CH. microRNA expression in erythropoiesis and erythroid disorders. Br. J. Haematol. 2010;150:144–151. doi: 10.1111/j.1365-2141.2009.07978.x. [DOI] [PubMed] [Google Scholar]
- 4.Kerenyi MA, Orkin SH. Networking erythropoiesis. J. Exp. Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, Jun-Shen HL, Olson EN. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O'Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dore LC, Amigo JD, Dos SC, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep. 2009;42:493–499. doi: 10.5483/bmbrep.2009.42.8.493. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Yu J, Yang GH, Wang XS, Zhang JW. Regulation of erythroid differentiation by miR-376a and its targets. Cell Res. 2011;21:1196–1209. doi: 10.1038/cr.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan JY, Wang F, Yu J, Yang GH, Liu XL, Zhang JW. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell. Mol. Med. 2009;13:4551–4559. doi: 10.1111/j.1582-4934.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 16.Yang GH, Wang F, Yu J, Wang XS, Yuan JY, Zhang JW. MicroRNAs are involved in erythroid differentiation control. J. Cell. Biochem. 2009;107:548–556. doi: 10.1002/jcb.22156. [DOI] [PubMed] [Google Scholar]
- 17.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 18.Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 20.Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl Acad. Sci. USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerfield M, Wegner J, Jegalian BG, DeRobertis EM, Puschel AW. Specific activation of mammalian Hox promoters in mosaic transgenic zebrafish. Genes Dev. 1992;6:591–598. doi: 10.1101/gad.6.4.591. [DOI] [PubMed] [Google Scholar]
- 22.Westerfield M, Liu DW, Kimmel CB, Walker C. Pathfinding and synapse formation in a zebrafish mutant lacking functional acetylcholine receptors. Neuron. 1990;4:867–874. doi: 10.1016/0896-6273(90)90139-7. [DOI] [PubMed] [Google Scholar]
- 23.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Moss EG. MicroRNAs: hidden in the genome. Curr. Biol. 2002;12:R138–R140. doi: 10.1016/s0960-9822(02)00708-x. [DOI] [PubMed] [Google Scholar]
- 25.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim. Biophys. Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem. Sci. 2008;33:474–481. doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 29.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc. Natl Acad. Sci. USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong KY, Owens KS, Rogers JH, Mullenix J, Velu CS, Grimes HL, Dahl R. MIR-23A microRNA cluster inhibits B-cell development. Exp. Hematol. 2010;38:629–640. doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–445. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian L, Van Laake LW, Huang Y, Liu S, Wendland MF, Srivastava D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 2011;208:549–560. doi: 10.1084/jem.20101547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl Acad. Sci. USA. 2009;106:12103–12108. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Lin ZQ, Long B, Li JH, Zhou J, Li PF. Cardiac hypertrophy is positively regulated by MicroRNA miR-23a. J. Biol. Chem. 2012;287:589–599. doi: 10.1074/jbc.M111.266940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Yang X, Zhou Y, Peng S, Wu L, Lin H, Wang SY. Differentially expressed plasma microRNAs in premature ovarian failure patients and the potential regulatory function of mir-23a in granulosa cell apoptosis. Reproduction. 2012;144:235–244. doi: 10.1530/REP-11-0371. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Wei W, Sarkar FH. miR-23a, a critical regulator of “migR"ation and metastasis in colorectal cancer. Cancer Discov. 2012;2:489–491. doi: 10.1158/2159-8290.CD-12-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathore MG, Saumet A, Rossi JF, de Bettignies C, Tempe D, Lecellier CH, Villalba M. The NF-kappaB member p65 controls glutamine metabolism through miR-23a. Int. J. Biochem. Cell Biol. 2012;44:1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 39.Tsai YS, Lin CS, Chiang SL, Lee CH, Lee KW, Ko YC. Areca nut induces miR-23a and inhibits repair of DNA double-strand breaks by targeting FANCG. Toxicol. Sci. 2011;123:480–490. doi: 10.1093/toxsci/kfr182. [DOI] [PubMed] [Google Scholar]
- 40.Chhabra R, Dubey R, Saini N. Gene expression profiling indicate role of ER stress in miR-23a∼27a∼24-2 cluster induced apoptosis in HEK293T cells. RNA Biol. 2011;8:648–664. doi: 10.4161/rna.8.4.15583. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y, Li Y, Ye F, Wang F, Wan X, Lu W, Xie X. Identification of miR-23a as a novel microRNA normalizer for relative quantification in human uterine cervical tissues. Exp. Mol. Med. 2011;43:358–366. doi: 10.3858/emm.2011.43.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegel C, Li J, Liu F, Benashski SE, McCullough LD. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proc. Natl Acad. Sci. USA. 2011;108:11662–11667. doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009;16:492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp. Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Emmrich S, Henke K, Hegermann J, Ochs M, Reinhardt D, Klusmann JH. miRNAs can increase the efficiency of ex vivo platelet generation. Ann. Hematol. 2012;91:1673–1684. doi: 10.1007/s00277-012-1517-z. [DOI] [PubMed] [Google Scholar]
- 46.Chan G, Cheung LS, Yang W, Milyavsky M, Sanders AD, Gu S, Hong WX, Liu AX, Wang X, Barbara M, et al. Essential role for Ptpn11 in survival of hematopoietic stem and progenitor cells. Blood. 2011;117:4253–4261. doi: 10.1182/blood-2010-11-319517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Li Y, Yin F, Chan RJ. Activating PTPN11 mutants promote hematopoietic progenitor cell-cycle progression and survival. Exp. Hematol. 2008;36:1285–1296. doi: 10.1016/j.exphem.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu D, Wang S, Yu WM, Chan G, Araki T, Bunting KD, Neel BG, Qu CK. A germline gain-of-function mutation in Ptpn11 (Shp-2) phosphatase induces myeloproliferative disease by aberrant activation of hematopoietic stem cells. Blood. 2010;116:3611–3621. doi: 10.1182/blood-2010-01-265652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan G, Kalaitzidis D, Usenko T, Kutok JL, Yang W, Mohi MG, Neel BG. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 2009;113:4414–4424. doi: 10.1182/blood-2008-10-182626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Modi H, McDonald T, Rossi J, Yee JK, Bhatia R. A critical role for SHP2 in STAT5 activation and growth factor-mediated proliferation, survival, and differentiation of human CD34+ cells. Blood. 2011;118:1504–1515. doi: 10.1182/blood-2010-06-288910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.