Abstract
Ligands targeting telomeric G-quadruplexs are considered good candidates for anticancer drugs. However, current studies on G-quadruplex ligands focus exclusively on the interactions of ligands and monomeric G-quadruplexes under dilute conditions. Living cells are crowded with biomacromolecules, and the ∼200-nucleotide G-rich single-stranded overhang of human telomeric DNA has the potential to fold into multimeric G-quadruplex structures containing several G-quadruplex units. Studies on interactions between ligands and multimeric G-quadruplexes under molecular crowding conditions could provide a new route for screening specific telomeric G-quadruplex-targeting ligands. Herein, TMPipEOPP, a cationic porphyrin derivative designed by us, was demonstrated as a promising multimeric telomeric G-quadruplex ligand under molecular crowding conditions. It could highly specifically recognize G-quadruplexes. It could also promote the formation of G-quadruplexes and stabilize them. Detailed studies showed that TMPipEOPP interacted with monomeric G-quadruplexes in sandwich-like end-stacking mode of quadruplex/TMPipEOPP/quadruplex and interacted with multimeric human telomeric G-quadruplexes by intercalating into the pocket between two adjacent G-quadruplex units. The pocket size greatly affected TMPipEOPP binding. A larger pocket was advantageous for the intercalation of TMPipEOPP. This work provides new insights into the ligand-binding properties of multimeric G-quadruplexes under molecular crowding conditions and introduces a new route for screening anticancer drugs targeting telomeric G-quadruplexes.
INTRODUCTION
G-quadruplexes are special secondary structures adopted in some guanine (G)-rich DNA sequences (1). DNA sequences with a high potential to form G-quadruplexes are found in many genomic regions with biological significance (2–4). A well-known example is the telomeric sequence located at the termini of the linear chromosomes of most eukaryotic organisms. Human telomeric DNA consists of thousands of d(TTAGGG) repeats, ending in a ∼200-nucleotide G-rich single-stranded overhang. This overhang may fold into G-quadruplex structures in the presence of specific ligands, thus leading to inhibition of telomerase activity and interference with telomere biology (5). As a result, the telomeric G-quadruplex is considered an attractive target for cancer therapeutic intervention (6), and the ligands that can promote telomeric G-quadruplex formation and stabilize telomeric G-quadruplex are considered good candidates for anticancer agents (7).
To date, a large number of G-quadruplex ligands have been reported (8,9). However, most studies have two limitations. One is that most studies focused on the interactions between the tested ligands and monomeric G-quadruplexes that have only one G-quadruplex unit. As aforementioned, the ∼200-nucleotide telomeric G-rich single-stranded overhang has the potential to fold into consecutive G-quadruplex structures (multimeric G-quadruplexes) containing several units (10–12). Ligands that can selectively bind to the pocket between two adjacent G-quadruplex units might be more suitable ligands for telomeric G-quadruplex and better telomerase inhibitors. The computational molecular modelling studies showed that small-molecule ligands could bind into such pocket sites and increase the stabilities of multimeric G-quadruplexes by making them less flexible (13,14), whereas very few ligands have been reported that target multimeric G-quadruplex structures (15,16). Studies on interactions between ligands and multimeric G-quadruplexes could provide new possibilities for anticancer drug design (16).
The other limitation of current G-quadruplex ligand studies is that most studies are carried out under dilute conditions. However, living cells are crowded with many biomacromolecules including nucleic acids, polysaccharides and proteins, as well as soluble and insoluble components (17). These crowding conditions can greatly affect the structure and stability of G-quadruplexes (17–19). More importantly, some ligands are significantly less effective or even lose the ability to stabilize G-quadruplexes under crowding conditions (20). Based on these considerations, studies on interactions between ligands and multimeric G-quadruplexes under molecular crowding conditions could be more suitable for selecting telomere-binding G-quadruplex ligands. To mimic molecular crowding conditions, several crowding reagents, such as poly(ethylene glycol) (PEG), polysaccharides, ethanol, glycerol, haemoglobin, dimethyl sulfoxide, acetonitrile, ficoll, dextrans and betaine are used as cosolutes (21). The commonly used one is PEG, as it is inert to most molecules (22).
The size of the porphyrin core is close to the size of the G-quartet unit of G-quadruplexes, and four positively charged side arm substituents can be easily introduced around the core; therefore, porphyrin derivatives are important candidates in G-quadruplex ligand studies (23). Although the widely studied G-quadruplex ligand 5,10,15,20-tetrakis(N-methylpyridinium-4-yl)-21H,23H-porphyrin (TMPyP4, Scheme 1) shows attractive G-quadruplex-stabilizing ability, it lacks selectivity against duplex DNA (24). Recently, by changing the small side arm methylpyridine substituents of TMPyP4 to larger [2-(1-methyl-1-piperidinyl) ethoxy] phenyl substituents, our group synthesized a new cationic porphyrin derivative 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy] phenyl} porphyrin (TMPipEOPP, Scheme 1) and found that it could be used as a highly specific optical probe for discriminating monomeric G-quadruplexes from duplex and single-stranded DNAs under dilute conditions (25). We also demonstrated that the binding stoichiometry of TMPipEOPP to monomeric G-quadruplexes changed with the G-quadruplex/TMPipEOPP concentration ratio (25). At high ratios, a single TMPipEOPP molecule stacked between the ends of two monomeric G-quadruplexes like a sandwich, implying that TMPipEOPP might be developed into a ligand targeting the multimeric G-quadruplex structure (25). Therefore, in this work, two important features of TMPipEOPP were studied: whether TMPipEOPP could discriminate G-quadruplexes from duplex, single-stranded and triplex DNAs under molecular crowding conditions, and whether TMPipEOPP could be used as a specific ligand of multimeric telomeric G-quadruplexes under molecular crowding conditions.
Scheme 1.
The chemical structures of TMPyP4 and TMPipEOPP.
MATERIALS AND METHODS
Materials and reagents
The oligonucleotides listed in Table 1 were purchased from Sangon Biotech. Co. Ltd (Shanghai, China). The concentrations of the oligonucleotides were represented as single-stranded concentrations. Single-stranded concentration was determined by measuring the absorbance at 260 nm. Molar extinction coefficient was determined using a nearest neighbour approximation (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer), and the calculated molar extinction coefficients of these oligonucleotides were listed in Table 1. Bovine serum albumin (BSA) was obtained from Sigma, and its concentration was calculated based on the molecular weight of 65 000 (26). The 5,10,15,20-tetra-{4-[2-(1-methyl-1-piperidinyl)ethoxy] phenyl} porphyrin (TMPipEOPP) was synthesized according to the method reported by our group recently (25). Na2EDTA (Disodium ethylenediamine tetraacetic acid), Tris [tris(hydroxymethyl)aminomethane], KCl, PEG 200, CH3OH, HCl were obtained from Sigma. Deionized and sterilized water (resistance > 18 MΩ/cm) was used throughout the experiments.
Table 1.
The oligonucleotides used in this work
| Abbreviation | Sequence (5′→3′) | Extinction coefficient [L·mol−1·cm−1] | Structure |
|---|---|---|---|
| Hum24a | (TTAGGG)4 | 244 600 | G-quadruplex (monomeric) |
| C-MYCb | TGAGGGTGGGGAGGGTGGGGAA | 229 900 | G-quadruplex (monomeric) |
| KRASc | AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | 341 000 | G-quadruplex (monomeric) |
| Hum48 | (TTAGGG)8 | 489 000 | G-quadruplex (multimeric) |
| Hum54 | (TTAGGG)9 | 550 100 | G-quadruplex (multimeric) |
| Hum59 | TAGGG(TTAGGG)9 | 603 100 | G-quadruplex (multimeric) |
| AT | (AT)6 | 133 300 | Duplex |
| GC | (GC)6 | 101 100 | Duplex |
| LD | GCGCAATTGCGC | 108 700 | Duplex |
| T*AT | 5′-TTTTTTTTTTTTTTTTTT | 146 400 | Triplex |
| 5′-AAAAAAAAAAAAAAAAAA | 219 400 | ||
| 3′-TTTTTTTTTTTTTTTTTT | 146 400 | ||
| Hum24-cd | (CCCTAA)4 | 220 400 | Single-stranded |
| Hum24-Mute | (TTAGAG)4 | 257 000 | Single-stranded |
| C-MYC-cf | TTCCCCACCCTCCCCACCCTCA | 179 900 | Single-stranded |
| ssDNA1 | GAGCTCTCGAAAGAGCTCCGATTA | 235 800 | Single-stranded |
| ssDNA2 | TAGAGCACACCTGTCCGTG | 189 400 | Single-stranded |
aHum24 is a 24-nucleotide G-rich oligonucleotide with the repeated subunit of human telomere.
bC-MYC is a 22-nucleotide G-rich sequence, which is a regulator gene that codes for a transcription factor.
cKRAS is a 32-nucleotide G-rich sequence, which is located in the promoter of the human KRAS gene.
dHum24-c is a C-rich oligonucleotide with a complementary sequence to Hum24.
eHum24-Mut is a variant of Hum24, it cannot fold into G-quadruplex structure.
fC-MYC-c is a C-rich oligonucleotide with a complementary sequence to C-MYC.
UV-vis absorption spectroscopy
Absorption spectra were measured on a TU-1901 UV-vis spectrophotometer with 1 cm-path-length micro quartz cell (40 μL, Starna Brand, England). Solutions containing individual DNAs or BSA with designated concentration, 10 mM Tris–HCl buffer (pH = 7.4), 150 mM KCl, 1 mM Na2EDTA and 400 mL/L PEG 200 were prepared. Each solution was heated to 95°C for 5 min to remove any aggregates, then cooled rapidly to 25°C and was allowed to incubate at 25°C for 30 min. After overnight incubation at 4°C, 5 μM of TMPipEOPP was added, and the absorption spectra in the range of 350–800 nm were recorded.
Absorption titration experiments were carried out by varying the DNA concentration and maintaining the TMPipEOPP concentration constant. The sample solutions were prepared as aforementioned, and the absorption spectra in the range of 350–800 nm were recorded.
The Job plot analysis was performed by systematic variation of the molar fraction of TMPipEOPP and individual G-quadruplexes while keeping a constant total concentration of TMPipEOPP and G-quadruplex at 5 μM. The mixtures of TMPipEOPP and G-quadruplex were prepared as aforementioned, and the absorption signals at 421, 454 and 695 nm were recorded.
Melting temperature (T1/2) detection of G-quadruplexes
Melting temperature (T1/2) detection of G-quadruplexes was carried out on a Cary-60 UV-vis spectrophotometer equipped with a single cell peltier temperature control accessory. The G-quadruplex (2.5 μM) solution were prepared in 10 mM Tris–HCl buffer (pH = 7.4) containing 150 mM KCl, 1 mM Na2EDTA and 400 mL/L PEG 200. The solution was heated to 95°C for 5 min, then cooled rapidly to 25°C and was allowed to incubate at 25°C for 30 min. After overnight incubation at 4°C, 0 or 5 μM of TMPipEOPP was added. After a sufficient mixing, the absorption signal at 295 nm (400 nm as control wavelength) was recorded at ∼10°C. When the absorption signal became constant, the temperature was increased in steps of 1°C, and the absorption signal was recorded at each temperature until the signal did not decrease any more. At each temperature, the mixture was leaved to equilibrate for 1 min before absorption signal was recorded.
Fluorescence spectroscopy
Fluorescence spectra were measured on a SHIMADZU RF-5301PC spectrofluorimeter with 1 cm-path-length micro quartz cell (40 μL, Starna Brand, England). Solutions containing 10 μM individual oligonucleotides, 10 mM Tris–HCl buffer (pH = 7.4), 150 mM KCl, 1 mM Na2EDTA and 400 mL/L PEG 200 were prepared. Each solution was heated to 95°C for 5 min to remove any aggregates, then cooled rapidly to 25°C and was allowed to incubate at 25°C for 30 min. After overnight incubation at 4°C, 5 μM of TMPipEOPP was added. Fixing the excitation or emission wavelengths, corresponding emission or excitation spectra were collected at room temperature.
The Job plot analysis was performed by systematic variation of the molar fraction of TMPipEOPP and individual G-quadruplexes while keeping a constant total concentration of TMPipEOPP and G-quadruplex at 5 μM. The mixtures of TMPipEOPP and G-quadruplex were prepared as aforementioned. Setting the excitation wavelength at 700 nm, the fluorescence signal at 719 nm was recorded.
Circular dichroism spectroscopy
In all, 3 mL reaction mixture was prepared in 10 mM Tris–HCl buffer (pH = 7.4) containing 1 μM individual DNA oligonculeotides, 1 mM Na2EDTA, 400 mL/L PEG 200, 0 or 150 mM KCl. The mixture was heated at 95°C for 5 min, cooled slowly to 25°C and then incubated at 4°C overnight. After addition of 0 or 5 μM TMPipEOPP, circular dichroism (CD) spectrum of the mixture was recorded between 230 and 320 nm in 1-mm path length cuvettes on a Jasco J-715 spectropolarimeter. Spectra were averaged from three scans, which were recorded at 100 nm/min with a response time of 1 s and a bandwith of 1.0 nm.
RESULTS
G-quadruplex discrimination from duplex, single-stranded and triplex DNAs under molecular crowding conditions
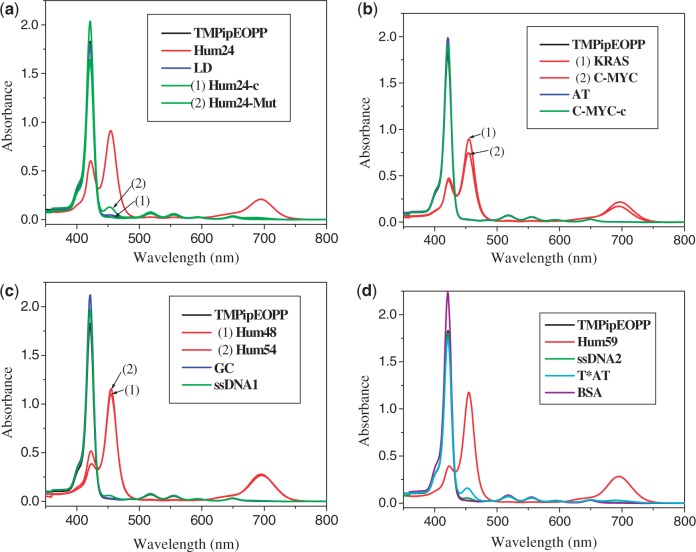
To test whether TMPipEOPP could specifically recognize G-quadruplexes under molecular crowding conditions, PEG with an average molecular weight of 200 daltons (PEG 200) was used as a molecular crowding agent. Interactions between TMPipEOPP and 15 DNA molecules with different structures (Table 1) were investigated by following the effects of the DNAs on the ultraviolet-visible (UV-vis) absorption spectrum of TMPipEOPP. As shown in Figure 1, in the presence of 40% (V/V) PEG 200, the UV-vis absorption spectrum of free TMPipEOPP had a strong Soret band centered at 421 nm, and four weak absorption bands centred at 518, 555, 595 and 648 nm. The presence of G-quadruplexes caused substantial hypochromicity of the Soret band and disappearance of the absorption bands at 518 nm and 555 nm, accompanied by the appearance of two new bands centred at 454 and 695 nm, indicating strong interactions between TMPipEOPP and the G-quadruplexes. Duplex and single-stranded DNAs had almost no effect on the TMPipEOPP absorption spectrum. For the tested duplex and single-stranded DNAs, only the single-stranded DNA, Hum24-Mut, led to the appearance of the new band at 454 nm. However, this new band was much weaker than the Soret band at 421 nm, and the Soret band remained the most prominent. In addition, the two bands at 518 and 555 nm were almost unaffected. Hum24-Mut is a variant of Hum24 that cannot fold into a G-quadruplex structure. The distinct effects of Hum24-Mut and Hum24 on TMPipEOPP absorption spectrum demonstrated that the substantial absorption spectrum changes in Figure 1 were caused by the interactions of TMPipEOPP with G-quadruplex structures. The results supported the hypothesis that TMPipEOPP could be an attractive G-quadruplex ligand with high selectivity against duplex and single-stranded DNAs under molecular crowding conditions. Of note, the three multimeric G-quadruplexes (Hum48, Hum54 and Hum59) also strongly affected the absorption spectrum of TMPipEOPP with even larger effects than the monomeric G-quadruplexes (Hum24, KRAS and C-MYC). This suggested that strong interactions also occurred between TMPipEOPP and multimeric G-quadruplexes. Triplex is another non-canonical DNA secondary structure (27). It has been reported that TMPyP4 has almost no binding selectively for G-quadruplex and triplex DNAs (28). As Hum24-Mut, the DNA triplex T*AT could also lead to the appearance of a weak band at 454 nm and had almost no effects on the previous absorption bands at 421, 518 and 555 nm, suggesting that TMPipEOPP could also well discriminate G-quadruplexes from triplex DNAs. TMPipEOPP also showed good binding selectivity for G-quadruplexes against other biomacromolecules, specifically proteins. With the addition of BSA, the absorption spectrum of TMPipEOPP showed almost no change. This further supported the possibility of using TMPipEOPP as a specific multimeric G-quadruplex ligand under molecular crowding conditions.
Figure 1.
UV-vis absorption spectra of TMPipEOPP in absence or presence of different DNAs. Free TMPipEOPP (black line); G-quadruplex (red line); duplex DNA (blue line); single-stranded DNA (green line); triplex DNA (cyan line); BSA (purple line). TMPipEOPP = 5 μM; multimeric quadruplex = 10 μM; monomeric quadruplex = duplex DNA = single-stranded DNA = triplex DNA = 20 μM; BSA = 8 μM.
Visible G-quadruplex recognition
Compared with duplex, single-stranded and triplex DNAs, G-quadruplexes had substantial effects on the TMPipEOPP absorption spectrum. The effects were large enough for G-quadruplexes to be visually discriminated from duplex, single-stranded and triplex DNAs by solution colour. As shown in Figure 2 and Supplementary Figure S1, a solution of free TMPipEOPP was pink. The addition of duplex, single-stranded or triplex DNAs or the biomacromolecule BSA had little or no effect on the solution colour. However, in the presence of G-quadruplexes, TMPipEOPP solutions changed to a distinct green-yellow colour. These results confirmed the high G-quadruplex-binding selectivity of TMPipEOPP and indicated that TMPipEOPP might be used as a G-quadruplex probe for visual recognition of G-quadruplexes.
Figure 2.
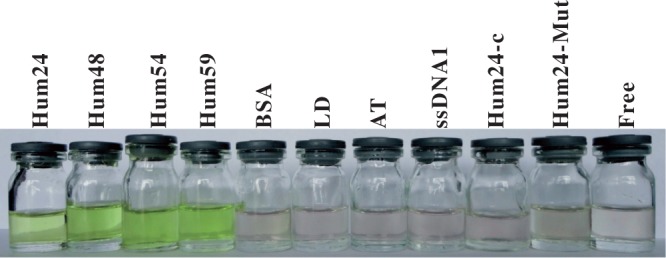
G-quadruplex discrimination from duplex DNAs, single-stranded DNAs and BSA by the naked eyes. The DNA used in each tube is labelled at the top of the figure. TMPipEOPP = 5 μM. DNA = 10 μM. BSA = 10 μM. Discrimination results of other DNAs are shown in Supplementary Figure S1.
Fluorescent recognition of G-quadruplexes
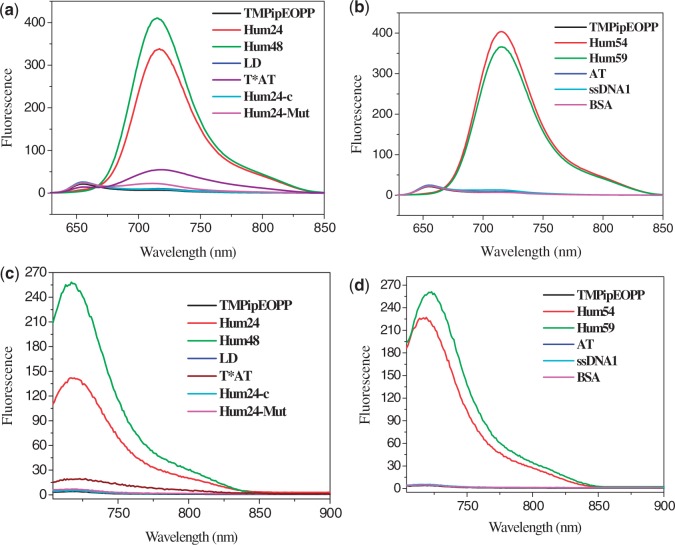
As G-quadruplexes had significant effects on the UV-vis absorption spectrum of TMPipEOPP, we tested whether they could also affect its fluorescence spectrum. Under molecular crowding conditions, free TMPipEOPP had a steady fluorescence with maximum emission wavelength of 655 nm when excited in its Soret band (421 nm). The addition of duplex or single-stranded DNAs had no effect on the maximum excitation or emission wavelengths (Supplementary Figures S2 and S3). However, in the presence of G-quadruplexes, the excitation and emission wavelengths are changed from 421 to 454 nm and from 655 to 719 nm, respectively, and a new excitation wavelength at 700 nm became the strongest excitation wavelength (Supplementary Figure S3). When excited the TMPipEOPP solution at 454 nm or 700 nm, the results in Figure 3 and Supplementary Figure S4 were seen. TMPipEOPP either free or mixed with duplex or single-stranded DNA was nearly non-fluorescent. However, dramatically increased fluorescence signals were observed in the presence of G-quadruplexes, no matter monomeric or multimeric G-quadruplexes. These data indicated that perfect discrimination of G-quadruplexes from duplex and single-stranded DNAs can also be achieved according to the fluorescence signal of TMPipEOPP under molecular crowding conditions. Although the presence of triplex T*AT could also increase the fluorescence of TMPipEOPP to some extent, the fluorescence intensities were much lower than those in the presence of G-quadruplexes. Considering double-stranded structure is the most fundamental structural configuration for DNA in human genome, the facts that BSA protein had no effect on the fluorescence of TMPipEOPP and G-quadruplexes could also cause great fluorescence signal increase even in the presence of duplex DNAs (Supplementary Figure S5) suggested TMPipEOPP could be developed as a specific G-quadruplex fluorescent probe for G-quadruplex detection in vivo.
Figure 3.
Fluorescence spectra of TMPipEOPP in the absence or presence of different DNAs or BSA when excited at (a) and (b) 454 nm or (c) and (d) 700 nm. TMPipEOPP = 5 μM. DNA = 10 μM (strand concentration). When excited at 454 nm, the excitation and emission silts were set to 5 nm. When excited at 700 nm, the excitation and emission silts were set to 3 nm.
Binding interactions between TMPipEOPP and monomeric G-quadruplexes
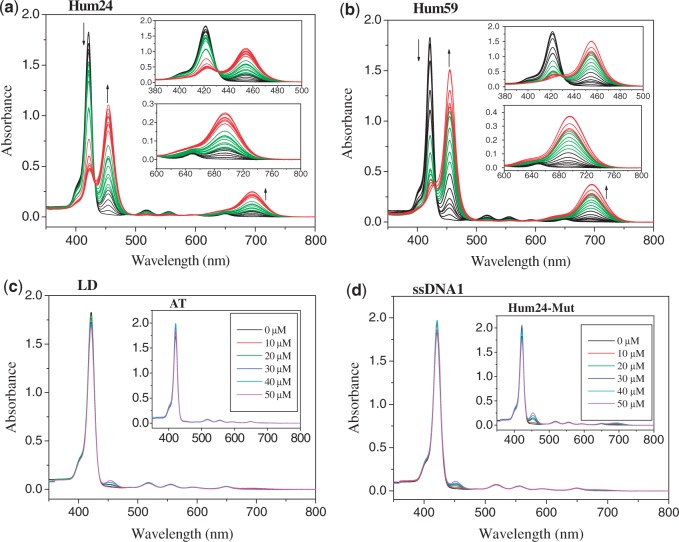
As TMPipEOPP recognized G-quadruplexes with high specificity, we investigated their binding interactions. First, the interactions of TMPipEOPP and monomeric G-quadruplexes were studied. Under dilute conditions, TMPipEOPP bound to monomeric G-quadruplexes with a complicated mode (25). The binding stoichiometry of TMPipEOPP to G-quadruplex changed from 2:1 to 1:2 as the ratio of [G-quadruplex]/[TMPipEOPP] increased (25). To investigate whether the TMPipEOPP–G-quadruplex interaction under molecular crowding conditions was similar to that under dilute conditions, the UV-vis titration spectra of TMPipEOPP with the three monomeric G-quadruplexes (Hum24, KRAS and C-MYC) were investigated in the presence of 40% PEG 200. Similar results were obtained for the three G-quadruplexes. Figure 4a shows a representative result with Hum24. As Hum24 concentration increased, absorption signal intensity at 421 nm decreased continuously, accompanied by the appearance of two new bands (centred at 454 and 695 nm) with continuous increase of their signal intensities. A noteworthy result was an isosbetic point observed at ∼430 nm, suggesting possible two-state transition processes (29). This isosbetic point was not observed under dilute conditions (25), implying that the interaction between TMPipEOPP and monomeric G-quadruplexes under molecular crowding conditions might be simpler than under dilute conditions. Titration with duplex, single-stranded, triplex DNAs or BSA, even at high concentrations, did not lead to obvious changes in the absorption spectrum of TMPipEOPP (Figure 4c and d, Supplementary Figures S8–S11). This further demonstrated the high G-quadruplex-binding selectivity of TMPipEOPP against duplex, single-stranded and triplex DNAs. To further demonstrate the G-quadruplex-recognition specificity of TMPipEOPP against duplex DNAs, the effects of G-quadruplexes, including monomeric and multimeric G-quadruplexes, on the absorption spectrum of TMPipEOPP were investigated in the presence of duplex DNAs. Similar results to those in the absence of duplex DNAs were obtained, thus suggesting that strong interactions also occurred between TMPipEOPP and G-quadruplexes in the presence of duplex DNAs (Supplementary Figure S12).
Figure 4.
Absorption titration of TMPipEOPP with (a) monomeric G-quadruplex Hum24, (b) multimeric G-quadruplex Hum59, (c) duplex LD or AT (insert) (d) single-stranded ssDNA1 or Hum24-Mut (insert). The Hum24 concentration increases from 0 to 50 μM. The Hum59 concentration increases from 0 to 25 μM. Arrows indicate the increasing G-quadruplex concentrations. The concentrations of LD, AT, ssDNA1 and Hum24-Mut are labelled in the figures. Absorption titration spectra of TMPipEOPP with other DNAs and BSA are shown in Supplementary Figures S6–S11.
To determine the number of molecules of bound TMPipEOPP per Hum24, continuous variation analysis (Job plot) was performed. Because substantial absorbance changes were observed at three wavelengths (421, 454 and 695 nm), the absorption signals at these three wavelengths were used to construct Job plots. Under dilute conditions, Job plot analysis using the absorption signals at different wavelengths gave different results because the binding modes of TMPipEOPP and monomeric G-quadruplexes changed with the (TMPipEOPP)/(G-quadruplex) ratio (25). However, under molecular crowding conditions, identical results were obtained at the three wavelengths. Specifically, TMPipEOPP bound to Hum24 with a stoichiometry of 1:2 (Table 2, Supplementary Figure S13). These results suggested that only a single binding mode occurred between TMPipEOPP and Hum24, further demonstrating a simpler TMPipEOPP–Hum24 binding interaction under molecular crowding conditions than under dilute conditions. Generally, ligands interact with G-quadruplexes in three modes: end-stacking, intercalation and outside binding. The large hypochromicity (∼75%, Table 2) of the porphyrin Soret band excluded the possibility of external binding interaction (30–32). Therefore, the possible binding mode between TMPipEOPP and Hum24 was an end-stacking or intercalative mode. In a computational study, Heddi and Phan (33) simulated the solvent distribution around human telomeric G-quadruplexes in a crowded solution using ethanol as the cosolvent. Enrichment of ethanol molecules at the two ends of the parallel G-quadruplex structure was observed. That is to say, the two ends of G-quadruplexes are relatively hydrophobic environments, facilitating accessibility of the large hydrophobic porphyrin core of TMPipEOPP. Considering the binding stoichiometry of two Hum24s per TMPipEOPP, TMPipEOPP and Hum24 might adopt a sandwich-like Hum24/TMPipEOPP/Hum24 end-stacking mode (Scheme 2). Such a binding mode can be used to explain fluorescence experimental results aforementioned. In the Hum24/TMPipEOPP/Hum24 complex, TMPipEOPP was upheld in a rigid and more planar form. The possible non-radiative torsional relaxation channel was severely restricted, thus resulting in the enhancement of TMPipEOPP fluorescence on G-quadruplexes addition (34).
Table 2.
Binding parameters for TMPipEOPP–G-quadruplexes interactions
| G-quadruplexes | Binding stoichiometry of TMPipEOPP to G-quadruplex | Ka (×10−6 M−1)a | Hypochro-micity of the Soret band |
|---|---|---|---|
| Hum24 | 1:2 | 1.1 | 75% |
| KRAS | 1:2 | 2.6 | 86% |
| C-MYC | 1:2 | 1.4 | 81% |
| Hum48 | 1:1.6 | 83% | |
| Hum54 | 1:1.4 | 80% | |
| Hum59 | 1:1 | 0.62 | 80% |
aThe mean value of Scatchard analysis results using the absorption signals at 454 and 695 nm.
Scheme 2.
The proposed binding modes between TMPipEOPP and G-quadruplexes formed by human telomeric sequences with different lengths.
Martino et al. (35) investigated the interactions between TMPyP4 and monomeric G-quadruplexes under both dilute and molecular crowding conditions. Identical binding stoichiometry of four TMPyP4 molecules per monomeric G-quadruplex was obtained under the both conditions. They proposed that the binding process comprised two sequential events: an end-stacking-mode interaction followed by simultaneous binding of three TMPyP4 molecules to the quadruplex structure. They also investigated the binding model of TMPyP4 to bimolecular human telomeric quadruplex with the sequence d(TAGGGTTAGGG) and proposed two binding models: one TMPyP4 molecule stacks onto a A•T base pair and adopts the end-stacking binding model to quadruplex, and the second TMPyP4 molecule is perpendicular to the quadruplex stacks and interacts with loops (36). Compared with TMPyP4, TMPipEOPP binds to monomeric G-quadruplexes in a much simpler way. The particular sandwich-like binding mode implies that TMPipEOPP may be used as a specific multimeric G-quadruplex ligand by intercalating into the interfaces of two G-quadruplex units in multimeric G-quadruplexes. Such a binding mode may also provide TMPipEOPP with better G-quadruplex-recognition specificity against duplex, single-stranded and triplex DNAs than TMPyP4 under molecular crowding conditions.
To further investigate the interaction between TMPipEOPP and monomeric G-quadruplexes, the UV-vis titration spectra obtained earlier in the text were used to construct Scatchard plots [Equation (1)] (31,32):
| (1) |
where r is the number of moles of bound TMPipEOPP per mole of G-quadruplex; n is the number of ligand-binding sites on the G-quadruplex; Ka is the binding constant; and Cf is the free TMPipEOPP in the TMPipEOPP/G-quadruplex mixture (37).
A linearity relationship was seen between r/Cf and r (Figure 5a and Supplementary Figure S19). Linear fitting gave an n value of 0.54, indicating a binding stoichiometry of one TMPipEOPP per two Hum24, which was consistent with the Job plot analysis. The binding constant Ka was determined to be ∼1.1 × 10−6 M−1 (Table 2).
Figure 5.
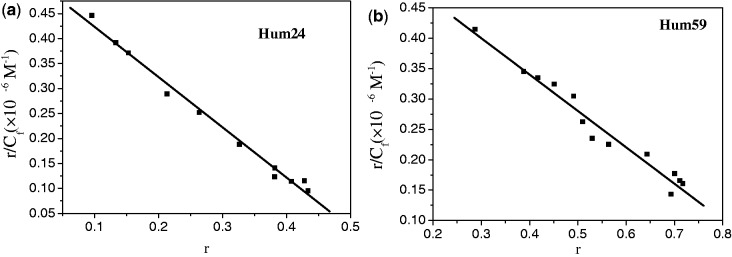
Scatchard plots for TMPipEOPP with (a) Hum24 or (b) Hum59 in the presence of 40% PEG 200. The signals at 454 nm are used to construct the Scatchard plots. The Scatchard plots constructed by the signals at 695 nm are shown Supplementary Figures S19 and S22.
Two other monomeric G-quadruplexes, KRAS and C-MYC, gave results similar to Hum24 (Supplementary Figures S14, S15, S20 and S21). This indicated that the sandwich-like end-stacking mode was not a special case for Hum24.
Binding interactions between TMPipEOPP and multimeric telomeric G-quadruplexes
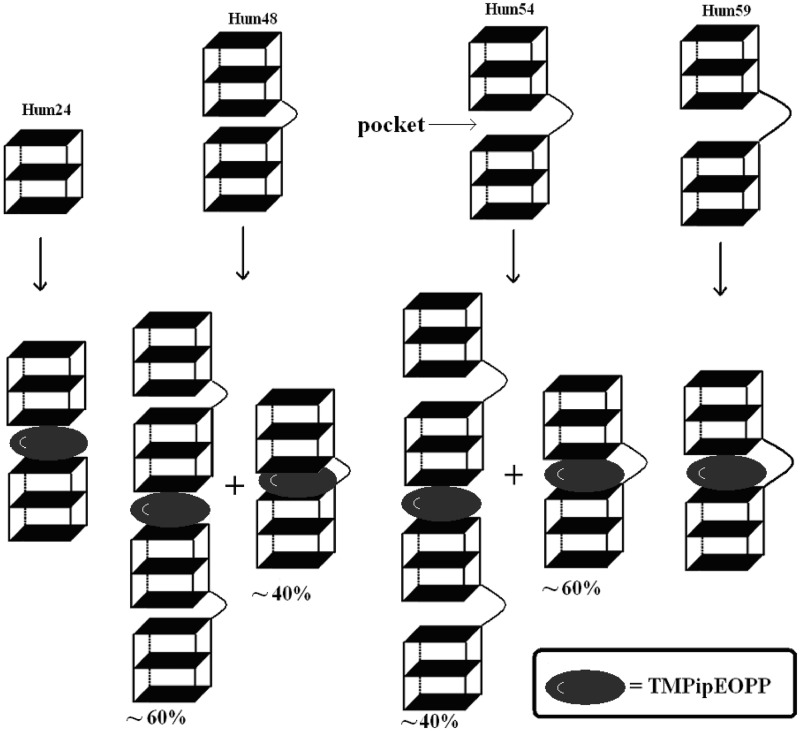
The aforementioned results indicated that TMPipEOPP bound to two monomeric G-quadruplexes in a sandwich-like mode. Thus, we assumed that TMPipEOPP could bind to a multimeric G-quadruplex structure by intercalating into the pockets between two adjacent G-quadruplex units (Scheme 2). To investigate this possibility, three human telomeric DNA oligonucleotides (Hum48, Hum54 and Hum59, Table 1) containing 8, 9 and 10 GGG repeats were used. Because the formation of one G-quadruplex unit requires four GGG repeats, these three oligonucleotides can fold into two G-quadruplex units, but these two units might be linked by loops of different lengths. Thus, the pocket sizes between two adjacent G-quadruplex units were different for the three multimeric G-quadruplexes. Hum48 had the smallest pocket, and Hum59 had the largest one.
The results in Figure 1 demonstrated strong interactions between TMPipEOPP and the three multimeric G-quadruplexes. Titration of TMPipEOPP with individual multimeric G-quadruplexes led to absorption spectrum changes of TMPipEOPP similar to those seen with Hum24 (Figure 4b and Supplementary Figure S7). The ≥80% hypochromicities (Table 2) of the porphyrin Soret band indicated that environments provided by the three multimeric G-quadruplexes were similar to Hum24. TMPipEOPP very possibly binds pockets between two adjacent G-quadruplex units. Such a binding mode is also consistent with the fluorescence experimental results and the computational model of multimeric G-quadruplex–ligand interactions constructed by Haider et al. (13,14).
To test this hypothesis, we performed Job plot analysis (Supplementary Figures S16–S18), which showed maxima at mole fractions of 0.38 for Hum48, 0.42 for Hum54 and 0.49 for Hum59. This result indicated that only Hum59 bound to TMPipEOPP with a stoichiometry of 1:1. The binding stoichiometries for TMPipEOPP were 1:1.6 for Hum48 and 1:1.4 for Hum54 (Table 2). These were in the range of 1:2–1:1, indicating that two binding modes might coexist, with one the sandwich-like end-stacking mode and the other the pocket intercalating mode (Scheme 2). For Hum48, the first mode was ∼60%, and the second one was ∼40%. For Hum54, the first one was ∼40%, and the second was ∼60%. These results suggested that TMPipEOPP interacted with multimeric G-quadruplexes, and the size of the pocket between two adjacent G-quadruplex units strongly influenced the interaction, with a large pocket favouring the binding interaction. A loop connecting two adjacent G-quadruplex units of 15 nucleotides (Hum59) provided a pocket large enough for TMPipEOPP binding.
Considering that TMPipEOPP–Hum59 interaction adopts a single-binding mode (pocket-intercalating mode), the UV-vis titration spectra of TMPipEOPP should be suitable for Scatchard analysis (Figure 5b and Supplementary Figure S22). As expected, a linearity relationship was observed between r/Cf and r. Linear curve fitting with Equation (1) gave an n of 0.93, further confirming a binding stoichiometry of 1:1. The binding constant, Ka, was determined to be around 0.62 × 106 M−1.
Effects of TMPipEOPP on circular dichroism spectra of G-quadruplexes
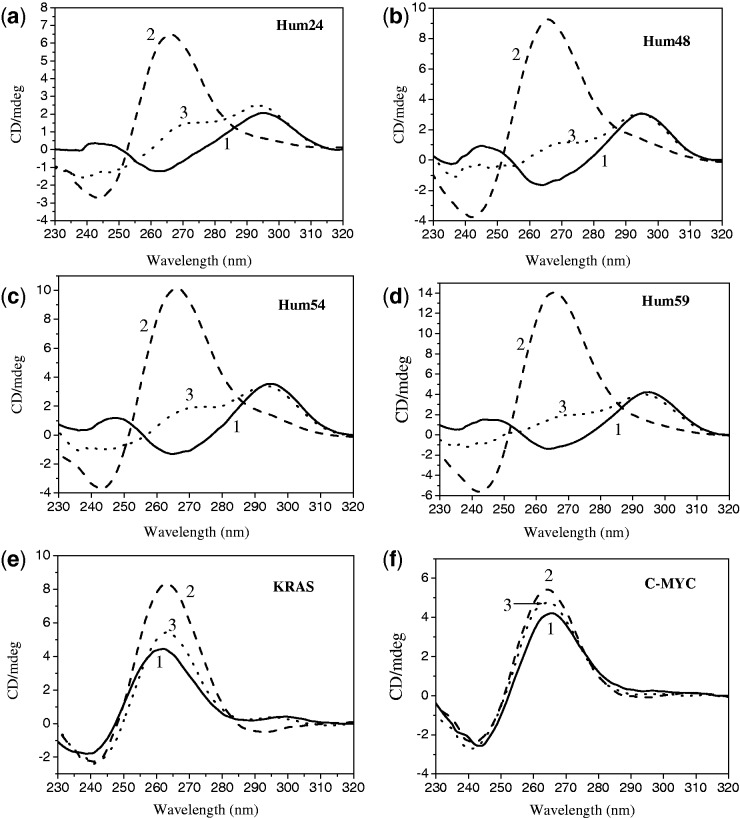
CD spectroscopy is an important tool for studying G-quadruplexes (7). CD is widely used to investigate the conformation of G-quadruplexes and sometimes to study the interactions between ligands and G-quadruplexes (7). Under molecular crowding conditions, the short telomeric sequence Hum24 and the long telomeric sequences Hum48, Hum54 and Hum59 showed a positive peak at ∼295 nm and a negative peak at ∼265 nm even without K+ ions (Figure 6). This suggested that these G-rich sequences partly folded into an antiparallel G-quadruplex structure under molecular crowding conditions. In the presence of 150 mM K+, the positive peak at 295 nm disappeared. A new positive peak with much higher CD signal intensity was observed at ∼265 nm, and the negative peak shifted from 265 nm to 245 nm. This was typical of parallel G-quadruplex structures (38). This result was comparable with previous reports that G-rich telomeric sequences fold into parallel G-quadruplexes in the presence of K+ under molecular crowding conditions (39,40). Although the addition of 5 μM TMPipEOPP instead of 150 mM K+ did not lead to the disappearance of the positive peak at 295 nm, a shoulder peak was observed at ∼265 nm. This peak indicated that the addition of TMPipEOPP promoted the partial conversion of human telomeric sequence from an antiparallel structure to a parallel structure. In contrast to G-rich telomeric sequences, KRAS and C-MYC can partly fold into parallel G-quadruplexes in the absence of K+, showing a positive CD peak at 265 nm and a negative CD peak at 245 nm. The addition of K+ or TMPipEOPP further promoted the formation of parallel G-quadruplexes, accompanied by the increase in positive CD signal. These results indicated that TMPipEOPP not only promoted the conversion of G-quadruplexes from antiparallel to parallel structure but also promoted the formation of parallel G-quadruplexes. This gives another evidence of the existence of binding interactions between TMPipEOPP and monomeric or multimeric G-quadruplexes. TMPipEOPP possibly preferentially binds to G-quadruplexes with parallel structure. The formation of parallel telomeric G-quadruplexes under conditions that mimic cells (a molecular crowding environment with a high K+ concentration) might be suitable for TMPipEOPP binding.
Figure 6.
CD spectra of (a) Hum24, (b) Hum48, (c) Hum54, (d) Hum59, (e) KRAS and (f) C-MYC in the absence (1, solid line) and presence of 150 mM K+ (2, dashed line) or 5 μM TMPipEOPP (3, dotted line).
Stabilization of G-quadruplexes by TMPipEOPP
A good G-quadruplex ligand should have two important characteristics: high G-quadruplex-recognition specificity and high G-quadruplex stabilization. Our experiments demonstrated that TMPipEOPP specifically recognized G-quadruplexes, but not duplex or single-stranded DNAs. We next investigated whether TMPipEOPP stabilized G-quadruplex structures. The stabilizing abilities of ligands to G-quadruplexes can be reduced under molecular crowding conditions. Some ligands, e.g. Hoechst dye, lose their G-quadruplex-stabilizing abilities under these conditions (20). To test the ability of TMPipEOPP to stabilize G-quadruplexes, the melting temperatures (T1/2) of G-quadruplexes in the presence and absence of TMPipEOPP were compared. Destruction of the G-quadruplex structure at high temperature led to a UV-absorption signal decrease at 295 nm (41). In the temperature-absorbance profile, the T1/2 is the temperature of the absorption signal midway between the minimal and maximal levels (7). With the increase of temperature, typical reverse S-shape G-quadruplex melting curves were observed for the six G-rich sequences (Supplementary Figure S23). Using a two-state transition model, thermodynamic parameters, including the enthalpy change (ΔH°), entropy change (ΔS°) and free energy change at 37°C (ΔG°37) of these six G-rich sequences, were evaluated by analysing the melting curves (Table 3). In the presence 150 mM K+, all six G-rich sequences gave negative ΔG°37 values, demonstrating that stable G-quadruplexes were formed under molecular crowding conditions, which are consistent with the results of CD spectroscopy. TMPipEOPP was highly effective at stabilizing monomeric or multimeric G-quadruplexes. For example, 5 μM TMPipEOPP increased the T1/2 of Hum24 from 51.4°C to 60.6°C for a net increase of 9.2°C and increased the T1/2 of Hum59 by 9.8°C (Table 3, Supplementary Figure S23). These results demonstrated the interactions between TMPipEOPP and G-quadruplexes and showed that these interactions can increase the stabilities of G-quadruplexes.
Table 3.
Thermodynamic parameters for G-rich oligonucleotides and the stabilizing abilities of TMPipEOPP to G-quadruplexes
| G-quadruplexes | ΔH° (kJ/mol) | ΔS° (J/mol·K) | ΔG°37 (kJ/mol) |
T1/2 (°C) |
|
|---|---|---|---|---|---|
| [TMPipEOPP] (μM) |
|||||
| 0 | 5 | ||||
| Hum24 | −67.45 | −208.0 | −2.94 | 51.4 | 60.6 |
| Hum48 | −67.94 | −214.8 | −1.32 | 43.5 | 56.5 |
| Hum54 | −72.96 | −229.3 | −1.83 | 45.5 | 57.7 |
| Hum59 | −82.79 | −258.4 | −2.65 | 47.3 | 58.1 |
| KRAS | −73.26 | −229.0 | −2.24 | 47.8 | 54.9 |
| C-MYC | −82.21 | −264.7 | −0.11 | 37.6 | 47.1 |
CONCLUSION
In summary, using a combination of spectroscopic (UV, fluorescence and CD) techniques, we showed that the cationic porphyrin TMPipEOPP, developed by our group, discriminated G-quadruplexes from duplex, single-stranded and triplex DNAs under molecular crowding conditions. The dramatic influence of G-quadruplexes on TMPipEOPP fluorescence makes TMPipEOPP a promising fluorescent probe for in vivo detection of G-quadruplexes. TMPipEOPP interacted with multimeric human telomeric G-quadruplexes by intercalating into the pockets between two adjacent G-quadruplex units. This finding is important because growing evidence indicates that multimeric G-quadruplex structures are formed by telomeric single-stranded overhang regions. Also, because of the large number of other quadruplex-forming sequences besides telomeric sequences in the human genome (16,42,43), screening ligands that can specifically recognize quadruplex–quadruplex interfaces are considered important steps for the design of ligands that target telomeric quadruplexes but have low side effects to other potential G-quadruplex-forming regions (16). In this work, we demonstrated for the first time that the pocket size between two adjacent G-quadruplex units strongly affects interactions between ligands and multimeric G-quadruplexes. This work provides new insights into the ligand-binding properties of multimeric quadruplex structures and a new route for screening anticancer drug targeting telomeric G-quadruplexes. It was reported that acetonitrile could be used as a non-hydrogen-bonding molecular crowding reagent (44). To fully exploit the possibility of TMPipEOPP as multimeric G-quadruplex ligand under molecular crowding conditions, our next work is to investigate the interaction between TMPipEOPP and G-quadruplexes using acetonitrile as cosolute and compare the results with those obtained in this work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–23.
FUNDING
Funding for open access charge: Natural Science Foundation of China [No. 20801040]; the National Basic Research Program of China [No. 2011CB707703]; the National Natural Science Foundation of Tianjin [No. 12JCYBJC13300].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ou TM, Lu YJ, Tan JH, Huang ZS, Wong KY, Gu LQ. G-quadruplexes: targets in anticancer drug design. ChemMedChem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- 2.Yang DZ, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji X, Sun H, Zhou H, Xiang J, Tang Y, Zhao C. Research progress of RNA quadruplex. Nucleic Acid Ther. 2011;21:185–200. doi: 10.1089/nat.2010.0272. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y, Brosh RM., Jr G-quadruplex nucleic acids and human disease. FEBS J. 2010;277:3470–3488. doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y. Chemistry in human telomere biology: structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 2011;40:2719–2740. doi: 10.1039/c0cs00134a. [DOI] [PubMed] [Google Scholar]
- 6.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;20:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 7.Murat P, Singh Y, Defrancq E. Methods for investigating G-quadruplex DNA/ligand interactions. Chem. Soc. Rev. 2011;40:5293–5307. doi: 10.1039/c1cs15117g. [DOI] [PubMed] [Google Scholar]
- 8.Arola A, Vilar R. Stabilisation of G-quadruplex DNA by small molecules. Curr. Top. Med. Chem. 2008;8:1405–1415. doi: 10.2174/156802608786141106. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Zhang F, Li H, Liu C, Xia J, Ma L, Chu W, Zhang Z, Chen C, Li S, et al. Recent progress and future potential for metal complexes as anticancer drugs targets G-quadruplex DNA. Curr. Med. Chem. 2012;19:2957–2975. doi: 10.2174/092986712800672067. [DOI] [PubMed] [Google Scholar]
- 10.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Ishizuka T, Kurabayashi K, Komiyama M. Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: a protective capping structure for telomere ends. Angew. Chem. Int. Ed. Engl. 2009;48:7833–7836. doi: 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- 12.Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J. Am. Chem. Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 13.Haider SM, Neidle S. A molecular model for drug binding to tandem repeats of telomeric G-quadruplexes. Biochem. Soc. Trans. 2009;37:583–588. doi: 10.1042/BST0370583. [DOI] [PubMed] [Google Scholar]
- 14.Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinohara K, Sannohe Y, Kaieda S, Tanaka K, Osuga H, Tahara H, Xu Y, Kawase T, Bando T. A chiral wedge molecule inhibits telomerase activity. J. Am. Chem. Soc. 2010;132:3778–3782. doi: 10.1021/ja908897j. [DOI] [PubMed] [Google Scholar]
- 16.Cummaro A, Fottichia I, Franceschin M, Giancola C, Petraccone L. Binding properties of human telomeric quadruplex multimers: a new route for drug design. Biochimie. 2011;93:1392–1400. doi: 10.1016/j.biochi.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. J. Phys. Chem. B. 2009;113:8784–8792. doi: 10.1021/jp809486g. [DOI] [PubMed] [Google Scholar]
- 18.Miyoshi D, Karimata H, Sugimoto N. Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem. Int. Ed. Engl. 2005;29:3740–3744. doi: 10.1002/anie.200462667. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Zheng KW, Hao YH, Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. J. Am. Chem. Soc. 2009;131:10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- 21.Petraccone L, Pagano B, Giancola C. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods. 2012;57:76–83. doi: 10.1016/j.ymeth.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi D, Fujimoto T, Sugimoto N. Molecular crowding and hydration regulating of G-quadruplex formation. Top. Curr. Chem. 2013;330:87–110. doi: 10.1007/128_2012_335. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen MC, Ulven T. Macrocyclic G-quadruplex ligands. Curr. Med. Chem. 2010;17:3438–3448. doi: 10.2174/092986710793176320. [DOI] [PubMed] [Google Scholar]
- 24.Romera C, Bombarde O, Bonnet R, Gomez D, Dumy P, Calsou P, Gwan JF, Lin JH, Defrancg E, Pratviel G. Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie. 2011;93:1310–1317. doi: 10.1016/j.biochi.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Zhu LN, Zhao SJ, Wu B, Li XZ, Kong DM. A new cationic porphyrin derivative (TMPipEOPP) with large side arm substituents: a highly selective G-quadruplex optical probe. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035586. e35586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang L, Jiang X, Dong SJ. In vitro study on the binding of neutral red to bovine serum albumin by molecular spectroscopy. J. Photochem. Photobiol. A Chem. 2006;184:93–97. [Google Scholar]
- 27.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 28.Ren JS, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 29.Arora A, Maiti S. Stability and molecular recognition of quadruplexes with different loop length in the absence and presence of molecular crowding agents. J. Phys. Chem. 2009;113:8784–8792. doi: 10.1021/jp809486g. [DOI] [PubMed] [Google Scholar]
- 30.Pasternack RF, Gibbs EJ, Villafranca JJ. Interactions of porphyrins with nucleic acids. Biochemistry. 1983;22:2406–2414. doi: 10.1021/bi00279a016. [DOI] [PubMed] [Google Scholar]
- 31.Wei C, Jia G, Zhou J, Han G, Li C. Evidence for the binding mode of porphyrins to G-quadruplex DNA. Phys. Chem. Chem. Phys. 2009;11:4025–4032. doi: 10.1039/b901027k. [DOI] [PubMed] [Google Scholar]
- 32.Wei C, Wang J, Zhang M. Spectroscopic study on the binding of porphyrins to (G4T4G4)4 parallel G-quadruplex. Biophys. Chem. 2010;148:51–55. doi: 10.1016/j.bpc.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 34.Mohanty J, Barooah N, Dhamodharan V, Harikrishna S, Pradeepkumar PI, Bhasikuttan AC. Thioflavin T as an efficient inducer and selective fluorescent sensor for the human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2013;135:367–376. doi: 10.1021/ja309588h. [DOI] [PubMed] [Google Scholar]
- 35.Martino L, Pagano B, Fotticchia I, Neidle S, Giancola C. Shedding light on the interaction between TMPyP4 and human telomeric quadruplexes. J. Phys. Chem. B. 2009;113:14779–14786. doi: 10.1021/jp9066394. [DOI] [PubMed] [Google Scholar]
- 36.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 37.Keating LR, Szalai VA. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry. 2004;43:15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 38.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Xu L, Feng S, Zhou X. Human telomeric G-quadruplexes undergo dynamic conversion in a molecular crowding environment. Chem. Commun. 2011;47:3517–3519. doi: 10.1039/c0cc05242f. [DOI] [PubMed] [Google Scholar]
- 40.Yu HQ, Gu X, Nakano S-i, Miyoshi D, Sugimoto N. Beads-on-a-string structure of long telomeric DNAs under molecular crowding conditions. J. Am. Chem. Soc. 2012;134:20060–20069. doi: 10.1021/ja305384c. [DOI] [PubMed] [Google Scholar]
- 41.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 42.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG3(TTAG3)3) J. Am. Chem. Soc. 2010;132:17105–17017. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.