Abstract
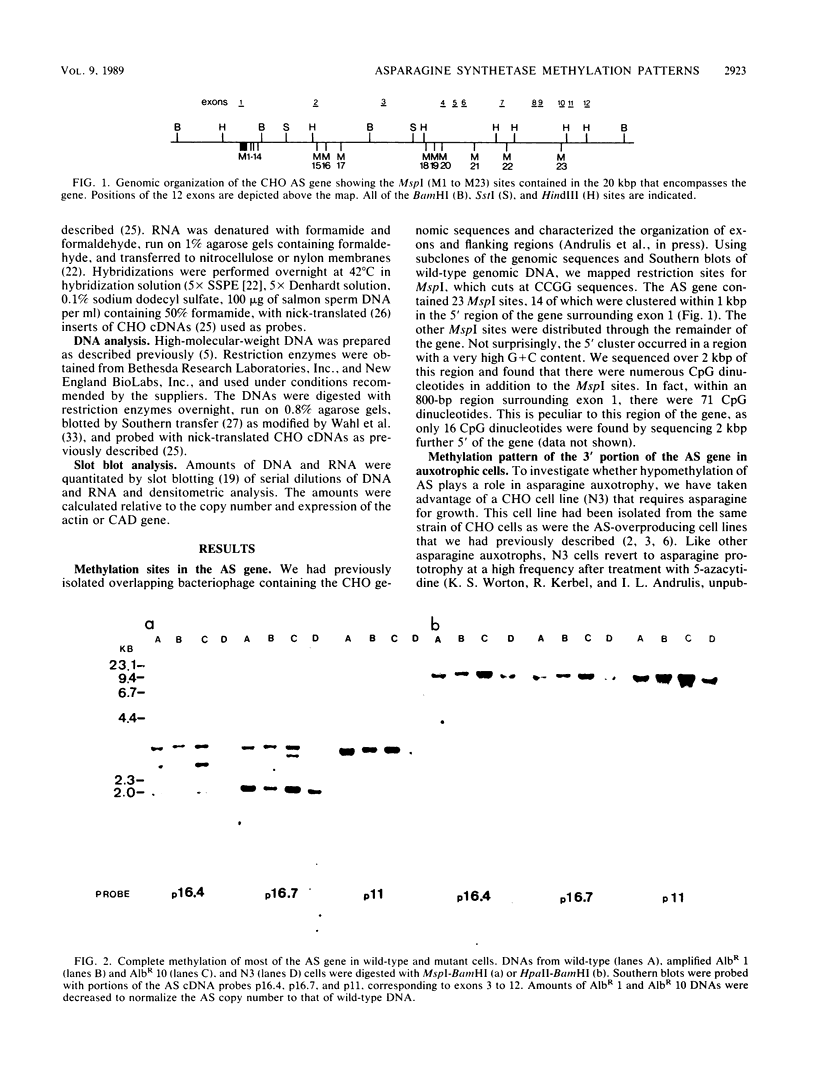
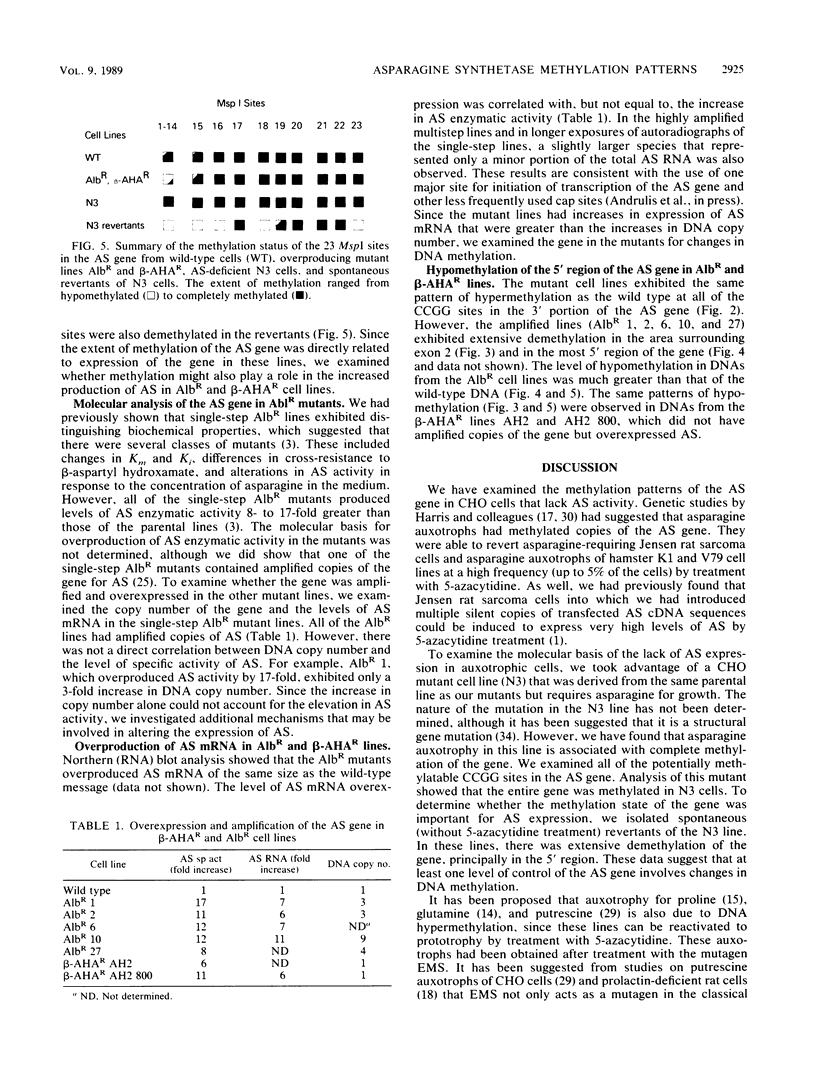
In Chinese hamster ovary cells, the gene for asparagine synthetase, which spans 20 kilobase pairs, was found to contain a cluster of potential sites for CpG methylation in a 1-kilobase-pair region surrounding the first exon. Fourteen of the sites that could be assayed for methylation by MspI-HpaII digestions were found in this region, with an additional nine MspI sites spread throughout the remainder of the gene. The methylation status of the gene was analyzed in a series of cell lines that differed in the amount of asparagine synthetase activity. The level of expression showed a direct correlation with the extent of methylation of a subset of the MspI sites found in the 5' region of the gene. The rest of the gene was completely methylated in most cell lines. Wild-type cells, which expressed a basal level of asparagine synthetase activity, were partially demethylated in the 5' region. In contrast, asparagine-requiring N3 cells, which lacked detectable mRNA for asparagine synthetase, were methylated throughout the entire gene. Spontaneous revertants of strain N3, selected for growth in asparagine-free medium, exhibited extensive hypomethylation of the asparagine synthetase gene. The methylation pattern of the gene in cell lines that overproduced the enzyme was also examined. Albizziin-resistant cell lines, which had amplified copies of the gene, were extensively demethylated in the 5' region. Overexpression of asparagine synthetase in beta-aspartyl hydroxamate-resistant lines without amplified copies of the gene was also correlated with DNA hypomethylation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrulis I. L., Chen J., Ray P. N. Isolation of human cDNAs for asparagine synthetase and expression in Jensen rat sarcoma cells. Mol Cell Biol. 1987 Jul;7(7):2435–2443. doi: 10.1128/mcb.7.7.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis I. L., Duff C., Evans-Blackler S., Worton R., Siminovitch L. Chromosomal alterations associated with overproduction of asparagine synthetase in albizziin-resistant Chinese hamster ovary cells. Mol Cell Biol. 1983 Mar;3(3):391–398. doi: 10.1128/mcb.3.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis I. L., Evans-Blackler S., Siminovitch L. Characterization of single step albizziin-resistant Chinese hamster ovary cell lines with elevated levels of asparagine synthetase activity. J Biol Chem. 1985 Jun 25;260(12):7523–7527. [PubMed] [Google Scholar]
- Andrulis I. L., Hatfield G. W., Arfin S. M. Asparaginyl-tRNA aminoacylation levels and asparagine synthetase expression in cultured Chinese hamster ovary cells. J Biol Chem. 1979 Nov 10;254(21):10629–10633. [PMC free article] [PubMed] [Google Scholar]
- Andrulis I. L., Siminovitch L. DNA-mediated gene transfer of beta-aspartylhydroxamate resistance into Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5724–5728. doi: 10.1073/pnas.78.9.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis I. L., Siminovitch L. Isolation and characterization of Chinese hamster ovary cell mutants resistant to the amino acid analog beta-aspartyl hydroxamate. Somatic Cell Genet. 1982 Jul;8(4):533–545. doi: 10.1007/BF01538713. [DOI] [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Gantt J. S., Chiang C. S., Hatfield G. W., Arfin S. M. Chinese hamster ovary cells resistant to beta-aspartylhydroxamate contain increased levels of asparagine synthetase. J Biol Chem. 1980 May 25;255(10):4808–4813. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington M. A., Jones P. A., Imagawa M., Karin M. Cytosine methylation does not affect binding of transcription factor Sp1. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2066–2070. doi: 10.1073/pnas.85.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. High-frequency induction by 5-azacytidine of proline independence in CHO-K1 cells. Somat Cell Mol Genet. 1984 Nov;10(6):615–624. doi: 10.1007/BF01535227. [DOI] [PubMed] [Google Scholar]
- Harris M. Induction and reversion of asparagine auxotrophs in CHO-K1 and V79 cells. Somat Cell Mol Genet. 1986 Sep;12(5):459–466. doi: 10.1007/BF01539917. [DOI] [PubMed] [Google Scholar]
- Harris M. Simultaneous induction of epigenetic variants. Somat Cell Mol Genet. 1986 Nov;12(6):567–573. doi: 10.1007/BF01671942. [DOI] [PubMed] [Google Scholar]
- Harris M. Variants inducible for glutamine synthetase in V79-56 cells. Somat Cell Mol Genet. 1984 May;10(3):275–281. doi: 10.1007/BF01535249. [DOI] [PubMed] [Google Scholar]
- Ivarie R. D., Morris J. A. Induction of prolactin-deficient variants of GH3 rat pituitary tumor cells by ethyl methanesulfonate: reversion by 5-azacytidine, a DNA methylation inhibitor. Proc Natl Acad Sci U S A. 1982 May;79(9):2967–2970. doi: 10.1073/pnas.79.9.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith D. H., Singer-Sam J., Riggs A. D. Active X chromosome DNA is unmethylated at eight CCGG sites clustered in a guanine-plus-cytosine-rich island at the 5' end of the gene for phosphoglycerate kinase. Mol Cell Biol. 1986 Nov;6(11):4122–4125. doi: 10.1128/mcb.6.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock L. F., Melton D. W., Caskey C. T., Martin G. R. Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol Cell Biol. 1986 Mar;6(3):914–924. doi: 10.1128/mcb.6.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. J., Grosveld F. Site specific demethylation in the promoter of human gamma-globin gene does not alleviate methylation mediated suppression. EMBO J. 1987 Aug;6(8):2329–2335. doi: 10.1002/j.1460-2075.1987.tb02508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M. K., Jr, Orr G. L-asparagine biosynthesis by nutritional variants of the Jensen sarcoma. Biochem Biophys Res Commun. 1967 Jan 23;26(2):228–233. doi: 10.1016/0006-291x(67)90239-2. [DOI] [PubMed] [Google Scholar]
- Ray P. N., Siminovitch L., Andrulis I. L. Molecular cloning of a cDNA for Chinese hamster ovary asparagine synthetase. Gene. 1984 Oct;30(1-3):1–9. doi: 10.1016/0378-1119(84)90098-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanners C. P., Eliceiri G. L., Green H. Two types of ribosome in mouse-hamster hybrid cells. Nat New Biol. 1971 Mar 10;230(10):52–54. doi: 10.1038/newbio230052a0. [DOI] [PubMed] [Google Scholar]
- Steglich C., Grens A., Scheffler I. E. Chinese hamster cells deficient in ornithine decarboxylase activity: reversion by gene amplification and by azacytidine treatment. Somat Cell Mol Genet. 1985 Jan;11(1):11–23. doi: 10.1007/BF01534730. [DOI] [PubMed] [Google Scholar]
- Sugiyama R. H., Arfin S. M., Harris M. Properties of asparagine synthetase in asparagine-independent variants of Jensen rat sarcoma cells induced by 5-azacytidine. Mol Cell Biol. 1983 Nov;3(11):1937–1942. doi: 10.1128/mcb.3.11.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M. A., Reik W., Allen N. D. Transgenes as molecular probes for genomic imprinting. Trends Genet. 1988 Mar;4(3):59–62. doi: 10.1016/0168-9525(88)90040-6. [DOI] [PubMed] [Google Scholar]
- Swain J. L., Stewart T. A., Leder P. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987 Aug 28;50(5):719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Waye M. M., Stanners C. P. Isolation and characterization of CHO cell mutants with altered asparagine synthetase. Somatic Cell Genet. 1979 Sep;5(5):625–639. doi: 10.1007/BF01542699. [DOI] [PubMed] [Google Scholar]