Abstract
The H1 linker histones are abundant chromatin-associated DNA-binding proteins. Recent evidence suggests that linker histones also may function through protein–protein interactions. To gain a better understanding of the scope of linker histone involvement in protein–protein interactions, we used a proteomics approach to identify H1-binding proteins in human nuclear extracts. Full-length H1.0 and H1.0 lacking its C-terminal domain (CTD) were used for protein pull-downs. A total of 107 candidate H1.0 binding proteins were identified by LC-MS/MS. About one-third of the H1.0-dependent interactions were mediated by the CTD, and two-thirds by the N-terminal domain-globular domain fragment. Many of the proteins pulled down by H1.0 were core splicing factors. Another group of H1-binding proteins functions in rRNA biogenesis. H1.0 also pulled down numerous ribosomal proteins and proteins involved in cellular transport. Strikingly, nearly all of the H1.0-binding proteins are found in the nucleolus. Quantitative biophysical studies with recombinant proteins confirmed that H1.0 directly binds to FACT and the splicing factors SF2/ASF and U2AF65. Our results demonstrate that H1.0 interacts with an extensive network of proteins that function in RNA metabolism in the nucleolus, and suggest that a new paradigm for linker histone action is in order.
INTRODUCTION
The chromatin of most mammalian tissues consists of arrays of nucleosomes spaced at ∼160–210-bp intervals, together with 0.6–1.0 H1 linker histones per nucleosome (1). Six major linker histone isoforms are expressed in human somatic cells (2). The linker histones of higher eukaryotes have a short (13–40 residue) unstructured N-terminal domain (NTD), a central well-ordered globular domain (GD) and a ∼100 residue unstructured C-terminal domain (CTD). Stabilization of the condensed states of chromatin is the most common function attributed to linker histones (3,4). The role of the NTD in chromatin condensation is unknown. The GD is a winged helix DNA-binding domain (5,6) that mediates the interaction of linker histones with nucleosomes (7). The intrinsically disordered CTD (8) interacts with the linker DNA that connects adjacent nucleosomes in a chromatin fiber (9) and is required for the chromatin condensing functions of linker histones (10,11).
In addition to their role in regulating chromatin architecture via protein–DNA interactions, linker histones interact with non-histone proteins. A recent literature survey uncovered reports scattered during 20-year period for 16 linker histone-binding proteins (4). In most of these cases, the interactions were identified using co-IP and/or pull-down assays for the specific linker histone–protein interaction in question. Two recent studies used a more systematic approach to probe for linker histone–protein interactions. In one case (12), anti-H1 antibodies were used to co-IP H1–protein complexes from Drosophila Kc cells. A number of ribosomal proteins were identified that, together with H1, were involved in specific gene repression. In the second study (13), a cell line expressing a tandem tagged version of the H1.2 isoform was used together with chromatography and glycerol gradient ultracentrifugation to isolate a stable complex containing H1.2, specific ribosomal proteins, transcriptional repressors (e.g. YB1, PARP1 and PURα) and transcriptional activators (e.g. WDR5, CAPERα and nucleolin).
The scope of linker histone participation in protein–protein interactions currently is unknown, i.e. are H1–protein interactions relatively isolated occurrences, or are they more common than currently believed? Moreover, the extent to which each of the linker histone domains mediates protein–protein interactions is unclear. This is an intriguing question given that the GD and the CTD both are well-established DNA-binding domains. Here, we have used the HaloTag methodology (14) together with mass spectrometry to identify candidate H1-binding proteins in nuclear extracts prepared from human cell lines. Both full-length H1.0 and H1.0 lacking its CTD were used as the ligands in these experiments, allowing determination of interactions that were dependent on the CTD and NTD-GD fragment. We have identified 107 proteins that were pulled down by H1.0 under the conditions of our experiments. The CTD and the NTD-GD fragment each were found to mediate multiple protein–protein interactions. Thirty-three of the proteins pulled down by H1.0 were splicing factors. H1.0 also pulled down numerous proteins involved in rRNA biogenesis, ribosome function/translation and cellular transport. Ninety-four of the proteins identified in our studies are found in the nucleolus. The interactions between H1.0 and U2AF65, SF2/ASF and FACT were validated by quantitative binding experiments with pure recombinant proteins. Our results indicate that histone H1.0 is a central component of a large network of protein–protein interactions involved in RNA metabolism in the nucleolus.
MATERIALS AND METHODS
Protein expression and purification
To prepare the HaloTag fusion proteins, cDNAs encoding full-length mouse H1.0 (residues 1–194) and H1ΔCTD (residues 1–97) were cloned into the pFN18K T7 Flexi vector (HaloTag 7, Promega) via the PmeI and SgfI restriction sites. The HaloTag-only control expression vector was constructed following manufacturer’s instructions, using annealed oligonucleotides 5′-CGCGTAAGGGTAGGTTT and 5′-AAACCTACCCTTACGCGAT to ligate the digested pFN18 plasmid. All constructs were confirmed by DNA sequencing. pFN18-Halo-H1.0, pFN18-Halo-H1ΔCTD and pFN18-Halo were transformed into Escherichia coli BL21(DE3) pLysS-competent cells. The transformed cells were grown at 37°C in LB medium to an OD600 of 0.6. After induction with 0.4 mM isopropyl-1-thio-d-galactopyranoside, cells were grown for 30 min, rifampicin was added to a concentration of 50 µg/ml and cells were grown for ≥2 h. The cells were then harvested and washed with 10 mM Tris, pH 8.0, and 100 mM NaCl. Cell pellets were sonicated in three volumes of Halo buffer [25 mM Tris, pH 8.0, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 100 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1.0 mM benzamidine and protease inhibitor cocktails II and III (Calbiochem)], and the HaloTag proteins were purified on HiTrap SP-FF followed by MonoQ (for HaloTag alone) (GE Healthcare). The proteins were dialyzed overnight against Halo buffer and stored at 4°C. Concentration was defined using a BCA protein assay kit (Thermo scientific).
pET30a-U2AF65 and pET24b-SF2/ASF (encoding for residues 11–196 of SF2/ASF) were transformed into E. coli BL21(DE3) pLysS-competent cells and expressed as described earlier in the text. Cell pellets were isolated as described previously. Cell pellets were resuspended in 50 mM potassium phosphate buffer (pH 7.4), 900 mM NaCl, 10 mM imidazole and loaded onto a 5-ml HisTrap HP column (GE Healthcare). Proteins were eluted from the HisTrap column with a 10–500 mM linear imidazole gradient. Peak fractions were loaded on a 15% sodium dodecyl sulfate (SDS) polyacrylamide gel to analyze for purity and/or degradation, then pooled and dialyzed into 10 mM Tris, pH 7.5, 300 mM NaCl and 0.1 mM PMSF. FACT was expressed purified as described in the study conducted by Winkler et al. (15).
Preparation of nuclear extracts
Nuclear extracts were prepared using modified Dignam protocol (16). Four different human cell lines were used as follows: CEM, HeLa, U2OS and RPE-1. Cells were grown, harvested, spun and washed in phosphate-buffered saline (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, and a pH of 7.4), resuspended in five packed cell volumes of buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2 and 10 mM KCl), incubated on ice for 10 min and spun at 2000 rpm for 10 min at 4°C. The cell pellet was resuspended in two original packed cell volumes, transferred to a Dounce homogenizer and lysed with 20–60 strokes depending on cell type. Aliquots of cells were stained with DAPI (6-diamidino-2-phenylindole) and checked for >90% lysis using light microscopy. Lysed cells were spun at 2000 rpm for 10 min to pellet nuclei, the supernatant was removed and pellet was re-spun at 17 000 rpm for 1 min to isolate remained cytoplasm. The pellet was resuspended in one packed nuclear volume of high-salt buffer (20 mM HEPES, pH 7.9, 25% v/v glycerol, 0.42 M NaCl, 1.5 mM MgCl2 and 0.2 mM EDTA), the mixture stirred gently and rotated for 30 min in a cold room. Aliquots of nuclei were stained with DAPI and checked for complete lysis. The lysed nuclei were spun at 17 000 rpm for 30 min at 4°C and the resulting nuclear extract dialyzed against buffer D (20 mM HEPES, pH 7.9, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF) for 5 h. The protein concentrations of the extracts were determined using a BCA protein assay kit (Thermo scientific). Nuclear extracts were flash-frozen in liquid nitrogen and kept at −80°C.
Pull-downs
A slurry (300 µl) of HaloTag beads (Promega) was pre-equilibrated with Halo buffer and then incubated with 0.4 mg of HaloTag-H1.0 fusion proteins or the HaloTag protein alone in the presence of 0.05% Igepal-40 and protease inhibitors [PIC Sets II and III (Calbiochem), 0.5 mM benzamidine, 10 µg/ml of tosyl phenylalanyl chloromethyl ketone (TPCK) and 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF)] with rocking overnight at 4°C. The beads were extensively washed with Halo buffer containing 250 mM NaCl and then equilibrated with Halo buffer containing 150 mM NaCl. Nuclear extract (0.5 mg of total protein) was added to the beads, the volume adjusted to 500 μl with Halo buffer, followed by incubation overnight. For the RNAse experiments, a portion of the nuclear extracts was incubated with 0.1 mg/ml of RNAse A (Thermo Scientific) and 7.5 U of exonuclease T (New England Biolabs) in the presence of 10 mM MgCl2 and 1 mM DTT for 30 min at room temperature, before incubation with the beads. The unbound proteins were removed by washing three times with Halo buffer containing 250 mM NaCl. Bound proteins were eluted from the beads by boiling in 1 M NaCl and 1% SDS. The pulled down proteins were precipitated with 20% trichloroacetic acid, followed by centrifugation for 30 min at 14 000 rpm. The protein pellet was washed with 100% acetone twice and dissolved in Laemmli loading buffer (60 mM Tris–Cl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol and 0.01% bromophenol blue). For LC-MS/MS analysis, proteins were electrophoresed on a 4–12% pre-cast polyacrylamide gel (Invitrogen) and stained with Imperial Blue (Thermo Scientific). Proteins were run ∼4 mm into the resolving gel, and the entire stained region was excised and split into three fractions. Gel fractions were submitted to the Proteomics and Metabolomics Facility at Colorado State University for in-gel digestion and LC-MS/MS analysis. For western blots, the proteins were electrophoresed and transferred to a nitrocellulose membrane as described later in the text.
In-gel digestion
Gel fractions were washed with water and destained with 200 ml of acetonitrile (ACN) and 50 mM ammonium bicarbonate (AmBIC) (1:1 v/v) at 60°C. Destained gel pieces were then dehydrated with 200 μl of 100% ACN with subsequent rehydration in 200 μl of 25 mM DTT in 50 mM AmBic (incubated for 20 min at 60°C). Alkylation of reduced cysteine was performed by the addition of 200 μl of 55 mM iodoacetamide in 50 mM AmBic (incubated for 20 min in dark at room temperature). Reduced and alkylated gel pieces were washed and dehydrated again with 100% ACN followed by rehydration with 20 μl of 12 ng/ml of trypsin in 0.01% ProteaseMAX™ surfactant (Promega Corporation). After 10 min, an additional 30 μl of 0.01% ProteaseMAX surfactant was added and digestion allowed proceeding for 1 h at 50°C. Digestion was stopped with the addition of 2.5 μl of 10% triflouroacetic acid. The supernatant containing the peptides was dried in a SpeedVac vacuum centrifuge and reconstituted in 10 μl of 3% ACN and 0.1% formic acid.
Mass spectrometry
Peptides from the digested gel fractions were purified and concentrated using an on-line enrichment column (Agilent Zorbax C18, 5 μm, 5 × 0.3 mm). Subsequent chromatographic separation was performed on a reverse phase nanospray column (Agilent 1100 nanoHPLC, Zorbax C18, 5 μm, 75 μm ID × 150-mm column) using a 90 min linear gradient from 25–55% buffer B (90% ACN and 0.1% formic acid) at a flow rate of 300 nl/min. Peptides were eluted directly into the mass spectrometer (Thermo Scientific LTQ linear ion trap), and spectra were collected over a m/z range of 200–2000 Da using a dynamic exclusion limit of two MS/MS spectra of a given ion for 30 s (exclusion duration of 90 s). Compound lists of the resulting spectra were generated using Bioworks 3.0 software (Thermo Scientific) with an intensity threshold of 5000 and 1 scan/group. MS/MS spectra were searched against the Uniprot protein database (2 September 2012) using a taxonomy filter for ‘Homo sapiens’ concatenated with reverse sequences for determination of the peptide FDR (148 254 sequence entries) using the Mascot database search engine (version 2.3). Search parameters were as follows: average mass, parent ion mass tolerance of 2 Da, fragment ion mass tolerance of 1.5 Da, fully tryptic peptides with one missed cleavage, variable modification of oxidation of M and fixed modification of carbamidomethylation of C.
Mascot search results for each independently analyzed gel fraction were imported and combined for each pull-down using probabilistic protein identification algorithms implemented in Scaffold software (Proteome Software, Portland, OR, USA). Peptide and protein probability thresholds of 95 and 99%, respectively, and a minimum of two unique peptides, were applied to the results (≤0.2% peptide FDR as calculated by Scaffold based on matches to reverse hits). Proteins containing shared peptides were grouped by Scaffold to satisfy the laws of parsimony. Manual validation of MS/MS spectra was performed for all protein identifications above the probability thresholds that were based on only two unique peptides. Criteria for manual validation included the following: (i) minimum of 80% coverage of theoretical y or b ions (at least five in consecutive order); (ii) absence of prominent unassigned peaks >5% of the maximum intensity; and (iii) indicative residue specific fragmentation, such as intense ions N-terminal to proline and immediately C-terminal to aspartate and glutamate (used as additional parameters of confirmation).
Data analysis was performed separately for each experimental replicate. Proteins were determined to be candidate H1-binding partners if they were observed in at least one cell type in both experimental replicates and were not observed in any replicate of the HaloTag controls. Proteins were further determined to be CTD dependent if they were observed in at least one cell type, specifically in wild-type H1 pull-downs, in both experimental replicates and were not observed in any replicate of either the Halo controls or in CTD mutant pull-downs. Common contaminant proteins, including keratins and trypsin (autolysis), were discarded. We have also excluded H1.0 because it might result from recombinant proteins eluted from beads during pull-downs.
Western blotting
H1.0-binding proteins were pulled down from CEM nuclei as for the proteomics analysis. The sample was electrophoresed on a 4–12% gradient SDS polyacrylamide gel and transferred to a nitrocellulose membrane. Membranes were incubated in 5% non-fat milk in a phosphate-buffered saline solution containing 0.05% Tween (PBS-T) for 1 h and then probed with rabbit anti-RbAp48 antibodies (Novus, 1:2500 56483) overnight. After a series of washes in PBS-T, the membranes were re-probed with IRDye 680 goat anti-rabbit antibodies (1:20000; 926-32221, LI-COR) for 1 h. The proteins were visualized using an Odyssey (LI-COR). Western blots were performed at least three times. CEM nuclear extract was used as a positive control.
Fluorescence quenching microplate titration assay
Clear bottom 384-well microplates were prepared as previously described (17). Binding experiments were set-up by diluting a high-concentration stock of the unlabeled titrant (U2AF65, SF2/ASF or FACT) into a series of concentrations ranging from 1 nM to 10 μM. The reaction conditions consisted of 20 mM Tris, pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM Tris(2-carboxyethyl)phosphine (TCEP) and 0.01% of both 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and octylglucoside detergents. The presence of low amounts of both ionic and non-ionic detergents minimizes ‘sticking issues’ common at low concentrations and significantly increased reproducibility. Histone H1.0 was conjugated with Oregon Green 488 via an iodoacetimide linkage at residue 19. Labeled H1.0 was kept at a constant concentration between 1 and 10 nM. The mixtures (40 μl) were allowed to equilibrate for 30 min at room temperature in the dark before scanning at the appropriate wavelength using a Typhoon 8600 variable mode fluoroimager. The fluorescence intensities were quantified using the program ImageQuant TL, and data were analyzed using Kaleidagraph version 3.6 (Synergy software). All experiments were performed in replicative quadruplicate.
RESULTS
Proteomics identification of candidate H1.0-binding proteins
To catalog the H1-binding proteins present in human nuclear extracts, we performed pull-down experiments using the HaloTag system followed by mass spectrometry to identify the isolated proteins. The HaloTag protein is a mutant dehalogenase DhaA from Rhodococcus rhodochrous, which forms a covalent non-hydrolyzable intermediate on incubation with a modified hydrocarbon substrate (14). We first engineered chimeric HaloTag-H1.0 proteins by cloning the cDNAs encoding full-length mouse H1.0 (residues 1–194) and mouse H1.0 lacking its CTD (residues 1–97, designated H1ΔCTD) into the HaloTag expression vector. Residues 1–97 (corresponding to the NTD-GD fragment) of mouse H1.0 are 100% identical to human H1.0, whereas residues 98–194 (corresponding to the CTD) are 94% identical. Chimeric proteins were purified from E. coli and incubated with a sepharose-bound HaloTag substrate to covalently attach the proteins to sepharose beads.
Pull-downs were performed in parallel with nuclear extracts prepared from four different human cell lines (HeLa, CEM, RPE-1 and U2OS). Nuclear extracts were incubated with sepharose-bound HaloTag-H1.0 proteins or the HaloTag protein alone in buffer containing 150 mM NaCl. After extensive washing with buffer containing 250 mM NaCl, bound proteins were eluted from the sepharose beads and run on a SDS–polyacrylamide gel. Excised gel slices were digested with trypsin, and the proteolyzed samples were analyzed by LC-MS/MS. Candidate H1.0-binding proteins were identified in one of two ways. In our initial analysis, two pull-downs were performed for each of the four cell types. To be classified as a H1.0-binding protein, at least two unique peptides had to be observed in both replicates for at least one cell type, but not in the HaloTag only samples. Seventy-three proteins were identified in this manner as potential H1.0-binding partners (Table 1). Table 1 also includes 36 proteins that were identified by the RNAse analysis described later in the text (Supplementary Table S1). The calculated pI of human is H1.0 (10.9), and those of the candidate H1-binding proteins ranged from 3.8 to 11.9, suggesting that the observed H1.0–protein interactions do not result from non-specific electrostatic interactions. Most of the proteins pulled down by H1.0 were observed in one or two cell types (Table 1). Small nuclear ribonucleoprotein (snRNP) SmD1, splicing factor 3B subunits 1 and 3, U5 snRNP 116 kDa subunit, PUF60, nucleolin, FACT subunit SSRP1, PABPC1, ribosomal proteins L4 and L7 and transitional endoplasmic reticulum ATPase are notable as being observed in three or four cell types.
Table 1.
Candidate H1.0-binding proteins grouped by common function
| Proteina | Accession number | Identifier | Cell type | pI | Nucleolarb | CTD- dependent |
|---|---|---|---|---|---|---|
| mRNA Splicing | ||||||
| Small nuclear ribonucleoprotein Sm D1 | P62314 | SNRPD1 | C, R, U | 11.6 | + | − |
| Small nuclear ribonucleoprotein Sm D3 | P62318 | SNRPD3 | U | 10.3 | + | − |
| Small nuclear ribonucleoprotein E | P62304 | SNRPE | C, U | 9.5 | + | − |
| Small nuclear ribonucleoprotein-associated protein N | P63162 | SNRPN | U | − | ND | |
| Splicing factor 3 A subunit 3 | Q12874 | SF3A3 | C | 5.3 | + | + |
| Splicing factor 3B subunit 1 | O75533 | SF3B1 | C, R, U | 6.7 | + | − |
| Splicing factor 3B subunit 2 | Q13435 | SF3B2 | C | 5.5 | + | + |
| Splicing factor 3B subunit 3 | Q15393 | SF3B3 | C, H, U | 5.1 | + | − |
| Pre-mRNA branch site protein p14 | Q9Y3B4 | SF3B14 | C | 9.4 | + | − |
| Splicing factor U2AF 35 kDa subunit | Q01081 | U2AF1 | C, U | 8.9 | + | − |
| Splicing factor U2AF 65 kDa subunit, isoform 2 | P26368 | U2AF2 | C, U | 9.2 | + | − |
| U2 small nuclear ribonucleoprotein A′ | P09661 | SNRPA1 | U | 8.7 | + | ND |
| U5 small nuclear ribonucleoprotein 116 kDa component | Q15029 | EFTUD2 | C, H, R, U | 5.1 | + | − |
| U5 small nuclear ribonucleoprotein 200 kDa helicase | O75643 | SNRNP200 | C, U | 5.7 | + | − |
| Pre-mRNA-processing-splicing factor 8 | Q6P2Q9 | PRP8 | C, U | 9.0 | + | − |
| Serine/arginine-rich splicing factor 1 (SF2/ASF) | Q07955 | SRSF1 | C, U | 10.4 | + | − |
| Serine/arginine-rich splicing factor 2 | Q01130 | SRSF2 | C | 11.9 | + | + |
| Poly(U)-binding-splicing factor PUF60, isoform 2 | Q9UHX1 | PUF60 | C, R, U | 5.3 | + | − |
| Heterogeneous nuclear ribonucleoprotein A2/B1, isoform A2 | Q9BUJ2 | HNRNPA2B1 | C | 8.7 | + | − |
| Heterogeneous nuclear ribonucleoproteins C1/C2, isoform C1 | P07910-2 | HNRNPC | U | 4.9 | + | ND |
| Heterogeneous nuclear ribonucleoprotein D | B4DTC3 | HNRNPD | C | 7.6 | + | − |
| Heterogeneous nuclear ribonucleoprotein F | P52597 | HNRNPF | U | 5.4 | + | ND |
| Heterogeneous nuclear ribonucleoprotein G | P38159 | RBMX | C | 10.1 | + | − |
| Heterogeneous nuclear ribonucleoprotein H | P31943 | HNRNPH1 | C, U | 5.9 | + | − |
| Heterogeneous nuclear ribonucleoprotein M | P52272 | HNRNPM | C | 8.8 | + | − |
| Heterogeneous nuclear ribonucleoprotein U | Q00839 | HNRNPU | U | 5.8 | + | ND |
| Heterogeneous nuclear ribonucleoprotein U-like 1, isoform 2 | Q9BUJ2 | HNRPUL1 | U | 8.9 | + | − |
| Nuclease-sensitive element-binding protein 1 | P67809 | YBX1 | U | 9.9 | + | ND |
| ATP-dependent RNA helicase DDX1 | Q92499 | DDX1 | U | 6.8 | + | ND |
| Interleukin enhancer-binding factor 2 | Q12905 | ILF2 | C | 5.2 | + | − |
| Poly(rC)-binding protein 1 | Q15365 | PCBP1 | C | 6.7 | + | + |
| Nucleoprotein TPR | P12270 | TPR | C | 5.0 | + | + |
| RNA binding protein 39, isoform 2 | Q14498 | RBM39 | C | 10.1 | + | − |
| rRNA biogenesis | ||||||
| Nucleolin | P19338 | NCL | C, H, R, U | 4.6 | + | − |
| Nucleophosmin, isoform 2 | P06748-2 | NPM1 | U | 4.5 | + | ND |
| FACT complex subunit SPT16 | Q9Y5B9 | SUPT16H | C, U | 5.5 | + | − |
| FACT complex subunit SSRP1 | Q08945 | SSRP1 | C, H, U | 6.4 | + | − |
| Casein kinase II subunit α' | P19784 | CSNK2A2 | U | 8.7 | + | − |
| Casein kinase II subunit α1 | E7EU96 | CSNK2A1 | H, U | 7.3 | + | − |
| Histone deacetylase 2 | Q92769 | HDAC2 | C, U | 5.6 | + | + |
| Nucleolar and coiled-body phosphoprotein 1, isoform β | Q14978 | NOLC1 | C | 9.5 | + | − |
| ATP-dependent RNA helicase DDX5 | P17844 | DDX5 | C | 9.1 | + | − |
| Nucleolar RNA helicase 2 | Q9NR30 | DDX21 | C | 9.3 | + | − |
| Lupus La | P05455 | SSB | R, U | 6.7 | + | − |
| Matrin | A8MXP9 | MATR3 | C, U | 5.9 | + | − |
| Ribosome/translation | ||||||
| 40S ribosomal protein S2 | P15880 | RPS2 | R, U | 10.3 | + | − |
| 40S ribosomal protein S3a | P61247 | RPS3A | U | 9.8 | + | ND |
| 40S ribosomal protein S8 | P62241 | RPS8 | U | 10.3 | + | ND |
| 40S ribosomal protein S9 | P46781 | RPS9 | R, U | 10.7 | + | − |
| 40S ribosomal protein S13 | P62277 | RPS13 | U | 10.5 | + | ND |
| 40S ribosomal protein S15 | P62841 | RPS15 | U | 10.4 | + | ND |
| 40S ribosomal protein S23 | P62266 | RPS23 | U | 10.5 | + | ND |
| 40S ribosomal protein S24 | P62847 | RPS24 | U | 10.9 | + | ND |
| 60S ribosomal protein P0 | P05388 | RPLP0 | U | 5.7 | + | ND |
| 60S ribosomal protein L3 | P39023 | RPL3 | U | 10.2 | + | ND |
| 60S ribosomal protein L4 | P36578 | RPL4 | H, R, U | 11.1 | + | − |
| 60S ribosomal protein L5 | P46777 | RPL5 | U | 9.7 | + | ND |
| 60S ribosomal protein L6 | Q02878 | RPL6 | U | 10.6 | + | ND |
| 60S ribosomal protein L7 | P18124 | RPL7 | H, R, U | 10.7 | + | − |
| 60S ribosomal protein L7a | P62424 | RPL7A | U | 10.6 | + | ND |
| 60S ribosomal protein L8 | P62917 | RPL8 | H, U | 11.0 | + | − |
| 60S ribosomal protein L10 | P27635 | RPL10 | U | 10.1 | + | ND |
| 60S ribosomal protein L12 | P30050 | RPL12 | U | 9.5 | + | ND |
| 60S ribosomal protein L17 | P18621 | RPL17 | U | 10.2 | + | − |
| 60S ribosomal protein L18 | Q07020 | RPL18 | H, U | 11.7 | + | − |
| 60S ribosomal protein L21 | P46778 | RPL21 | U | 10.5 | + | ND |
| 60S ribosomal protein L22 | P35268 | RPL22 | U | 9.2 | + | ND |
| 60S ribosomal protein L27a | P46776 | RPL27A | U | 11.0 | + | ND |
| 60S ribosomal protein L30 | E5RI99 | RPL30 | U | 9.7 | + | ND |
| Eukaryotic translation initiation factor 3 subunit A | Q14152 | EIF3A | R, U | 6.4 | − | + |
| Eukaryotic translation initiation factor 3 subunit B, isoform 2 | P55884 | EIF3B | R, U | 4.9 | − | + |
| Eukaryotic translation initiation factor 3 subunit F | O00303 | EIF3F | R, U | 5.2 | − | + |
| Eukaryotic translation initiation factor 3 subunit I | Q13347 | EIF3I | U | 5.4 | − | + |
| Eukaryotic translation initiation factor 3, subunit L | B0QY89 | EIF3L | R | 5.9 | − | + |
| Eukaryotic translation initiation factor 5B | O60841 | EIF5B | U | 5.4 | + | ND |
| Polyadenylate-binding protein 1, isoform 2 | P11940 | PABPC1 | H, R, U | 9.5 | + | − |
| Polyadenylate-binding protein 4, isoform 2 | Q13310-2 | PABPC4 | H | 9.3 | − | − |
| Signal recognition particle 19 kDa protein | P09132 | SRP19 | U | 9.9 | + | ND |
| Signal recognition particle 68 kDa protein | Q9UHB9 | SRP68 | C, U | 8.8 | + | + |
| Signal recognition particle 72 kDa protein | O76094 | SRP72 | U | 9.3 | + | ND |
| Transport | ||||||
| Importin subunit α-7 | O60684 | KPNA6 | U | 4.9 | + | + |
| Importin subunit β-1 | Q14974 | KPNB1 | U | 4.7 | + | ND |
| Ran GTPase-activating protein 1 | P46060 | RANGAP1 | C | 4.6 | − | + |
| Major vault protein | Q14764 | MVP | R, U | 5.3 | − | − |
| Coatomer subunit α, isoform 2 | P53621 | COPA | R, U | 7.5 | + | + |
| Coatomer subunit β | P53618 | COPB1 | U | 5.7 | − | ND |
| Miscellaneous | ||||||
| Histone H1.5 | P16401 | HIST1H1B | C | 10.9 | + | − |
| Histone H2A type 2-C | Q16777 | HIST2H2AC | H, U | 10.9 | + | − |
| Histone H4 | P62805 | HIST1H4A | H | 11.4 | + | − |
| Nucleosome assembly protein 1-like 1 | P55209 | NAP1L1 | U | 4.4 | + | ND |
| Heterochromatin protein 1-binding protein 3 | Q5SSJ5 | HP1BP3 | C | 9.7 | + | + |
| TATA-binding protein-associated factor 2 N, isoform short | Q92804 | TAF15 | C | 8.0 | + | − |
| Acidic leucine-rich nuclear phosphoprotein 32 family member A | P39687 | ANP32A | U | 4.0 | − | ND |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B | Q92688-2 | ANP32B | U | 3.9 | + | ND |
| Acidic leucine-rich nuclear phosphoprotein 32 family member E | Q9BTT0 | ANP32E | U | 3.8 | + | ND |
| Protein virilizer homolog | Q69YN4 | KIAA1429 | U | 4.9 | − | ND |
| DNA damage-binding protein 1 | Q16531 | DDB1 | U | 5.1 | + | − |
| X-ray repair cross-complementing protein 5 | P13010 | XRCC5 | H | 5.6 | + | − |
| X-ray repair cross-complementing protein 6 | P12956 | XRCC6 | H | 6.2 | + | − |
| E3 ubiquitin-protein ligase HUWE1, isoform 2 | Q7Z6Z7 | HUWE1 | R, U | 5.1 | + | + |
| Transitional endoplasmic reticulum ATPase | P55072 | VCP | C, R, U | 5.1 | + | + |
| Putative RNA-binding protein Luc7-like 2 | Q9Y383 | LUC7L2 | C | 10.0 | − | + |
| Heat shock cognate 71 kDa protein | P11142 | HSPA8 | C, U | 5.4 | + | − |
| Heat shock protein β-1 | P04792 | HSPB1 | U | 6.0 | + | ND |
| Myosin regulatory light chain12B | O14950 | MYL12B | H | 4.7 | + | − |
| Myosin light polypeptide 6 | P60660 | MYL6 | H | 4.6 | + | − |
| Ribonuclease inhibitor | P13489 | RNH1 | R | 4.7 | + | + |
aProteins identified in at least two of the three control replicates in the RNAse analysis are shown in italics. The remainder of the proteins appeared in both replicates of at least one cell type in our initial analysis.
Most of the candidate H1-binding proteins can be grouped into four functional categories (Table 1). The first group comprises proteins involved in pre-mRNA splicing. Splicing is carried out by the spliceosome, which is composed of a host of core splicing factors and many other associated proteins (18,19). Thirty-three splicing factors were pulled down by H1.0, representing a significant fraction of the spliceosomal proteins and nearly one-third of the total H1-binding proteins identified in our studies. A second major group of H1.0-binding proteins functions in rRNA biogenesis, including nucleolin, nucleophosmin, FACT, casein kinase II, La and NolC1 (Nopp140). The third category was related to translation and included 24 ribosomal proteins, eukaryotic translation initiation factor 3 and signal recognition particle subunits. The fourth grouping involved proteins that function in cellular transport, and included importin subunits, major vault protein and coatomer subunits. The remaining proteins were grouped as miscellaneous. Remarkably, 94 of the 107 H1.0-binding proteins from Table 1 have been observed in proteomics analyses of the human nucleolus (20,21), suggesting a role for H1.0 in ribosome biogenesis/function.
We speculated that the disordered H1.0 CTD may mediate many of the observed protein–protein interactions. To address this question, we determined which proteins were pulled down by the H1ΔCTD construct. The interactions that were observed in at least one of the two H1ΔCTD replicates were classified as CTD-independent, whereas the interactions that were absent from both of the H1ΔCTD replicates were classified as CTD-dependent. We found that 20 of the 73 H1.0–protein interactions identified in this experiment were dependent on the CTD (Table 1). These findings indicate that in addition to their well-established roles in DNA binding, the H1.0 CTD and NTD-GD both can serve as combinatorial protein–protein interaction modules.
Many of the candidate H1.0-binding proteins detected in our studies interact with RNA or are components of ribonucleoprotein particles. Thus, it is possible that the H1.0–protein interactions may be indirect and RNA-mediated. To address this question, H1.0-binding proteins were identified in three separate pull-downs from control and RNAse-treated U2OS nuclear extracts. Based on the criterion that two or more unique peptides were observed in at least two of the three repetitions, 79 proteins were identified (Supplementary Table S1). Of these, 43 were observed in our initial analysis, and 36 were new and added to Table 1. Fifty-eight of the 79 proteins were detected in at least one of the three RNAse-treated samples, and they were scored as RNA-independent (Supplementary Table S1). U2AF65, PRP8, PUF60, SRSF1, nucleolin and heterogenous nuclear RNPs (hnRNPs) C1/C2, F and M all are RRM domain-containing RNA-binding proteins that were present in two or more of the RNAse-treated replicates. We conclude that there was no wholesale loss of H1.0–protein interactions because of RNAse treatment, and that H1.0 interacts directly with many of the RNA-binding proteins in the extracts. Twenty-one of the proteins detected in the control samples were absent from all three of the RNAse-treated extracts, suggesting that these interactions may be RNA-dependent. The RNAse-dependent interactions tended to involve known RNA-binding proteins and were distributed among all functional groupings. An interaction could be RNA-dependent if H1 directly bound RNA that was complexed with other proteins, or if H1 interacted with only one of the protein components of a multi-protein–RNA complex. Additional studies will be needed to determine the molecular basis of the RNAse-dependent interactions.
Table 1 only includes the proteins that were identified in multiple replicates in our experiments. If we consider those cases in which two unique peptides were observed in only one replicate of one cell type, an additional 191 potential H1-binding proteins were identified by LC-MS/MS (Supplementary Table S2). RbAp48 is a protein that was observed in one replicate and in only one cell type. Western blotting confirmed that RbAp48 was pulled down by H1.0, and that the interaction was abolished in the absence of the CTD (Figure 1). This result indicates that at least some of the proteins that were observed by mass spectrometry in only one replicate (Supplementary Table S2) also are legitimate H1-binding partners.
Figure 1.

RbAp48 interacts with the CTD of histone H1.0. Proteins were pulled down from isolated CEM nuclei and subjected to western blotting as described in ‘Material and Methods’ section.
Validation of candidate H1–protein interactions
A definitive test of whether an interaction identified by pull-downs is direct is provided by biophysical characterization in a purified system. We, therefore, examined whether purified recombinant H1.0 was able to bind to three candidate proteins from Table 1 using the recently developed high-throughput interactions by fluorescence intensity (HI-FI) system (17,22). For these fluorescence (de)quenching experiments, a fixed concentration of fluorescently labeled H1.0 was mixed with varying concentrations of the recombinant protein in question. Changes in the fluorescence intensity as a function of candidate protein concentration were fit to a one-site model, allowing determination of the apparent dissociation constant (KD) and Hill coefficient for the interaction.
U2AF65 is a splicing factor pulled down by H1.0. U2AF65 is a subunit of the U2AF complex, which binds both proteins and pre-mRNA to facilitate splice site recognition and the early stages of spliceosome assembly (23). When the binding of U2AF65 to H1.0 was examined by the fluorescence assay, the normalized fluorescence quenching as a function of U2AF65 concentration was well fit by a single-site model (Figure 2). The KD for the interaction was 0.82 μM, and the Hill coefficient was 1. These data indicate that H1.0 binding to U2AF65 was direct and of moderate affinity. Serine/arginine-rich splicing factor 1, more commonly known as splicing factor 2/alternative splicing factor (SF2/ASF), is another spliceosomal protein pulled down by H1.0. The fluorescence quenching data for SF2/ASF (residues 11–196) indicate that it also is capable of binding directly to H1.0 (Figure 2). The KD for the interaction was 2.4 μM, and the Hill coefficient was 2. The FACT complex is a histone chaperone that consists of the Spt16 and SSRP1 subunits (24). We observed that H1.0 bound to recombinant FACT with high affinity (KD = 0.032 μM) and a Hill coefficient of 1 (Figure 2). Taken together, the quantitative biophysical data from Figure 2 validated the proteomics analysis for the proteins examined. Although Spt16 has a highly acidic C-terminal domain implicated in core histone binding (24), SSRP1, U2AF65 and SF2/ASF all lack such negatively charged regions. Thus, the mechanism of H1 binding to these proteins does not seem to be purely electrostatic. Likewise, we could not find any obvious common sequence motifs in the four proteins. Determination of the mechanism(s) responsible for H1-mediated protein–protein interactions will be a productive area of future research.
Figure 2.
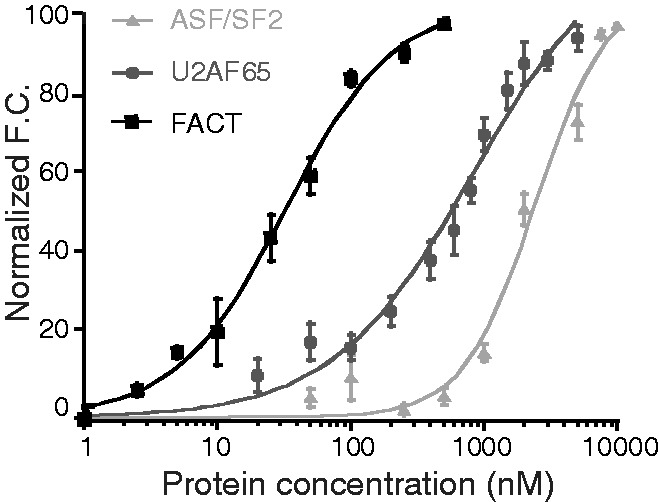
Interactions between histone H1.0 and U2AF65, SF2/ASF and FACT are direct. Shown are the normalized fluorescence changes on titration of recombinant U2AF65 (dark grey circle), SF2/ASF (grey triangle) and FACT (black square) into fluorescently labeled H1.0 as described in ‘Materials and Methods’ section. The error bars represent the standard error within individual data points.
DISCUSSION
Linker histones are ubiquitous chromatin-associated DNA-binding proteins (1,3). Although it is widely held that linker histones function by stabilizing the condensed states of chromatin (1,3,25,26), there is some evidence that they also act in part through specific protein–protein interactions (4). To define the scope of protein–protein interactions in linker histone function, we performed a systematic proteomic analysis of proteins that were pulled down from human nuclear extracts by full-length H1.0 and H1ΔCTD proteins. Our analyses identified 107 H1.0-binding proteins (Table 1). The number of H1.0-binding proteins probably is greater, as at least some of the 191 proteins observed in only one replicate are legitimate H1-binding partners (Figure 1). Most of the observed H1.0–protein interactions are RNA-independent (Supplementary Table S1). Although some of the H1.0-binding proteins may have been indirectly pulled down as part of multi-protein complexes, we observed direct interactions between H1.0 and all three of the proteins examined (U2AF65, SF2/ASF and FACT) (Figure 2). The unexpectedly large number of H1.0-binding proteins identified by our studies documents an important role for protein–protein interactions in linker histone action and suggests a new paradigm for H1 structure and function that extends well beyond its effects on chromatin architecture.
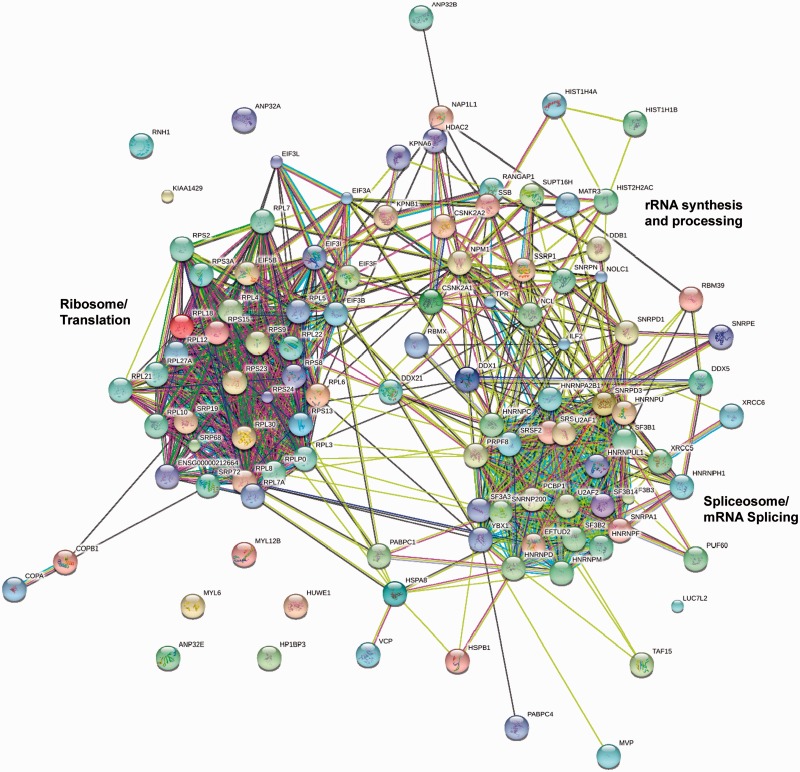
The nucleolus, the site of ribosome biogenesis (27,28), is composed of as many as 900 proteins in humans, including the linker histones (20,21). Strikingly, of the 107 proteins pulled down by H1.0, 94 have been identified as components of the nucleolus (Table 1). Most of the proteins detected in one replicate also are nucleolar (Supplementary Table S2). This suggests that the nucleolus may be a primary source of the H1-binding proteins in the nuclear extracts and raises the intriguing possibility that H1 is a key regulator of nucleolar function. In support of this notion, H1.0 pulled down numerous proteins involved in mRNA splicing, rRNA synthesis and processing and ribosome function. Interestingly, H1.0 previously has been shown to surround the nucleolus (29), whereas phosphorylated H1.2 and H1.4 localize to the nucleolus in vivo and are associated with increased RNA Pol I activity and rRNA biosynthesis (30). The involvement of rRNA synthesis/processing and ribosome biogenesis in nucleolar function is well established (27,28). The link to mRNA splicing may come from small nucleolar RNAs (snoRNAs), which are spliced out of the introns of host genes (31) and assembled into snoRNPs that modify and process rRNA (32). To determine whether the H1.0-binding proteins are functionally interrelated, the proteins from Table 1 were analyzed by the STRING engine (33). Results indicate that nearly all of the proteins pulled down by H1.0 are part of a well-defined interaction network (Figure 3). The analysis clearly identifies the spliceosome and the ribosome, reflecting the large number of splicing factors and ribosomal proteins pulled down by H1.0. Connecting the spliceosome and ribosome clusters are numerous proteins that function in rRNA synthesis and processing. Several proteins in particular stand out as central ‘hubs’, including nucleolin, nucleophosmin (B38), FACT, casein kinase II, La (SSB) and the RNA helicase DDX1 (Figure 3). Given that H1 interacts with so many multifunctional nucleolar proteins, the potential for regulation is enormous, as the interplay between protein concentrations and equilibrium constants will dictate which H1–protein interactions dominate under any given set of conditions in vivo. We note that FRAP studies have shown that there are multiple kinetic classes of H1 in the nucleus (34,35). In view of our findings, it seems reasonable to propose that the nucleolus may be the source of the slower exchanging fraction of H1.
Figure 3.
STRING analysis of the H1.0 interactome. The identifiers for the 107 proteins from Table 1 were entered into the STRING database (http://string.embl.de) (33). The confidence level was set to 0.4 (medium). Shown is the evidence view. Line colors represent the different types of evidence for the indicated association.
Examination of the specific spliceosomal proteins pulled down by H1.0 provides insight into how H1 may regulate spliceosome function. Recognition of 5′ and 3′ splice sites in the pre-mRNA is a key step in the splicing process. Splice site recognition involves recruitment of U2AF35 and U2AF65 to the 3′ splice site and the U1 snRNP particle to the 5′ splice site by the serine/arginine-rich splicing factors (SR proteins) (23). The SR proteins regulate both constitutive and alternative splicing (36). H1.0 pulled down U2AF35, U2AF65 and two SR proteins (Table 1). In addition, two U1 snRNP proteins and six other SRs were observed in one replicate. Direct interactions between H1.0 and both U2AF65 and SF2/ASF were rigorously confirmed by biophysical studies of pure recombinant proteins (Figure 2). The hnRNP proteins repress splice site recognition by competing for the same pre-RNA–binding sites as the SR proteins (23). H1.0 pulled down nine hnRNPs, and five more hnRNPs were observed in one replicate. Our results suggest that linker histones may regulate mRNA splice site recognition via interactions with SRs, U2AF and hnRNPs. H1.0 also pulled down two U2 snRNP components and four subunits of the U2-associated complex, SF3 (Table 1). This suggests that H1.0 also may be involved in U2 snRNP function.
In addition to the functional ramifications discussed earlier in the text, our studies have provided structural insight into H1-mediated protein–protein interactions. About 25% of the candidate H1–protein interactions were found to be dependent on the H1 CTD and 75% on the NTD-GD fragment (Table 1). The CTD is an intrinsically disordered domain whose function has been linked to its unique amino acid composition (the isoform CTDs consists of 38–42% lysine, 12–14% proline, 18–34% alanine and essentially no aromatic residues) (8,37). Despite having low-sequence complexity, intrinsically disordered protein domains often act as combinatorial protein–protein interaction modules, that is, the same domain is able to specifically interact with many different proteins (38). This has been termed ‘fuzziness’ (39). Our findings are entirely consistent with this concept. There is biochemical evidence for CTD-dependent protein–protein interactions as well (40,41). Perhaps more unexpected was the large number of interactions that were mediated by the NTD-GD fragment. The GD has a winged helix fold (5). Although most winged helix motifs are DNA-binding domains, several are known to mediate protein–protein interactions (42). The linker histone GD seems to fall into a unique class that can mediate both protein–protein and protein–DNA interactions. Finally, it is possible that the disordered NTD mediates some, if not many, of the H1.0–protein interactions identified in the H1ΔCTD samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3.
FUNDING
National Institutes of Health (NIH) [GM045916, GM066834 to J.C.H.; GM088409 to K.L. and J.C.H.; GM088371 to J.G.D.; F32GM096531 to D.D.W.]; International Rett Syndrome Foundation (to A.A.K.); Howard Hughes Medical Institute (to K.L.); Pew Scholars Program in the Biomedical Sciences (to J.G.D.). Funding for open access charge: NIH [GM045916 and GM066834].
Conflict of interest statement. None declared.
Supplementary Material
ACKOWLEDGEMENTS
Plasmid pET24b encoding SF2/ASF was a kind gift of Stuart Wilson, University of Sheffield. Plasmid pET30a encoding U2AF65 was a kind gift of Angus Lamond, University of Dundee. We thank Christine Krause for excellent technical assistance.
REFERENCES
- 1.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 2.Th’ng JPH, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 3.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 4.McBryant SJ, Lu X, Hansen JC. Multifunctionality of the linker histones: an emerging role for protein-protein interactions. Cell Res. 2010;20:519–528. doi: 10.1038/cr.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramakrishnan V, Finch J, Graziano V, Lee P, Sweet R. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 6.Clore GM, Gronenborn AM, Nilges M, Sukumaran DK, Zarbock J. The polypeptide fold of the globular domain of histone H5 in solution. A study using nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1987;6:1833–1842. doi: 10.1002/j.1460-2075.1987.tb02438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas J. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 8.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 2006;281:1853–1856. doi: 10.1074/jbc.R500022200. [DOI] [PubMed] [Google Scholar]
- 9.Caterino TL, Fang H, Hayes JJ. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol. 2011;31:2341–2348. doi: 10.1128/MCB.05145-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H10 C-terminal domain. J. Biol. Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 12.Ni J-Q, Liu L-P, Hess D, Rietdorf J, Sun F-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Choi J, Heo K, Kim H, Levens D, Kohno K, Johnson EM, Brock HW, An W. Isolation and characterization of a novel H1.2 complex that acts as a repressor of p53-mediated transcription. J. Biol. Chem. 2008;283:9113–9126. doi: 10.1074/jbc.M708205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Los G, Encell L. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 15.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011;286:41883–41892. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J, Martin P. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 17.Winkler D, Luger K, Hieb A. Quantifying chromatin-associated interactions: the HI-FI system. Methods Enzymol. 2012;512:243–274. doi: 10.1016/B978-0-12-391940-3.00011-1. [DOI] [PubMed] [Google Scholar]
- 18.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 20.Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 21.Jarboui MA, Wynne K, Elia G, Hall WW, Gautier VW. Proteomic profiling of the human T-cell nucleolus. Mol. Immunol. 2011;49:441–452. doi: 10.1016/j.molimm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Hieb A, D’Arcy S, Kramer M. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res. 2012;40:e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winkler DD, Luger K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011;286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DLJ. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip. Rev. RNA. 2010;1:415–431. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- 28.Pederson T. The nucleolus. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breneman J, Yau P, Teplitz R, Bradbury E. A light microscope study of linker histone distribution in rat metaphase chromosomes and interphase nuclei. Exp. Cell Res. 1993;206:16–26. doi: 10.1006/excr.1993.1115. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, Nardulli AM, Hager GL, Kelleher NL, Mizzen CA. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 2010;189:407–415. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiss T. SnoRNP biogenesis meets pre-mRNA splicing. Mol. Cell. 2006;23:775–776. doi: 10.1016/j.molcel.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb. Symp. Quant. Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 35.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 36.Sanford JR, Ellis J, Cáceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 2005;33:443–446. doi: 10.1042/BST0330443. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Hamkalo B, Parseghian M. Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry. 2009;48:164–172. doi: 10.1021/bi801636y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tompa P, Fersht A. Structure and Function of Intrinsically Disordered Proteins. Boca Raton, FL: Taylor and Francis Group, LLC; 2010. [Google Scholar]
- 39.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Widlak P, Kalinowska M, Parseghian MH, Lu X, Hansen JC, Garrard WT. The histone H1 C-terminal domain binds to the apoptotic nuclease, DNA fragmentation factor (DFF40/CAD) and stimulates DNA cleavage. Biochemistry. 2005;44:7871–7878. doi: 10.1021/bi050100n. [DOI] [PubMed] [Google Scholar]
- 41.Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- 42.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.