Abstract
Active regulator of SIRT1 (AROS) binds and upregulates SIRT1, an NAD+-dependent deacetylase. In addition, AROS binds RPS19, a structural ribosomal protein, which also functions in ribosome biogenesis and is implicated in multiple disease states. The significance of AROS in relation to ribosome biogenesis and function is unknown. Using human cells, we now show that AROS localizes to (i) the nucleolus and (ii) cytoplasmic ribosomes. Co-localization with nucleolar proteins was verified by confocal immunofluorescence of endogenous protein and confirmed by AROS depletion using RNAi. AROS association with cytoplasmic ribosomes was analysed by sucrose density fractionation and immunoprecipitation, revealing that AROS selectively associates with 40S ribosomal subunits and also with polysomes. RNAi-mediated depletion of AROS leads to deficient ribosome biogenesis with aberrant precursor ribosomal RNA processing, reduced 40S subunit ribosomal RNA and 40S ribosomal proteins (including RPS19). Together, this results in a reduction in 40S subunits and translating polysomes, correlating with reduced overall cellular protein synthesis. Interestingly, knockdown of AROS also results in a functionally significant increase in eIF2α phosphorylation. Overall, our results identify AROS as a factor with a role in both ribosome biogenesis and ribosomal function.
INTRODUCTION
Ribosomes catalyse the translation of the genetic code in mRNA into functional proteins. In addition, ribosomes are known to act in concert with eukaryotic initiation factors (eIFs) to regulate gene expression, allowing the cell to respond specifically and appropriately to internal and external stimuli (1,2). Perhaps unsurprisingly, given their important function, ribosomes have been implicated in human disease. For example, ribosome function can be deregulated in cancer, commonly via misregulation of eIF activity or increased ribosome biogenesis (3–6). More recently, mutations that restrict ribosome biogenesis or function have given rise to a range of diseases termed ribosomopathies (7,8).
Biogenesis of ribosomes requires the assembly of RNAs and proteins into two subunits, termed the small ‘40S’ and large ‘60S’. Mammalian ribosomes consist of four ribosomal RNAs (the 18S in the 40S subunit and the 28S, 5.8S and 5S in the 60S subunit) and 80 proteins (33 in the 40S and 47 in the 60S). These 84 molecules are not self-assembling, requiring hundreds of ribosome biogenesis factors, both RNA and protein, to produce competent ribosomes (9). The complex process of biogenesis begins in nucleoli, where the main ribosomal RNA (rRNA) is transcribed. This precursor rRNA (pre-rRNA) nucleolar transcript contributes mature rRNA to both ribosomal subunits, which are separated by spacer regions and therefore require processing by sequential nucleolytic cleavage [Figure 1A and (10)]. Despite the dedicated role of ribosome biogenesis factors, there is still a requirement for many of the ribosomal proteins in distinct stages of rRNA processing (11).
Figure 1.
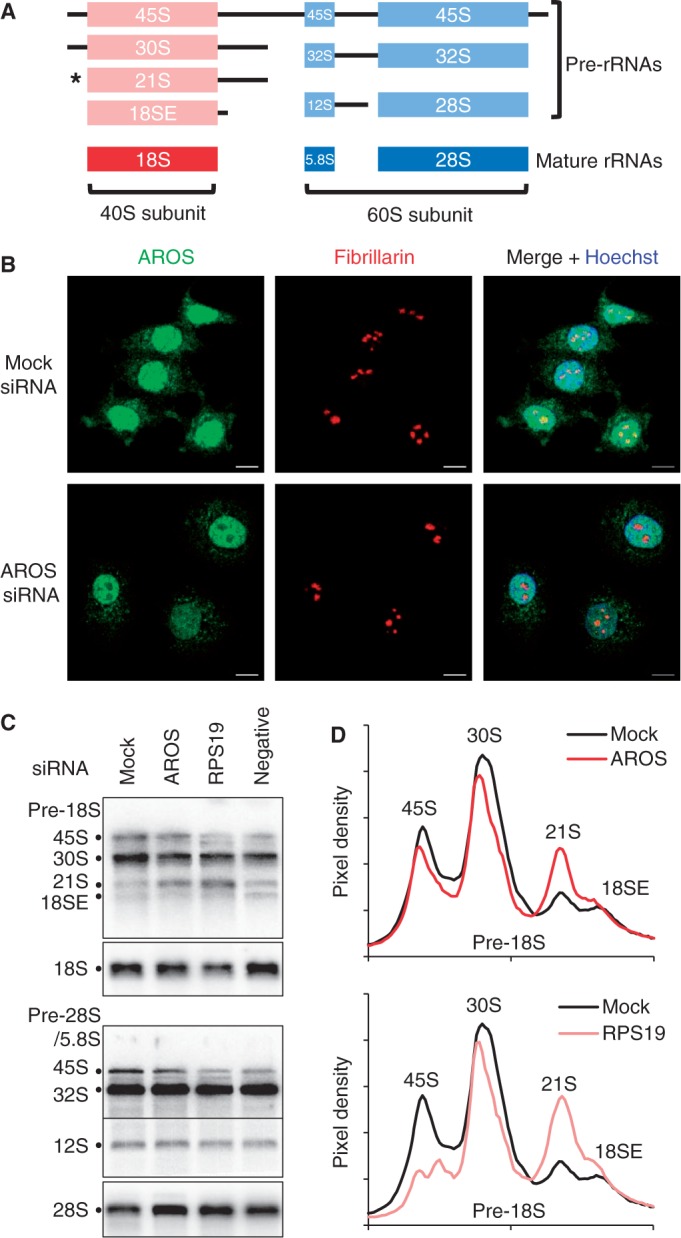
AROS is specifically required for 40S subunit rRNA processing. (A) Representation of ribosomal RNA maturation including pre-rRNA and rRNA nomenclature. Asterisk indicates the 21S pre-rRNA where processing is stalled by depletion of RPS19. (B) Immunofluorescent detection of AROS (green) and fibrillarin (red) localization in representative MCF7 cells with and without silencing of AROS. Bar is 10 µm. (C) Northern blotting for pre-rRNAs and18S and 28S mature rRNAs. Total RNA from HCT116 cells isolated following siRNA transfection were separated by electrophoresis and blotted using 32P labelled probes for each pre- or mature rRNA. The negative siRNA targets a splice form of SIRT1, SIRT1Δ8, and illustrates no effect on rRNA processing. (D) Pixel density from whole lanes in (C) calculated using Image J software (National Institutes of Health).
The role of the ribosomal protein RPS19 in ribosome biogenesis has been well defined, in part owing to its genetic association with the ribosomopathy Diamond–Blackfan anaemia (DBA) (12). RPS19 is required for the processing of the 40S subunit rRNA from the 21S to the 18SE final pre-rRNA form [Figure 1A and (13–15)]. Cells depleted of RPS19 exhibit reduced 40S abundance, a lower rate of protein synthesis and increased apoptosis. Active Regulator Of SIRT1 (AROS—also termed RPS19 binding protein 1) was identified as a direct binding partner for RPS19 in a yeast two-hybrid screen and found to be a widely expressed nuclear protein in the mouse (16). Subsequent analysis using the rat proteins illustrated that phosphorylation of RPS19 by CaM-kinase Iα enhanced its interaction with AROS in vitro (17). More recently, AROS was also identified as a direct interactant of the NAD+-dependent deacetylase SIRT1, promoting SIRT1-mediated suppression of p53 in human cancer cell lines (18). AROS protein has been localized to nuclei in both human and mouse cells including sub-nuclear foci presumed to be nucleoli—the site of ribosome biogenesis (16,18). As such, the subcellular location of AROS suggests a role in ribosome biogenesis, although this remains to be confirmed.
The present study examines the role of AROS in ribosome biogenesis and reports an unexpected cytoplasmic role for AROS, where it appears to regulate ribosome function. Initially, we confirm AROS localization within nucleoli and correlate this with a requirement for AROS in pre-18S rRNA processing, analogous with the function of RPS19. We show that AROS is also present in the cytoplasm of human cells and is a partially conserved nuclear/cytoplasmic shuttling protein, based on orthologue alignment and sequence analysis. Within the cytoplasm, AROS is shown to interact with free 40S subunits and also with translating polysomes, unexpectedly extending the role of AROS beyond ribosome biogenesis into ribosome function and protein synthesis. Finally, we highlight a role for AROS in maintaining the maximal rate of global protein synthesis, which appears to be in part linked to translation initiation via suppression of eIF2α phosphorylation.
MATERIALS AND METHODS
Cell culture and transient transfection
All cell lines were grown at 37°C and 5% CO2, as recommended by the ATCC. HCT116 colorectal adenocarcinoma cell line was a kind gift from Bert Vogelstein, Howard Hughes Medical Institute. Flag-tagged AROS (18) and Flag-tagged GADD34 (Growth arrest and DNA-damage inducible protein 34) (19) were transfected using Lipofectamine (Invitrogen). The siRNA (Dharmacon) transfection technique was as previously described (20). RPS19 siRNA sense sequence: 5′-AGAGCUUGCUCCCUACGAU(dTdT)-3′, as published (13). AROS siRNA sense sequence: 5′-GACCACCUCAGAGUAAACC(dTdT)-3′. RPSA, RPS6 and RPL7A siRNAs are On Target plus SmartPool from Dharmacon. Cells were harvested either 48 h (HCT116, HEK293 and MCF7) or 72 h (MCF10A) after transfection. All data are representative of three biological replicates.
Determination of protein synthesis rates
Cells were treated with 30 µCi/ml 35S-methionine label (Hartmann Analytic) for 30 min then harvested and lysed. Protein was precipitated onto filter paper (Whatmann) by addition of trichloroacetic acid to 12.5% and washed with 70% ethanol then acetone. Scintillation was read from dried filter paper in triplicate for each experimental condition (National Diagnostics). Total protein content was determined by BCA (bicinchoninic acid) assay (Pierce) and 35S-methionine incorporation standardized against a reference treatment set to 1.0 from three independent experiments. Error bars are standard deviation from the mean.
Quantification of RNA
RNA was isolated by the RNeasy protocol (Qiagen) or Trizol reagent (Invitrogen). Human tissue RNA samples were purchased from AMS Biotechnology. RNA was used in reverse transcriptase-polymerase chain reaction (RT-PCR) with specific primers as follows. AROS forward primer: 5′-GGAAGACGAAGGCAATTCAGGC-3′ and reverse primer: 5′-TCCTCGGTGAACACGGTGCC-3′. RPS19 forward primer: 5′-ACCAGCAGGAGTTCGTCAGAGC-3′ and reverse primer: 5′-CCACCTGTCCGGCGATTCTG-3′. β-actin forward primer: 5′-GCCAACAGAGAGAAGATGAC-3′ and reverse primer: 5′-CGCAAGATTCCATACCCAGG-3′. GAPDH forward primer: 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse primer: 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. For quantitative PCR, n ≥ 3 and error bars represent standard deviation from the mean. RT-PCR products were separated by electrophoresis through 1.5% agarose–TAE, using ethidium bromide and ultraviolet (UV) transillumination for visualization.
Northern blotting
RNA was resolved by 1% agarose formaldehyde-MOPS gel electrophoresis then passively transferred to zeta probe (BioRad) in 3 M sodium chloride, 0.3 M sodium citrate (pH 7.0) (SSC) overnight and UV cross-linked post-transfer by Stratalinker (Stratagene). Membranes were pre-hybridized in Church Gilbert’s for 30 min at 55°C. In all, 50 pmol of DNA oligonucleotide (Sigma-Aldrich) was labelled with 30 µCi 32P-ɣATP (Hartmann Analytic) using T4 PNK enzyme (New England Biosciences). Probes were selected by G25 column (GE Healthcare) and incubated with membranes overnight in Church Gilbert’s at 55°C. Membranes were washed in dilutions of SSC solution and exposed to a phosphorscreen (GE Healthcare). Oligonucleotide sequences as published (13).
Protein sample preparation and sodium dodecyl sulphate–polyacrylamide gel electrophoresis
Cells were lysed [10mM Tris at pH 8.0, 140 mM NaCl, 2 mM CaCl2, 0.5% v/v NP-40 and protease inhibitor cocktail (Roche)] and protein quantity assayed by the Pierce BCA method. Protein was denatured for sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS–PAGE) by addition of 5× Laemmli’s buffer. Cell fractionation was carried out according to the protocol described by Pierce, supplemented with protease inhibitor cocktail (Roche). Equivalent protein by mass was resolved by SDS–PAGE at 10–15% (w/v) acrylamide before electrophoretic transfer to nitrocellulose.
Immunoprecipitation and immunoblotting
Flag immunoprecipitation was carried out 24 h after transfection using pre-conjugated agarose beads and Flag-peptide elution (Sigma-Aldrich). Nitrocellulose membranes (Whatmann) were pre-blocked for 1 h, then incubated in primary antibody overnight at 4°C. Antibodies were purchased from Cell Signaling (RPS6, RPL7A, Histone H3, eIF4E, eIF2α, eIF2α S51-P, eIF4E S209-P, 4E-BP1, 4E-BP1-T37/46P, β-tubulin, SIRT1), Abcam (RPS19, RPS23, RPL27A), Santa Cruz (RPSA, RPL3, Lamin AC, GADD34), Alexis (AROS), Genetex (RPS2), Abgent (RPS8), Epitomics (LDH), Dako (p53) and Millipore (β-actin). HRP-conjugated secondary antibody (Dako) incubation was followed by visualization of antibodies by chemiluminescence (Roche/GE Healthcare).
Immunofluorescence
Cells were cultured on 13 mm uncoated glass cover slips (VWR). At harvest, cells were washed twice in phosphate buffered saline (PBS) and fixed in 4% (w/v) paraformaldehyde for 10 min, washed again in PBS then permeablized with 0.1% (v/v) Triton X-100 and finally blocked in 3% (w/v) BSA in PBS for 30 min, all at room temperature. Cells were incubated with AROS antibody at 1:100 (Alexis) diluted in 3% BSA PBS at 4°C overnight or for 2 h with fibrillarin antibody at 1:500 (Abcam) or nucleolin antibody at 1:500 (Abcam). Alexa-Fluor-conjugated secondary antibodies (Invitrogen) were incubated following removal of primary for 1 h at room temperature and DNA stained with Hoechst (Sigma-Aldrich). Slips were mounted to slides (Polysicences) and visualized using a Zeiss LSM510 Meta laser scanning confocal microscope.
Sucrose density ultracentrifugation
Before harvesting, cell cultures were treated with 100 µg/ml cycloheximide (Sigma-Aldrich) for 3 min or 2 µg/ml Harringtonine (Santa Cruz) for 5 min followed by cycloheximide. Cells were scraped in ice cold PBS, then lysed in ice cold 300 mM NaCl, 15 mM MgCl2, 15 mM Tris (pH 7.5) containing 1 mg/ml heparin sulphate and 0.1 mg/ml cycloheximide supplemented with 0.1% (v/v) Triton X-100. Post-nuclear lysates were layered on ∼10 ml 10–50% (w/v) sucrose gradients of the same buffer omitting Triton X-100. Gradients were centrifuged at 38 000 rpm for 3 h at 4°C in a SW40Ti rotor (Beckman Coulter) and separated through a live OD254nm UV spectrometer (Isco). Comparison of peak abundance was based on the area under the curve. In all, 1 ml fractions were collected (Foxy) and protein precipitated by addition of 150 µg/ml sodium deoxycholate and trichloroacetic acid to 10%. Precipitated protein was washed twice [50 mM Tris (pH 8) containing 70% acetone and 20% ethanol] and dissolved in 1× Laemmli’s buffer.
RESULTS
Ribosome biogenesis is stalled following AROS knockdown
The primary location for ribosome biogenesis is in the nucleoli (10). Thus, a role for AROS in ribosome biogenesis will require nucleolar localization, which has previously been assumed from immunofluorescent analyses (16,18). To assess the localization of AROS, MCF7 cells were stained using a polyclonal antibody raised against the full AROS protein (green) in parallel to a monoclonal antibody against the nucleolar protein fibrillarin (red) and visualized by immunofluorescence (Figure 1B). AROS exhibits diffuse expression across the nucleus, including co-localization with fibrillarin within nucleoli (Figure 1B). AROS protein also co-localizes with a second nucleolar marker, nucleolin, by immunofluorescence (Supplementary Figure S1A), confirming the nucleolar localization for AROS. Importantly, the nucleolar staining attributable to AROS was depleted by RNAi, confirming the specificity of the antibody (Figure 1B and Supplementary Figure S1). AROS staining was dramatically depleted from nucleoli but was also reduced in the cytoplasm and to a lesser extent in the nucleoplasm.
Given the nucleolar localization of AROS and the previously reported interaction between AROS and RPS19 (16), the role of AROS in ribosome biogenesis was investigated. RPS19 is known to promote 40S subunit biogenesis, being required for the nucleolar cleavage of 21S pre-rRNA into the final pre-rRNA termed the 18SE [Figure 1A and (13–15)]. AROS expression was reduced by siRNA in HCT116 cells and the abundance of pre-rRNA species assessed for both pre-18S (40S subunit) and pre-28S/5.8S (60S subunit) by northern blotting (Figure 1C). Following RNAi-induced knockdown of AROS pre-18S rRNA processing is stalled at the 21S stage, with little effect on the processing of pre-60S subunit rRNA (Figure 1C). A similar effect was observed following parallel RPS19 silencing, agreeing with previous reports (13–15). Quantification of the pixel density across each lane illustrates this specific increase in 21S pre-rRNAs (Figure 1D). Importantly, a negative control siRNA did not alter the abundance of either 21S or 18SE pre-rRNAs (Figure 1C). There was a reduction in the 45S species following RPS19 silencing, visible with both pre-rRNA probes. This could represent a shutdown in rRNA transcription, which may be a consequence of nucleolar disruption by RPS19 knockdown. Consistent with stalling in pre-rRNA processing, the abundance of mature 18S rRNA appears reduced following silencing of AROS and to a greater extent RPS19 (Figure 1C), with 28S rRNA abundance slightly increased. This increase is likely to be the result of a higher proportion of 28S rRNA within the RNA sample as a result of the depletion in 18S rRNA, instead of an active increase in 28S synthesis. Similar data were obtained using the HEK293 cell line (Supplementary Figure S2), further supporting a functional role for AROS in 40S subunit pre-rRNA processing.
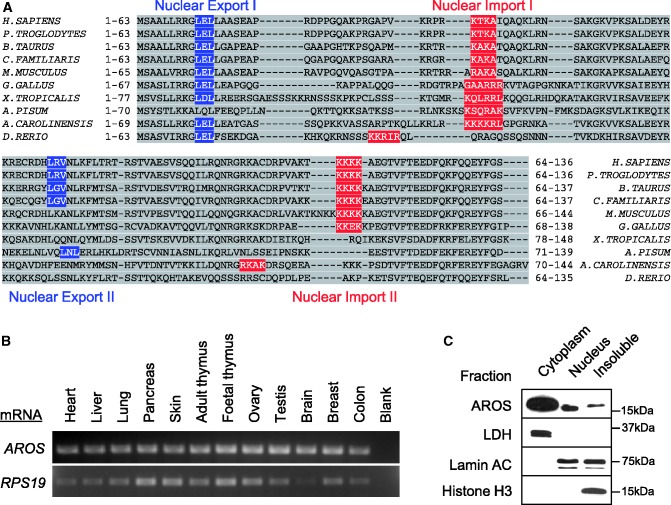
Next, we hypothesised that such a role for AROS in ribosome biogenesis would necessitate a degree of conservation and ubiquitous expression. Therefore, the evolutionary conservation and expression of AROS in human tissues were analysed. Interestingly, the AROS gene is conserved only within animalia, with orthologues in all families of vertebrates and some invertebrates (Figure 2A). These data suggest that AROS only has a role in ribosome biogenesis in animalia, which is either alternatively fulfilled in other species or that the function is not required. Taking human AROS as an example, we found that AROS mRNA could be detected in each of a representative sample of tissue extracts (Figure 2B). This is similar to RPS19 mRNA and is consistent with a ubiquitous function for the AROS gene in human cells. Similarly, wide expression of human and mouse AROS mRNAs have been previously reported (16,18).
Figure 2.
AROS is widely expressed in tissues and distributed within cells. (A) Sequence alignment [Clustal Omega (21)] of AROS from 10 species with blue nuclear export [NetNES—(22)] and red nuclear import [NLStradamus—(23)] sequences highlighted. Homo sapiens = human; Pan troglodytes = chimpanzee; Bos Taurus = cow; Canis familiaris = dog; Mus musculus = mouse (all mammal); Gallus gallus = chicken (bird); Xenopus tropicalis = western clawed frog (amphibian); Acyrthosiphon pisum = pea aphid (insect); Anolis carolinensis = Carolina anole (lizard); Danio rerio = zebrafish (fish). (B) Amplification of AROS and RPS19 mRNAs following reverse transcription from a panel of human tissues samples (AMS Biotechnology) were visualized using ethidium bromide staining of agarose TAE gels. (C) Western blotting following subcellular fractionation of HCT116 cells against indicated proteins. Lactate dehydrogenase (LDH) is used as a cytoplasmic control and Lamin AC and Histone H3 as nuclear markers.
AROS is a nuclear and cytoplasmic protein
AROS has been described as an exclusively nuclear protein (16,18), with the implication that AROS is occluded from the cytoplasm. However, our data suggest that a proportion of AROS is present in the cytoplasm (Figure 1B). To investigate this further, predictive tools (21–23) were used to search for nuclear import and export signals in the AROS protein sequence from a range of species. We found that AROS has two partially conserved predicted nuclear export (blue boxes) and import signals (red boxes) across the 10 species from six taxonomic classes analysed (Figure 2A). Importantly, AROS protein from each species contains at least one import and one export sequence. This suggests that AROS resides in both the cytoplasm and the nucleus and may be able to shuttle between each location.
To determine the relative abundance of AROS in the cytoplasm and nucleus, HCT116 cells were fractionated to obtain nuclear and cytoplasmic extracts (Figure 2C). Sequential biochemical lysis of the cytoplasm and then nucleus was carried out leaving an insoluble fraction that was also analysed. This insoluble fraction represents nuclear and cytoplasmic protein that resisted solubilization through the process. As expected, AROS is found in nuclei as previously reported (16,18), but there is a large proportion of AROS in the cytoplasm (Figure 2C). Both biochemical fractionation (Figure 2C) and immunofluorescent localization (Figure 1B) indicate nuclear and cytoplasmic populations of AROS. Immunofluorescence shows AROS protein diffuse across the cytoplasm but also in speckles, some of which appear peri-nuclear. Sub-cellular fractionation implies that the majority of AROS protein is cytoplasmic, whereas the immunofluorescence implies the inverse, that AROS is predominantly nuclear. There are a number of possible explanations for this; epitopes of the polyclonal antibody may be occluded in the cytoplasm of immunostained cells but can be detected on denaturation by SDS–PAGE, or the nuclear staining may be focused in comparison with the larger volume of cytoplasmic staining, giving the greater intensity of signal. Despite this difference, both methods demonstrate that AROS is present in both the nucleus and cytoplasm.
AROS associates with cytoplasmic ribosomes
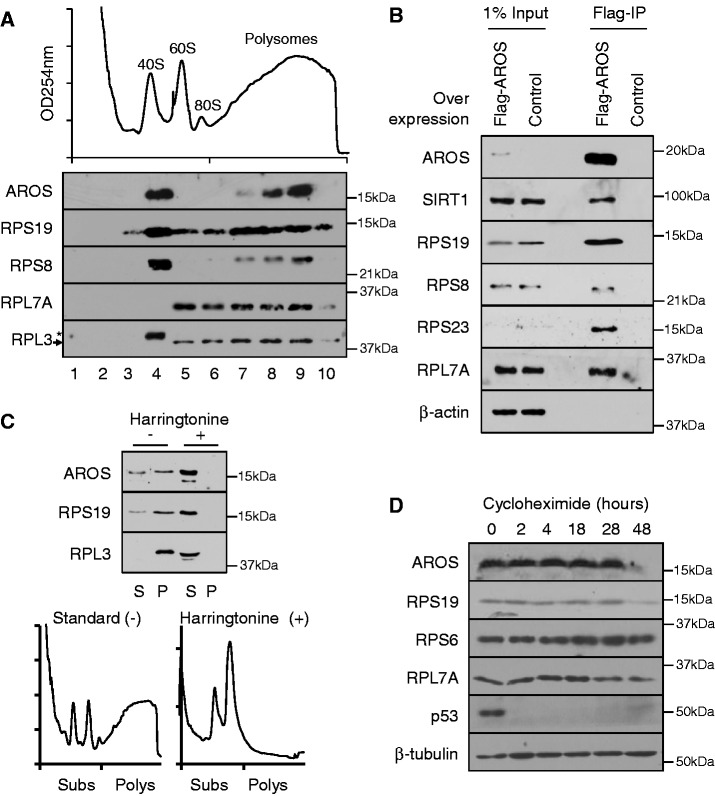
Given the direct interaction between AROS and RPS19 and the presence of AROS in the cytoplasm, we postulated that AROS may associate with ribosomes. To analyse this, post-nuclear extracts from HEK293 cells were generated, applied to a 10–50% sucrose density gradient and centrifuged to separate ribosomal components (Figure 3A). AROS sediments into two populations, fraction 4 corresponding to the 40S subunit and fractions 7–9 corresponding to actively translating ribosomes—the polysomes (Figure 3A). This pattern broadly resembles the distribution of the 40S proteins RPS8 and to a lesser extent RPS19, suggesting that AROS associates with 40S subunits. Similar data were produced with MCF7 cells (Supplementary Figure S3A).
Figure 3.
Association of AROS with 40 S subunits and polysomes. (A) OD254nm trace from 10–50% sucrose density gradient separation of post-nuclear HEK293 lysate, above, and analysis of purified protein from 10 equal fractions from the gradient by western blotting, below. Asterisk indicates a non-specific band with an arrow indicating RPL3 protein. (B) Immunoprecipitation of exogenous Flag-AROS in MCF7 cells using anti-Flag antibody conjugated to agarose beads. Equal protein was loaded by mass in the presence or absence of exogenous Flag-AROS and eluted using excess Flag peptide. Precipitates were analysed by western blotting compared with a 1% equivalent input. (C) Protein localization following inhibition of translation initiation and polysome formation by 5-min Harringtonine treatment (2 µg/ml) of HEK293 cells before sucrose density gradient separation. Purified protein from separated post-nuclear extracts analysed as sub-polysomal (S, Subs) or polysomal (P, Polys). OD254nm traces below illustrate a reduction in polysomes following Harringtonine treatment. (D) Analysis of protein expression following 100 µg/ml cycloheximide treatment of MCF7 cells for indicated times. P53 protein depletion is used as a control for cycloheximide efficacy and β-tubulin as a loading control.
To determine whether AROS interacts with ribosomal components immunoprecipitations were carried out from MCF7 cells transfected with Flag-tagged AROS or mock transfected cells (Figure 3B). Consistent with an interaction between AROS and the 40S ribosomal subunit, exogenous Flag-AROS was found to associate with the 40S proteins RPS19, RPS8 and RPS23 (Figure 3B). These three proteins are dispersed across the eukaryotic 40S subunit (24,25), suggesting an association of AROS with intact 40S subunits. Flag-AROS also associates with its known binding partner SIRT1, but not with the abundant negative control protein β-actin (Figure 3B). Together with the localization to the 40S peak on sucrose density ultracentrifugation, these data provide good evidence for an association of AROS with the 40S subunit.
Flag-AROS also co-immunoprecipitates RPL7A of the 60S subunit (Figure 3B), which is likely to result from the polysomal association of AROS (Figure 3A). To test this result further, HEK293 cells were treated with the translation initiation inhibitor Harringtonine for 5 min. This compound inhibits the joining of the 60S subunit to the 40S on mRNA, resulting in loss of polysomes owing to processive run off and limited re-initiation (26). Following Harringtonine treatment, there is a dramatic loss of polysomes, with a concomitant increase in sub-polysomes (Figure 3C). AROS protein could only be detected in polysomal fractions in the absence of Harringtonine, correlating well with ribosomal protein distribution. As such, sedimentation of AROS into the denser fractions of the gradient is dependent on polysomes. These data have been reproduced in the MCF7 cell line, and together with the co-immunoprecipitation data (Figure 3B), support the conclusion that AROS associates with polysomes in addition to 40S subunits.
Ribosomes and ribosomal proteins have long half-lives within the cell (27,28). We predicted that AROS protein may also have a long half-life, with kinetics similar to that of ribosomal proteins, allowing for a persistent and potentially functional association between AROS and the ribosome. To analyse protein half-life, synthesis was inhibited by cycloheximide treatment of MCF7 cells over a time course, and AROS protein levels were compared with that of ribosomal proteins and the rapidly degraded positive control protein p53 (Figure 3D). A fraction of AROS persists to 48 h post-treatment and is comparable with the stability of RPS19, and to a lesser extent RPS6 and RPL7A proteins. This was repeated in the HCT116 cell line giving data consistent with AROS being a persistent protein, akin to the ribosomal proteins in its kinetics following cycloheximide treatment (Supplementary Figure S3C).
AROS stability is linked to that of 40S proteins
Despite their direct interaction, the relationship between AROS and RPS19 is unknown, leading us to analyse whether the stability of AROS and RPS19 are linked. The expression of either AROS or RPS19 was reduced by siRNA in three human cell lines (HCT116, MCF7 and MCF10A) and subsequent whole cell extracts analysed for protein abundance by immunoblotting (Figure 4). Parallel samples were taken for RNA analysis. Silencing of AROS mRNA was effective in the three cell lines and resulted in AROS protein depletion (Figure 4A). Interestingly, depletion of AROS protein resulted in a dramatic reduction in the amount of RPS19 protein in each of the three cell lines. Importantly, this appears to be post-transcriptional regulation, as RPS19 mRNA was not reduced by AROS depletion in each cell line. The data also show a reciprocal regulation of AROS protein by RPS19; depletion of RPS19 by RNAi resulted in a reduction of AROS protein, but not AROS mRNA (Figure 4B). Despite the reduction in reciprocal protein following silencing of either AROS or RPS19, mRNA abundance increases in some instances, perhaps representing a compensatory mechanism attempting to redress the loss of protein. This reciprocal relationship was also observed in a fourth cell line, the HEK293 cells (Supplementary Figure S4A).
Figure 4.
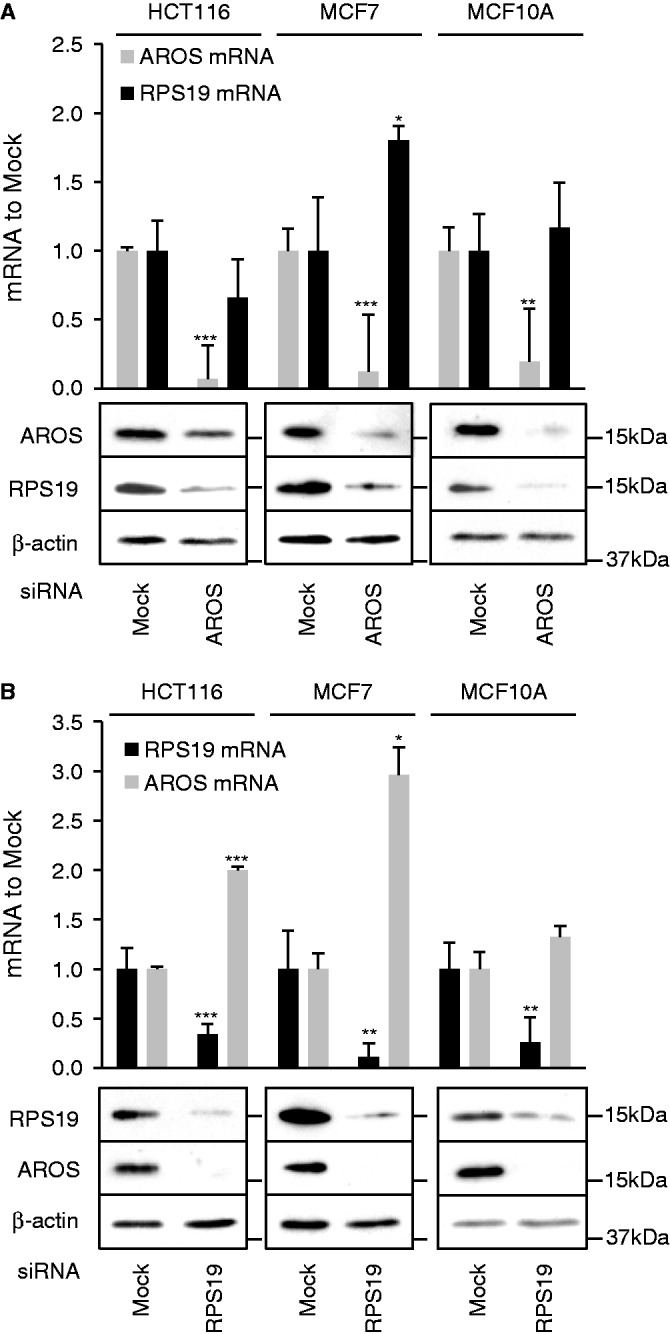
AROS and RPS19 form a positive post-transcriptional autoregulatory relationship. (A) qRT-PCR of mRNA and western blot of protein abundance following 48 h of RNAi against AROS in three cell lines. β-actin is used as a loading control for western blotting, and qRT-PCRs are standardized against β-actin (HCT116) or GAPDH mRNAs (MCF7 and MCF10A). (B) qRT-PCR and western blot data as for (A) following RNAi against RPS19. *P < 0.05, **P < 0.01, ***P < 0.001.
Reciprocal positive regulation of protein stability is often an indication of complex formation. In this instance, it seems likely that this complex is the 40S subunit. It is known that the stability of ribosomal proteins is coordinated such that a reduction in the expression of one ribosomal protein reduces the levels of other proteins in that subunit (11), and as such, AROS may be linked to further ribosomal proteins. To address this possibility, RNAi was again targeted against either AROS or RPS19, and lysates were immunoblotted for ribosomal proteins. A reduced abundance of RPS2, RPS6 and RPSA was observed following knockdown of either AROS or RPS19 in HCT116 cells (Figure 5A). This suggests that AROS is required to maintain the abundance of multiple 40S proteins, not just RPS19, an effect seen in two further cell lines—HEK293 and MCF7 cells (Supplementary Figure S4). Importantly, in each of the three cell lines, the expression of 60S proteins remained unchanged by knockdown of AROS or RPS19. This is consistent with a specific role for AROS in promoting 40S protein expression and also the 40S-specific role for AROS in ribosome biogenesis (Figure 1C).
Figure 5.
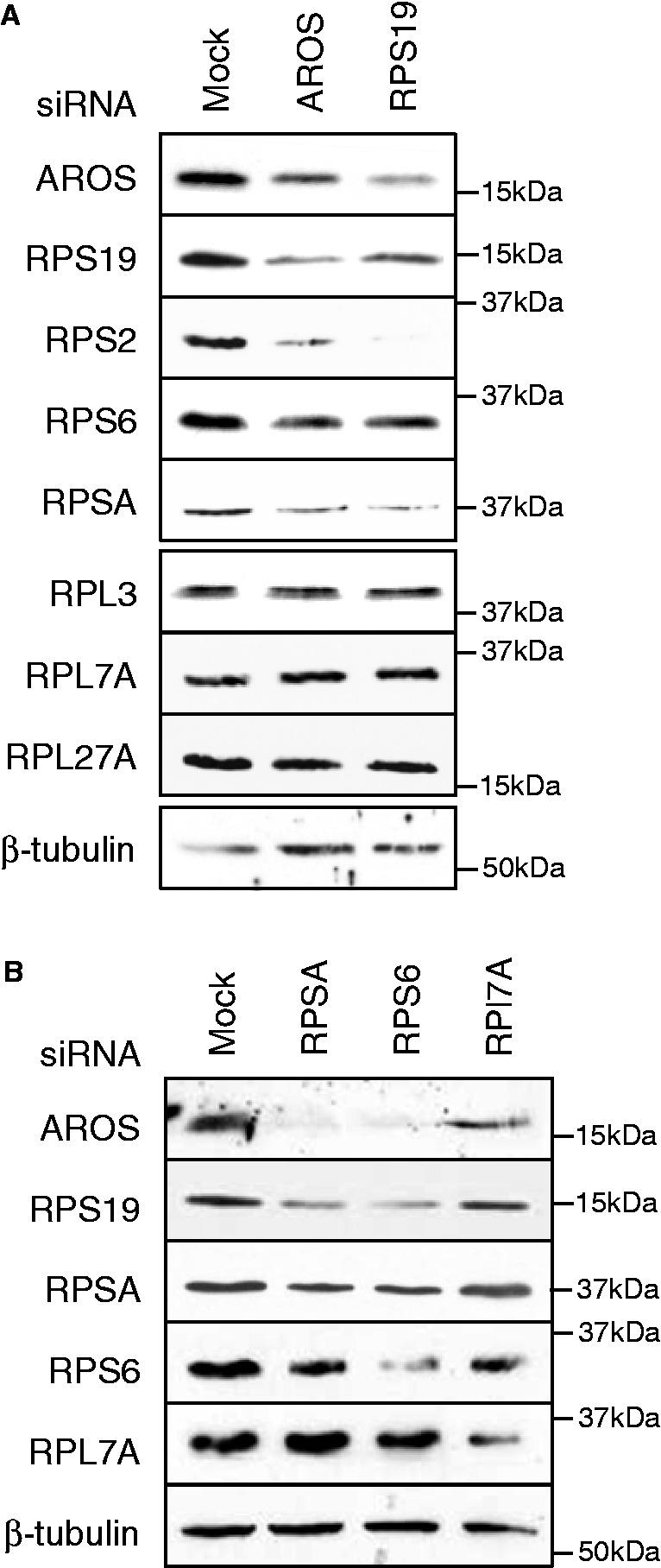
AROS is required for 40S subunit protein expression and vice versa. (A) Western blotting for ribosomal proteins following RNAi against AROS or RPS19 for 48 h in HCT116 cells. β-tubulin is used as a loading control. (B) Western blotting for AROS and ribosomal proteins following RNAi against small subunit (RPSA and RPS6) and large subunit (RPL7A) proteins in HCT116 cells. β-tubulin is used as a loading control.
Next, we analysed the reciprocal, to determine whether ribosomal proteins promote the abundance of AROS. RNAi was used to deplete HCT116 cells of ribosomal proteins from both the 40S and 60S subunits (Figure 5B). Depletion of RPSA or RPS6 from the 40S subunit greatly reduced the abundance of AROS (Figure 5B). In contrast, depletion of RPL7A, from the 60S subunit, resulted in only a slight reduction in the expression of AROS, which is perhaps comparable with the loss of RPS19 caused by RPL7A knockdown. This analysis was repeated in the HEK293 cell line, giving similar results (Supplementary Figure S4A). Taken together, these RNAi experiments indicate that the protein abundance of AROS is linked reciprocally with the abundance of 40S proteins, also supporting an association between AROS and 40S subunits.
Depletion of AROS reduces global protein synthesis in part via eIF2α
The specific reduction in 40S protein and rRNA following AROS silencing suggests that AROS is important for entire 40S subunit abundance. To test this hypothesis, sucrose density gradients were used to visualize ribosomal component abundance in cells depleted of AROS compared with control-treated cells (Figure 6A). There was a modest, but reproducible, 20% reduction in free 40S subunits following AROS silencing, with no effect on 60S subunit levels. Furthermore, the abundance of translating polysomes was also reduced by AROS depletion compared with control cells (Figure 6A). This is similar to (but not as dramatic as) previously published analyses of the effect of silencing RPS19 on ribosomal subunit abundances (13–15) and is consistent with the loss of 40S component rRNA and proteins (Figures 1C and 5A).
Figure 6.
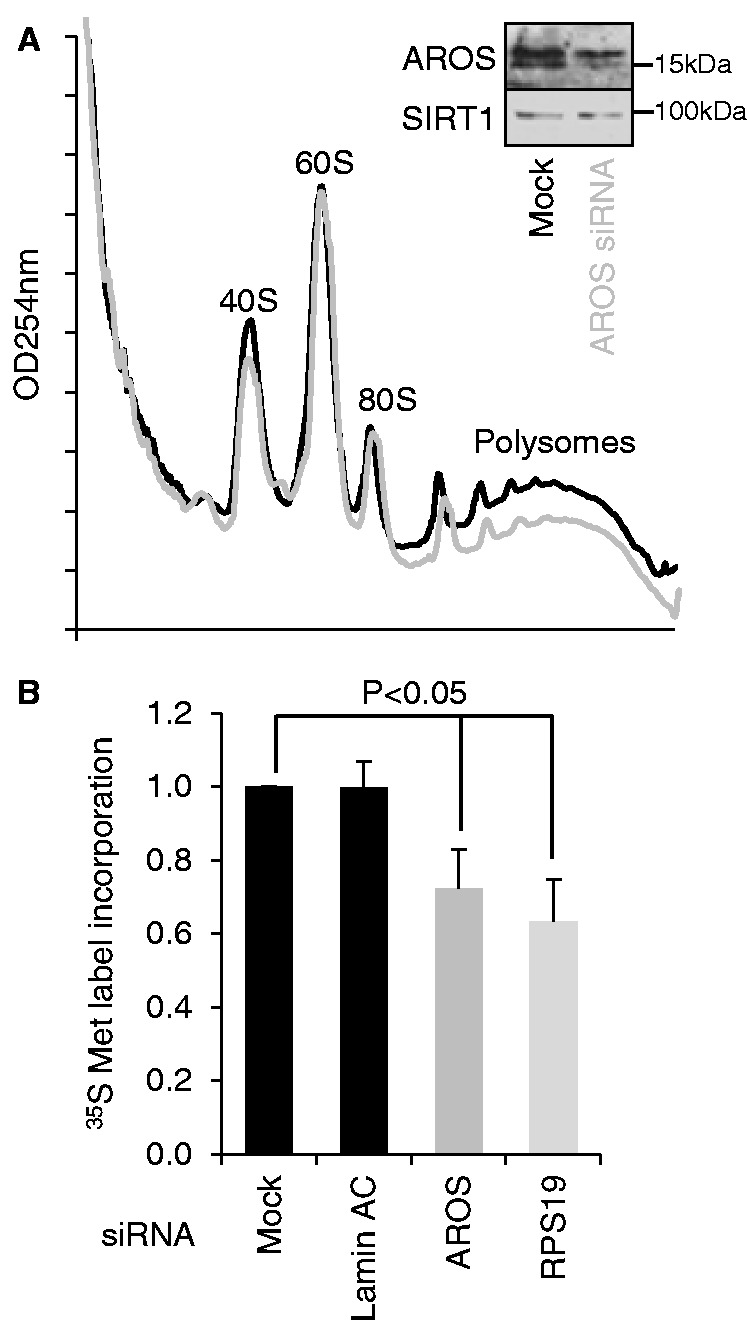
AROS maintains the maximal rate of global protein synthesis. (A) Sucrose density gradient (10–50%) separation of MCF7 cytoplasmic lysates treated with siRNA against AROS (red) compared with mock treatment (black). Traces represent optical density at 254 nm of gradients from lysates seeded with equivalent cells before RNAi. Insert shows depletion of AROS by RNAi. (B) Nascent protein synthesis in MCF7 cells during a 30-min incubation with 35 S-Met label. Protein synthesis analysis was carried out 48 h after RNAi and standardized against total protein content.
The reduction in polysome abundance following AROS depletion suggests that AROS knockdown reduces global protein synthesis. Thus, we asked whether depletion of AROS reduced the incorporation of 35S-methionine into protein for a 30-min period (Figure 6B). Targeting AROS by siRNA decreased the incorporation of radiolabelled methionine into protein by 24% compared with control treatments (Figure 6B). A similar reduction in global protein synthesis rate was observed following depletion of RPS19 (Figure 6B), which is not unexpected, given the requirement for RPS19 for 40S subunit biogenesis (Figure 1C). Together with the loss of polysomes on AROS knockdown (Figure 6A), the data support a positive role for AROS in regulating global protein synthesis, which parallels the role of RPS19.
Global protein synthesis rates are dependent on 40S subunit abundance, suggesting that the positive role of AROS in 40S subunit biogenesis contributes to cellular global protein synthesis. However, protein synthesis rates are also controlled by the bioavailability and phosphorylation status of the canonical initiation factors. Recently, SIRT1, which is activated by association with AROS, was reported to interact with and suppress the phosphorylation of the initiation factor eIF2α (29). EIF2α is a part of the ternary complex, which comprises the initiator tRNA loaded with methionine (tRNAiMet) and the eIF2 proteins bound to GTP (eIF2:GTP). This complex recruits tRNAiMet to the 40S subunit before joining with the 60S subunit and thus promotes translation initiation. Regulation of the ternary complex comes from modifying the phosphorylation of eIF2α (1,2,30). Phosphorylation at serine 51 promotes inhibitory association between inactive eIF2:GDP and its guanine exchange factor eIF2B, thereby reducing recycling to active GTP-bound ternary complex.
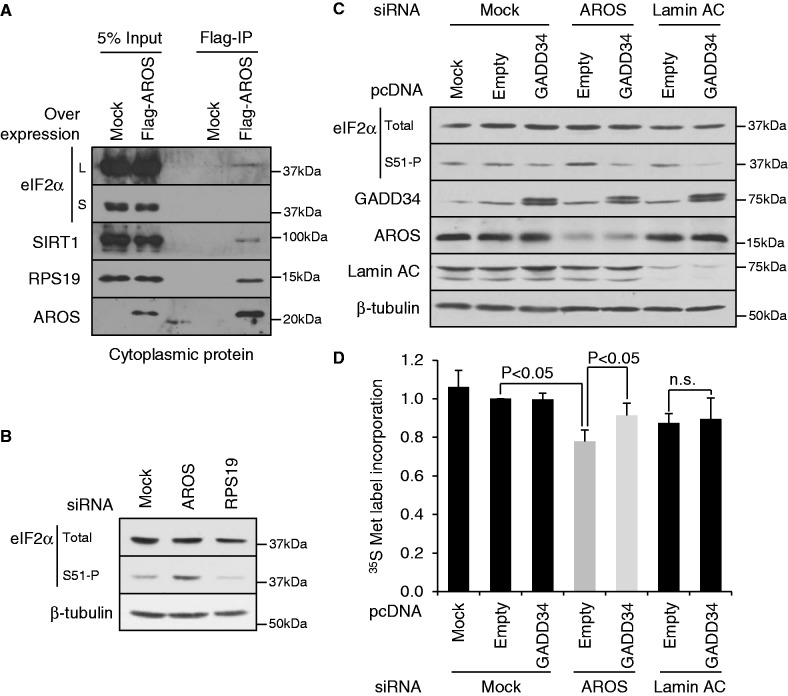
To determine whether AROS regulates protein synthesis beyond 40S subunit provision, we first asked whether, like SIRT1, AROS associates with eIF2α. Exogenous Flag-AROS was expressed in MCF7 cells and immunoprecipitated from cytoplasmic extracts to remove the influence of nuclear AROS from the analysis. A small proportion of cytoplasmic eIF2α specifically co-immunoprecipitated with Flag-AROS (Figure 7A—upper blot). The Flag-AROS:eIF2α association is not dissimilar to the established interaction between Flag-AROS and SIRT1 (18), which is only fractionally more abundant. This presents the possibility that AROS participates in regulation of eIF2α by SIRT1. Next, we asked whether AROS regulates the phosphorylation of eIF2α. Interestingly, we observe an increase in the phosphorylation of eIF2α on depletion of AROS (Figure 7B). The parallel silencing of RPS19 did not result in the same increase in eIF2α phosphorylation, suggesting that despite the close link between AROS and RPS19 in terms of ribosome biogenesis (Figure 4), there is a difference in their regulation of eIF2α, which appears to be selectively linked with AROS and not RPS19.
Figure 7.
AROS suppresses eIF2α phosphorylation. (A) Immunoprecipitation of Flag-AROS from cytoplasmic extracts of MCF7 cells. Equivalent protein was analysed by mass and detected using specific antibodies. L = long; S = short exposure. (B) Western blotting for abundance of total and phosphorylated eIF2α in HCT116 whole cell lysates following RNAi against AROS or RPS19. β-tubulin is used as a loading control. (C) HCT116 cells were depleted of AROS or control target Lamin AC by RNAi (Day 1), then transfected with cDNA to over-express GADD34, promoting eIF2α dephosphorylation, or control empty vector (Day 2). On day 3, whole cell lysates were collected and analysed by western blot after SDS–PAGE of protein. β-tubulin is used as a loading control. (D) Quantification of 35S-Met label incorporation into nascent protein during a 30-min incubation following RNAi and cDNA over-expression treatments as in (C). Label incorporation was measured by liquid scintillation and standardized to total cellular protein content. These data are expressed relative to mock siRNA plus empty vector treated cells. n.s. indicates not significant.
Little change was observed in the total protein or the phosphorylation status of eIF4E or 4EBP1 following knockdown of AROS (Supplementary Figure S5). The eIF4E is a constituent of, and 4E-BP1 a regulator of the eIF4F complex, which also controls global protein synthesis in response to alterations in phosphorylation status (1,2). A reduction in both total and phosphorylated 4E-BP1 was seen following RPS19 silencing. However, this seems unlikely to contribute to the reduction in global protein synthesis, as reduced 4E-BP1 should release eIF4E, allowing eIF4F formation and initiation of translation (31). Thus, it appears that regulation of eIF4F does not contribute to the decrease in protein synthesis rates following AROS knockdown.
The phosphorylation of eIF2α is suppressed by GADD34, which activates the phosphatase-dependent dephosphorylation of eIF2α (32). We used over-expression of GADD34 as a means to reduce eIF2α phosphorylation following AROS knockdown and monitored the subsequent effect on the rate of protein synthesis. The induction of eIF2α phosphorylation following AROS knockdown was repressed below control levels by over-expression of GADD34 (Figure 7C). In parallel RNAi treatments against Lamin AC or no target (Mock), the over-expression of GADD34 also reduced eIF2α phosphorylation compared with empty vector over-expression, illustrating the potent role of GADD34 in eIF2α dephosphorylation. Importantly, the reduced rate of global protein synthesis following AROS knockdown was significantly attenuated by over-expression of GADD34 (Figure 7D), increasing 35S-methionine incorporation from 78% of control treatment to 91%. Therefore, the increased phosphorylation of eIF2α following AROS knockdown is likely to contribute, at least in part, to the reduction in global protein synthesis rates.
DISCUSSION
In this study, we identify AROS as a ribosome biogenesis factor required for efficient cleavage of 21S pre-rRNA into 18SE pre-rRNA (Figure 1C). RPS19 is required for pre-rRNA processing at the same step [Figure 1C and (13–15)], and despite similar requirements for other 40S proteins (11), it seems likely that AROS promotes ribosome biogenesis via RPS19 (Figure 8). This is based on the direct interaction between the proteins (16) and the reciprocal dependence for protein stability discovered in this study (Figure 4). SIRT1 has been implicated in the regulation of ribosome biogenesis by epigenetically suppressing rRNA synthesis at the level of transcription. However, this results in suppression of biogenesis of both subunits (33).
Figure 8.
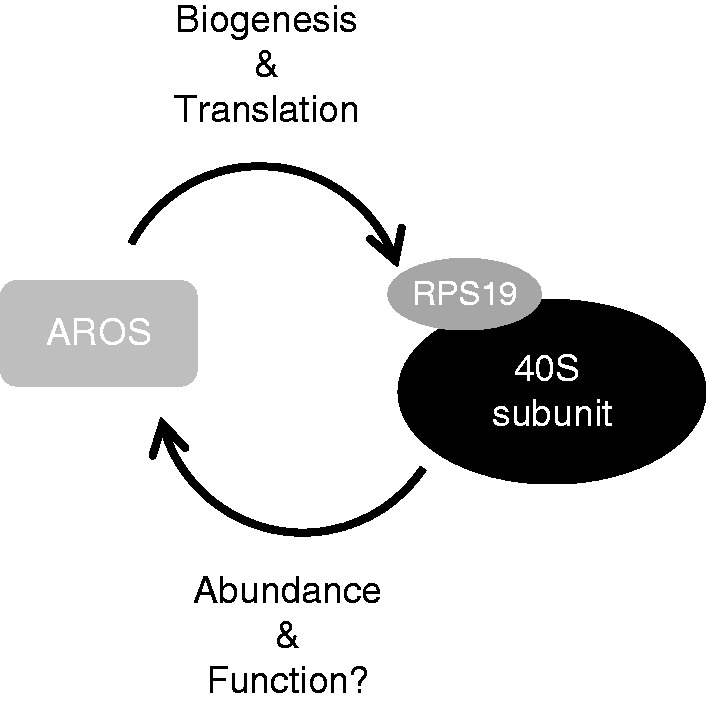
The relationship between AROS and the 40S ribosomal subunit. AROS protein abundance is closely related to that of the 40S subunit. AROS is required for the processing of pre-rRNA into the small subunit 18S rRNA and is linked to the protein levels of small ribosomal proteins. Furthermore, AROS appears to be required for translation, via the provision of 40S subunits and suppression of eIF2α phosphorylation. In reciprocal AROS, protein abundance is responsive to 40S protein levels, creating the possibility that the ribosome impacts on other functions of AROS. The schematic represents the model in our system of mammalian cell culture.
We show here that AROS associates with free 40S ribosomal subunits and translating polysomes (Figure 3). It is likely that AROS associates with the 40S subunit via RPS19, and in this context, it is relevant to note that RPS19 is a surface-accessible protein in the head region of the eukaryotic 40S subunit (24,25). We also suggest that AROS sedimentation in the polysome fractions is due to association with 40S subunits within polysomes, again via RPS19. It is difficult to test the assumed dependence on RPS19 for AROS association by biochemical means, as RPS19 cannot be removed from 40S subunits without affecting their biogenesis and abundance [Figure 1 and (13–15)]. However, we believe that the data presented here, namely (i) the sedimentation of AROS on sucrose gradient ultracentrifugation and (ii) AROS:ribosomal protein association by immunoprecipitation, are strong evidence for AROS association with the 40S subunit (Figure 3). It is possible that the association of AROS with polysomes is a result of AROS associating with 40S subunits initiating on mRNA already being translated downstream—i.e. independent of 80S formation. However, the association by immunoprecipitation with a 60S subunit protein suggests that AROS interacts with the 60S subunit within the 80S ribosome (Figure 3B).
The data prompt the question as to the extent of these AROS associations. Is AROS associated with all 40S subunits or just a sub-fraction? Conversely, is all cytoplasmic AROS associated with 40S subunits? AROS appears to be an abundant stable protein in the cytoplasm by western blotting and immunofluorescence (Figures 1B, 2C and 3D), and AROS could not be detected in lower density fractions of the gradient, where non-ribosome associated proteins would be expected (Figure 3A and Supplementary Figure S3A). This suggests that the cytoplasmic fraction of AROS constitutively associates with 40S subunits under the conditions analysed, but does not indicate whether all 40S subunits associate with an AROS protein. Despite similarities to ribosomal proteins, it seems unlikely that AROS is itself a ribosomal protein, given that it escaped characterization in the 1970s (34). As such, AROS can be termed a ribosome-associated protein, supplementary to its role as a ribosome biogenesis factor. This leads to a question regarding the dual functions of AROS. Why does a ribosome biogenesis factor remain bound to 40S subunits, and indeed polysomes, beyond their synthesis? The majority of ribosome biogenesis factors dissociate from the ribosome before it begins translation presumably to participate again in biogenesis (9). We see by the association of AROS with translating polysomes that these subunits are competent; yet, AROS remains associated.
The concept of linking ribosome biogenesis and translation has been discussed at length in the literature, most often in terms of co-regulation of participants in each pathway by upstream factors, such as the growth regulators c-myc and mTOR (35–37). However, these studies commonly focus on regulation of rRNA transcription, ribosomal protein and eIF synthesis. Recently, more direct links have been found that are not reliant on downstream transcriptional regulation. Most notably, eIF5B, as well as its role in translation initiation, has a crucial role during a cytoplasmic proofreading step in 40S biogenesis (38,39). Perhaps linked to this proofreading, where 40S and 60S subunits join in the absence of mRNA, two of the canonical translation termination factors also promote ribosome biogenesis. ABCE1 and Dom34 (Pelo in humans) are both required for 40S biogenesis via a mechanism, which is likely to relate to disassembly of 80S translation-like ribosomes (39–42), although it has also been documented that ABCE1 functions in pre-rRNA processing (40). Biogenesis of the 60S subunit also requires the activity of proteins with roles in translation. EIF6 is required for pre-rRNA processing for the 60S subunit, a process that then appears to link to its role as an initiation factor (43). Furthermore, the protein SBDS, which is implicated in the ribosomopathy Shwachman–Diamond syndrome, is required for both 60S biogenesis and translation initiation via displacement of the inhibitory eIF6 (44). The data presented here suggest that AROS can be added to the list of proteins involved in both ribosome biogenesis and translation. This is based on the likelihood that AROS is serving a function whilst associated with cytoplasmic 40S subunits, although full elucidation goes beyond the work here.
Depletion of AROS resulted in a reduction in the rate of protein synthesis (Figure 6B), which could represent a reduction in translation across all mRNAs or alternatively downregulation of specific messages following knockdown of AROS. We also report that knockdown of AROS results in a functional increase in phosphorylation of eIF2α (Figure 7B). This may be the downstream result of stress following loss of AROS, especially considering the role for AROS in ribosome biogenesis (Figure 1C). However, the increased eIF2α phosphorylation could also relate to SIRT1, which is bound and activated by AROS and known to suppress eIF2α phosphorylation (18,29). It will be interesting to determine whether the modulation of eIF2α following AROS knockdown involves SIRT1. The AROS:RPS19 interaction only requires the N-terminal region of AROS (16), allowing the possibility that the C-terminal is free to mediate association with other parties, such as SIRT1. Interestingly, SIRT1 has been implicated in various disease states such as cancer and neurodegeneration (20,45,46), both of which have been linked to aberrations in translation (3,4,47).
RPS19 has been implicated in the suppression of inflammation, either via formation of a post-apoptotic covalent dimer, which acts as a suppressor of acute inflammation (48), or via inhibition of the pro-inflammatory cytokine MIF (macrophage migration inhibitory factor) (49,50). Furthermore, RPS19 may play a role in an infectious disease; direct interaction between RPS19 and the hantavirus nucleocapsid protein is believed to subvert translation towards viral transcripts (51). It would also be interesting to evaluate the role of AROS in these functions of RPS19, given that AROS appears to associate with the 40S subunit and could presumably stabilize RPS19 within the ribosome or occlude other RPS19 binding partners.
Mutations in ribosomal proteins and biogenesis factors have been linked to a group of diseases termed ribosomopathies (7,8). First among these was the erythroblastopenia DBA, which has been linked to mutation in at least 10 ribosomal protein genes contributing to both ribosomal subunits but most frequently to mutations in RPS19 (52). Interestingly, reduced expression of RPS19 protein and defective ribosome biogenesis has been directly linked to the aetiology of DBA (13–15). Although mutation in AROS has not been identified in genetic screens of DBA patients, given the strong dependence on AROS for RPS19 protein expression and the overlapping function in 40S biogenesis reported here (Figures 1C and 4A), it would be interesting to analyse AROS in the emerging cellular and animal models of DBA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Yorkshire Cancer Research (to J.M.); Medical Research Council and Biotechnology and Biological Sciences Research Council (to A.E.W.). Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Xavier Pichon and David Read for assistance with confocal microscopy and Amandine Bastide for assistance with 35S-Met labelling and sucrose density ultracentrifugation analyses. They also thank Simon Allison for helpful discussion and assistance with experimental procedures.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 3.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 4.Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat. Rev. Clin. Oncol. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- 5.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 6.Montanaro L, Treré D, Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim. Biophys. Acta. 2012;1825:101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Luft F. The rise of a ribosomopathy and increased cancer risk. J. Mol. Med. 2010;88:1–3. doi: 10.1007/s00109-009-0570-0. [DOI] [PubMed] [Google Scholar]
- 8.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol. BioSyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadjiolova KV, Nicoloso M, Mazan S, Hadjiolov AA, Bachellerie JP. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. . Biochem. 1993;212:211–215. doi: 10.1111/j.1432-1033.1993.tb17652.x. [DOI] [PubMed] [Google Scholar]
- 11.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 13.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cell Mol. Dis. 2007;39:35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Maeda N, Toku S, Kenmochi N, Tanaka T. A novel nucleolar protein interacts with ribosomal protein S19. Biochem. Biophys. Res. Commun. 2006;339:41–46. doi: 10.1016/j.bbrc.2005.10.184. [DOI] [PubMed] [Google Scholar]
- 17.Maeda N, Toku S, Naito Y, Nishiura H, Tanaka T, Yamamoto H. Phosphorylation of ribosomal protein S19 at Ser59 by CaM kinase I alpha. J. Neurochem. 2009;109:393–402. doi: 10.1111/j.1471-4159.2009.05971.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Garrey JL, Lee YY, Au HHT, Bushell M, Jan E. Host and viral translational mechanisms during cricket paralysis virus infection. J. Virol. 2010;84:1124–1138. doi: 10.1128/JVI.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 21.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen,Ba A, Pogoutse A, Provart N, Moses A. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal Structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 25.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40s ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 26.Huang MT. Harringtonine, an inhibitor of initiation of protein biosynthesis. Mol. Pharmacol. 1975;11:511–519. [PubMed] [Google Scholar]
- 27.Nikolov EN, Dineva BB, Dabeva MD, Nikolov TK. Turnover of ribosomal proteins in regenerating rat liver after partial hepatectomy. Int. J. Biochem. 1987;19:159–163. doi: 10.1016/0020-711x(87)90326-0. [DOI] [PubMed] [Google Scholar]
- 28.Miller BG. The biological half-lives of ribosomal and transfer RNA in the mouse uterus. J. Endocrinol. 1973;59:81–85. doi: 10.1677/joe.0.0590081. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh HS, Reizis B, Robbins PD. SIRT1 associates with eIF2-alpha and regulates the cellular stress response. Sci. Rep. 2011;1 doi: 10.1038/srep00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell PJ, Balkow K, Hunt T, Jackson RJ, Trachsel H. Phosphorylation of initiation factor eIF-2 and the control of reticulocyte protein synthesis. Cell. 1977;11:187–200. doi: 10.1016/0092-8674(77)90330-0. [DOI] [PubMed] [Google Scholar]
- 31.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 32.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-Mediated Dephosphorylation of eIF2α. J. Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 34.McConkey EH, Bielka H, Gordon J, Lastick SM, Lin A, Ogata K, Reboud JP, Traugh JA, Traut RR, Warner JR, et al. Proposed uniform nomenclature for mammalian ribosomal proteins. Mol. Gen. Genet. 1979;169:1–6. doi: 10.1007/BF00267538. [DOI] [PubMed] [Google Scholar]
- 35.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 36.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J. Cell. Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 38.Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strunk BS, Novak MN, Young CL, Karbstein K. A Translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol. Cell. Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 45.Donmez G. The neurobiology of sirtuins and their role in neurodegeneration. Trends Pharmacol Sci. 2012;33:494–501. doi: 10.1016/j.tips.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Knight JRP, Milner J. SIRT1, metabolism and cancer. Curr. Opin. Oncol. 2012;24:68–75. doi: 10.1097/CCO.0b013e32834d813b. [DOI] [PubMed] [Google Scholar]
- 47.Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, Willis AE. RNA binding protein/RNA element interactions and the control of translation. Curr. Protein Peptide Science. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto T. Roles of the ribosomal protein S19 dimer and the C5a receptor in pathophysiological functions of phagocytic leukocytes. Pathol. Int. 2007;57:1–11. doi: 10.1111/j.1440-1827.2007.02049.x. [DOI] [PubMed] [Google Scholar]
- 49.Lv J, Huang XR, Klug J, Fröhlich S, Lacher P, Xu A, Meinhardt A, Yao Lan H. Ribosomal protein S19 is a novel therapeutic agent in inflammatory kidney disease. Clin. Sci. 2013;124:627–637. doi: 10.1042/CS20120526. [DOI] [PubMed] [Google Scholar]
- 50.Filip AM, Klug J, Cayli S, Frohlich S, Henke T, Lacher P, Eickhoff R, Bulau P, Linder M, Carlsson-Skwirut C, et al. Ribosomal protein S19 interacts with macrophage migration inhibitory factor and attenuates its pro-inflammatory function. J. Biol. Chem. 2009;284:7977–7985. doi: 10.1074/jbc.M808620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng E, Haque A, Rimmer MA, Hussein IT, Sheema S, Little A, Mir MA. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19) J. Biol. Chem. 2011;286:11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horos R, von Lindern M. Molecular mechanisms of pathology and treatment in Diamond Blackfan Anaemia. Br. J. Haematol. 2012;159:514–527. doi: 10.1111/bjh.12058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.