Abstract
Increasing attention has focused on the significance of RNA in sperm, in light of its contribution to the birth and long-term health of a child, role in sperm function and diagnostic potential. As the composition of sperm RNA is in flux, assigning specific roles to individual RNAs presents a significant challenge. For the first time RNA-seq was used to characterize the population of coding and non-coding transcripts in human sperm. Examining RNA representation as a function of multiple methods of library preparation revealed unique features indicative of very specific and stage-dependent maturation and regulation of sperm RNA, illuminating their various transitional roles. Correlation of sperm transcript abundance with epigenetic marks suggested roles for these elements in the pre- and post-fertilization genome. Several classes of non-coding RNAs including lncRNAs, CARs, pri-miRNAs, novel elements and mRNAs have been identified which, based on factors including relative abundance, integrity in sperm, available knockout data of embryonic effect and presence or absence in the unfertilized human oocyte, are likely to be essential male factors critical to early post-fertilization development. The diverse and unique attributes of sperm transcripts that were revealed provides the first detailed analysis of the biology and anticipated clinical significance of spermatozoal RNAs.
INTRODUCTION
Sperm structure and function appear fine-tuned towards a single purpose—the delivery of the paternal content to the oocyte in the most compact and accurate form. Our view of the presence and role of RNAs in human sperm has dramatically evolved (1). As sperm are generally considered transcriptionally inert, RNAs detected in the paternal gamete were initially assumed to be either content left behind after degradation and expulsion of the residual body, or simply contaminants from other surrounding cells (2,3). However, sperm retain specific coding (4–8) and non-coding RNAs (9,10). Irregularities in the levels of sperm RNAs have been recognized as markers and potential effectors of human male infertility (11–13). Their functional role on delivery to the oocyte has been suggested (14–16).
Previous studies have used microarray approaches to identify transcripts retained in sperm (12,17,18). Although informative, these analyses were unable to provide the enhanced view of this suite of RNAs afforded by RNA-seq [reviewed in (19)]. Apart from being recognized as a more accurate assessment of transcript levels than microarrays (20,21), RNA-seq has the added advantage that novel tissue isoforms and variants can be identified and quantitatively evaluated. Further, previously unrecognized coding and non-coding transcripts can be discovered and potential function ascribed (22–25). The population of sperm RNAs as revealed by RNA-seq provides a window into the developmental history, functional viability and potential elements delivered by sperm that are likely of significance upon fertilization. Together with the previous sncRNA analysis (10), this study provides a comprehensive snapshot of the transcript composition of human sperm among several individuals. Comparison of the many genomic regions encoding sperm RNAs with elements of the sperm epigenome has provided insight into the function and temporal action of these regulatory features. Highlighted in the analysis below is the use of exon profiles to identify full-length transcripts amongst a larger population of fragmented RNAs. Global analysis of this unique suite of paternal transcripts echoes past transcriptional history and late-stage spermatogenic function, yet projects its role on fertilization.
MATERIALS AND METHODS
RNA isolation and sequencing
To characterize various classes and features of sperm RNA, multiple sample preparation methods were used. The first set of samples was prepared as described (10,26,27) using RNA from a total of six sperm samples from three random and three proven fertile donors and two testes samples. Briefly, four of these sperm samples (henceforth referred to as T-Sperm samples)—Sp_D62[Tt], Sp_D64[Tt], Sp_D66[Tt] (individual donors) and Sp_P1[Tt] (mixed pool of three random donors)—were not subject to any of the commonly used RNA-selection methods to fully resolve the RNA population and reduce transcript expression profile bias. With the exception of the Pooled sample Sp_P1[Tt], all samples were subject to 50% PureSperm (Nidacon, Mölndal, Sweden) gradient to purify spermatozoa away from other contaminating cell-types. RNA from donor D62 was additionally separated into Poly(A+) (Sp_D62[A+]) and Poly(A−) (Sp_D62[A−]) fractions by oligo(dT) selection, with each fraction sequenced separately. Sperm samples were all sequenced in two lanes each on Illumina GAIIx Genome Analyzer. Two samples of commercially obtained pooled testes RNA—Te_PAm[A+] (Applied Biosystems/Ambion, Austin, TX, USA, Lot 054P010702031A) and Te_PCl[A+] (ClonTech, Mountain View, CA, USA, Lot 3090051)—were each sequenced in a single lane after Poly(A+) selection.
Single Primer Isothermal Amplification (SPIA) was used to prepare a second set of RNA-seq libraries from 20 ng of RNA from each of the three individual donors (Sp_D62[SP], Sp_D64[SP], Sp_D66[SP]), a single pooled sperm sample (Sp_P2[SP]) and a single pooled testes (Te_PAm[SP]) sample using the Nugen Ovation kit (Nugen Inc., San Carlos, CA, USA) for cDNA and initial linear amplification. The Nugen Encore system was used for adaptor ligation, end-repair and amplification. Samples were multiplexed at four samples per lane.
All samples were subjected to 2 × 36 cycles of paired-end sequencing on the Illumina GAIIx platform. Paired-end mapping was performed equivalently for all samples using Novoalign (V2.07.09, Novocraft Technologies, Selangor, Malaysia) with alignment to Human Genome build hg19, and also including ribosomal 18S and 28S and mitochondrial 12S and 16S indices for specific alignment of reads corresponding to these sequences. Reads mapping to multiple genomic locations were examined separately. Sequencing data are available at the NCBI GEO repository.
An SPIA-prepared RNA-seq Universal Human Reference (UHR; Nugen Ovation library, Nugen Inc.) sample composed of RNAs from pooled somatic tissues was used for comparison of the parallel SPIA-prepared sperm and testes RNAs with a wide representation of somatic RNAs. Data from this sequencing are publically viewable by including the text ‘track type = bigWig name = “rep123.uhr.7102” description = “rep123.uhr.7102” color = 5 20 198 autoScale = on visibility = 2 viewLimits = 1:500 bigDataUrl=http://ucscdatahosting.com/labuser/Tech_Support/rep123.uhr.7102.bw’ in a user track window of the UCSC genome browser at http://genome.ucsc.edu/cgi-bin/hgCustom?hgsid=322010991:
RT-PCR-transcript validation
Remaining total RNA from Sp_D62[Tt], Sp_D64[Tt] and Sp_D66 were reverse-transcribed using SuperScriptIII (Invitrogen, Carlsbad, CA, USA) with either oligo(dT) (Sp_D62 and Sp_D64 [Tt]) or gene-specific primers (Sp_D66 [Tt]) then amplified with HotStarTaq (Qiagen, Valencia, CA, USA), as described (28). PCR primer pairs were designed to query the entire transcript length (Supplementary Table S1). Products were visualized by agarose gel electrophoresis and images captured on the Typhoon 9210 scanner (Molecular Dynamics).
Bioinformatic analysis
Total transcript read counts from each sample were measured for 37 973 different isoforms (RefSeq annotation, HG19) from a total of 22 302 genes. Sperm RNAs contain a host of transcripts exhibiting multiple polyadenylation sites, intronic elements, or reflect differential processing (29). Accordingly to minimize this effect and to quantify transcript levels, a custom algorithm was developed to count exonic reads from all sequenced RNA samples across protein-coding regions only for all annotated transcript isoforms (See Supplementary Methods). The RPKM (Reads per Kilobase per Million reads) values were then calculated for both Total RNA and SPIA samples as summarized in Supplementary Tables S2 and S3. The aggregate of four T-sperm and SPIA-sperm samples was obtained by pooling all samples and renormalizing RPKM values based on total counts. The most abundant isoform of each gene (RPKM) was then used to rank each gene in each individual sample, allowing for intersample comparison by Spearman rank correlation. Transcript levels in sperm and testes were additionally assessed using the Genomatix RegionMiner (v2.41207), as this data source also includes a series of non-coding transcripts (e.g., RNU) in its assessment of transcript abundance. Ontological analysis of selected transcript groups was performed using GeneRanker (Genomatix v2.41103). Post-fertilization significance of sperm-delivered RNAs was examined using expression data from microarray analysis of human oocyte at MII, 2-, 4-, 6-, 10-cell, morula, blastocyte and stem-cell stage (30). Relative differential expression for each transcript across these developmental stages was assessed by calculating the change in reported expression (log 2) at each stage relative to MII baseline. Predicted targets of notable sperm-delivered miRNAs were calculated using Diana-microT algorithm (v3.0; 31,32) as described (10). High confidence targets (>0.95%) were selected for downstream analysis based on strict predicted binding score (miTG >25).
An intactness score was developed to assess the stability of total sperm RNAs with a length of at least 200 nt. To portray the relative level of coverage over transcript length, RNA-seq expression profiles for selected transcripts were obtained by computationally splicing read coverage corresponding to the transcript body (Supplementary Figure S1). Analysis was based on the annotated exon map of the most abundant isoform for each sperm transcript. Bias due to lack of read coverage at exon ends was minimized by excluding the last 15 nt of each exon. Normalized coverage profiles were calculated across transcripts in 100 bins. The sum of squares of actual deviation from the expected coverage of an ideal transcript (1% of total coverage/bin) was used as the intactness score. Transcripts were additionally and separately categorized and ordered based on positioning of underrepresented or absent regions as a function of the relative degree of 5′ versus 3′ coverage. A linear scaling function from the 5′-end maximum positive value to the maximum 3′-end negative value was applied to the normalized profiles, with subsequent ranked degree of variation from a level profile now indicative of the degree and site of end degradation.
To examine the association of RNA abundance in sperm with local epigenetic marks, 19 521 expression-enhanced promoter and exonic cluster regions from sperm sample Sp_D62[Tt] were identified using Genomatix RegionMiner tool (Audic-Claverie algorithm with P < 0.05, window size of 100 bp). External data sets were used for correlation analysis of H3K4me3 (34 912 regions), H3K27me3 (38 337 regions) and histone-enriched (25 114 regions; 33) and hypomethylated regions (79 124 regions; 34). Identified regions as reported by authors in hg18 genome build coordinates were converted to hg19 coordinates using the UCSC Genome Browser liftover tool. Minimum distance of center point of RNA clusters to closest local element of each histone region or hypomethylated DNA region set was calculated using GenomeInspector (Genomatix), yielding a correlation graph showing the distribution of clusters over a ±10 kb region relative to the center of each element.
RESULTS AND DISCUSSION
Two separate protocols were used to construct sequencing libraries from Total and poly(A) fractionated sperm and testes RNAs. Sequencing characteristics for all sample libraries are summarized in Supplementary Table S4. Reads not aligned (total reads − aligned reads) are primarily those rejected due to a low Quality Control score. Reads that could not be assigned to a unique genomic location despite paired-end mapping were considered separately in subsequent analyses (below). Although it is assumed that sequencing libraries prepared from Total RNA can more accurately portray the complete RNA profile of a cell, alternative methods of preparation are important, as they significantly reduce the contribution of the abundant ribosomal and mitochondrial and fragmented RNAs. In this regard, it has been shown that 28S and 18S ribosomal RNAs are essentially fragmented in the mature gamete ensuring translational silencing (26,35). This precludes their depletion by available technologies and hinders efficiently achieving adequate sequencing depth. Only those sperm RNAs of subsequent biological significance are expected to escape fragmentation. Acknowledging how sequencing library construction impacts transcript representation was essential in the characterization of the unique population of RNAs retained in sperm.
Library characteristics
Both RNA transcript abundance and read profile were to some degree dependent on the protocol used for sequencing library construction. Average fragment length of the SPIA-prepared sequencing libraries was ∼140 bp, while T-libraries (Total RNA) averaged between 70–90 bp. This increased the representation of transcripts of <100 nt in this set, enabling the resolution of sno and snaR RNAs. As expected, the poly(A+) sperm and testes samples showed a significant decrease in the relative abundance of both mitochondrial and ribosomal RNAs compared with the total RNA libraries. Although SPIA did not use a poly(A+) selection, the significant depletion of rRNA achieved by this method results from the use of a heterogeneous mixture of primers incapable of annealing to the highly structured ribosomal transcripts. The SPIA library preparation protocol provided an additional advantage over other strategies, as it required substantially less template RNA. This is of particular concern when investigating sperm RNAs (36). Briefly, total RNA was reverse transcribed using a proprietary mixture of chimeric RNA/DNA primers. Use of these hybrid oligonucleotides allows for continual priming and polymerization following second strand synthesis (37).
To assess library purity, a group of 523 transcripts were selected that were abundant in SPIA-testes (RPKM > 25, approximately 90th percentile), yet were relatively low or absent in the SPIA-sperm samples [RPKM (Aggregate) < 1; Supplementary Table S5]. RNAs that were depleted from mature sperm include those associated with Sertoli cells (22 genes, P = 1.2 e-04) and epididymis (22 genes, P = 4.1 e-06), confirming the absence of contaminating RNAs from surrounding cells. Other ontological groups of note include the absence of the large ribosomal subunit (11 genes, P = 1.7 e-06), and ribosome (16 genes, P = 1.9 e-04). This is consistent with the view that ribosomal fragmentation occurs earlier during the maturation phase of spermiogenesis, ensuring cessation of translation (26).
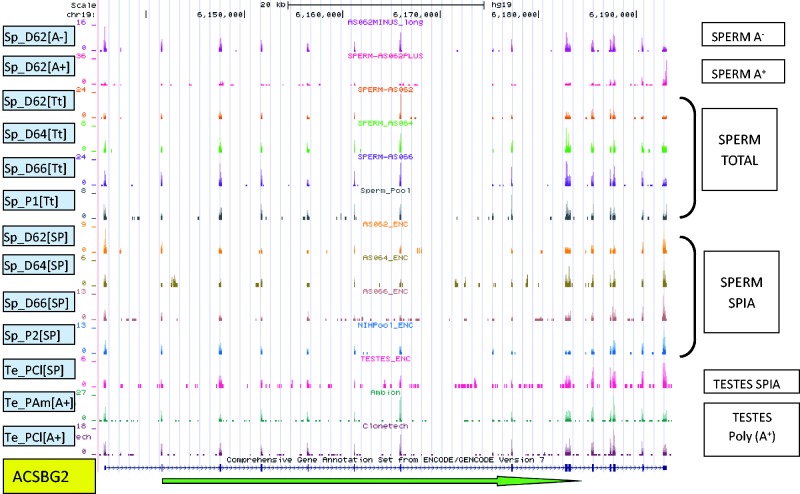
As exhibited by ACSBG2 and shown in Figure 1, the majority of T- and SPIA-library prepared transcripts displayed similar sequence-read profiles. However, clear differences were apparent for several transcripts, including PRM2 (Supplementary Figure S2), PRM1 and several non-coding RNAs. Analyses presented below for sperm transcript profiling used the T-libraries. Comparisons between sperm and other tissues used equivalently prepared SPIA-libraries.
Figure 1.
The distribution of sequencing reads across ACSBG2 in all libraries. The green arrow indicates transcript orientation. The number of reads corresponding to each read start at each base position is represented on the vertical axis. The scale for each sample is based on maximum read count within the displayed region. Four Total Sperm samples—Sp_D62[Tt], Sp_D64[Tt], Sp_D66[Tt] (individual donors) and Sp_P1[Tt] (mixed pool of three random donors)—were not subject to any of the commonly used RNA-selection methods. Sample D62 was additionally separated into Poly(A+) (Sp_D62[A+]) and Poly(A-) (Sp_D62[A−]) fractions by oligo(dT) selection. Commercially obtained pooled testes RNA—Te_PAm[A+] (Applied Biosystems/Ambion, Austin, TX, USA, Lot 054P010702031A) and Te_PCl[A+] (ClonTech, Mountain View, CA, USA, Lot 3090051)—were subject to Poly(A+) selection. Sequences from Single Primer Isothermal Amplification (SPIA—Nugen Ovation Nugen Inc., San Carlos, CA, USA) (Sp_D62[SP], Sp_D64[SP], Sp_D66[SP]), a single pooled sperm sample (Sp_P2[SP]) and a single pooled testes (Te_PAm[SP]) prepared libraries are compared.
Comparison of rank transcript abundance between samples as measured by RNA-seq yielded an average Spearman rank correlation between the four SPIA-sperm samples of ρ = 0.83 (0.76–0.89). The correlation between individual SPIA-sperm samples to SPIA-testes was ρ = 0.57 (0.54–0.61), while the correlation of SPIA-sperm with a somatic UHR sample was ρ = 0.38 (0.35–0.40) and SPIA-testes with UHR was ρ = 0.67. These values reflect both the unique composition of sperm transcripts and their stability compared with somatic cells. Although the correlation between sperm sample transcript rank was acceptable, the absolute measure of abundance of many transcripts varied between sperm samples. This may reflect the wide-ranging degrees of fragmentation and/or the undirected expulsion of many RNAs during maturation.
Multiple-mapped reads
One issue uniquely encountered in RNA-seq as compared with microarray analysis is the treatment of reads mapping to multiple genomic locations (38,39). Many RNA-seq studies simply ignore these reads, assuming that within a coding sequence, ‘repetitive’ elements are of marginal significance. However, there are many genes for which identical sequences of the complete or partial transcript are annotated in multiple genomic locations. Sequence reads derived from these transcripts would map equivilently to all these locations. For example, transcripts like, SPANXB1/B2/F1 appear replicated across multiple locations. In comparison, others exhibit limited reiteration of sequence spans, e.g., the 3′ exon of STK19, across multiple locations. Accordingly, if multiple alignment reads were discarded, transcript levels would be severely underestimated as highlighted by the sperm-associated SPANX transcript (40). Accordingly, all multiple-mapped reads were separately retained for transcripts exceeding RPKM > 2, and data are summarized in Supplementary Table S6; sample Sp_D62[Tt].
In comparison with the above, transcript levels determined by probe hybridization are not affected by multiple genomic origins of individual RNAs, as the level of each is estimated as an aggregate for that probe. For example, when comparing the recent microarray-based survey of human round spermatid RNAs (41) several of these transcripts are virtually absent from the RNA-Seq-processed data sets when multiple-mapped reads are ignored (Supplementary Tables S2, S3 and S6). This apparent discordance is resolved when multiple reads are correctly assigned, e.g., SPANX genes, histone HIST2HAA3/4, CXorf51, RIMBP3/3B/3C and HSFY1/2. This independent validation suggests that other multiple-mapped transcripts, e.g., snaRs, require consideration.
Alternative polyadenylation and isoforms
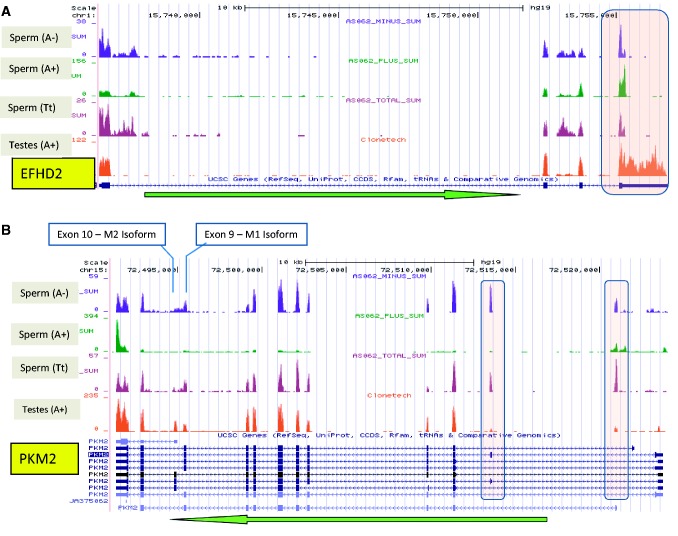
Alternative polyadenylation (APA) is a transcriptional modification whereby the 3′UTR is cleaved at a secondary poly(A) recognition site, giving rise to a transcript possessing an unaltered coding sequence and a significantly shortened cell-specific 3′ UTR (42–45). EFHD2 RNAs in the male reproductive tract offer an example of APA. This transcript is represented in all sperm RNA-seq data sets, and its coding sequence shown to be intact by RT-PCR (Supplementary Figure S3). But unlike that observed in testes, the UTR of this RNA is truncated in sperm relative to annotated transcripts (Figure 2). Approximately 20% of sperm transcripts exhibit clear APA sites relative to the full-length 3′ UTR of the primary annotated transcripts. Interestingly, more than one-half are sperm-specific and ontologically nondescript (Supplementary Table S7). This modification has been shown to influence mRNA stability and would impact relative abundance (46,47).
Figure 2.
Novel sperm transcript expression profiles: The green arrow indicates transcript orientation. The number of reads corresponding to each base position is represented on the vertical axis. The scale for each sample is based on maximum read count within the displayed region. (A) The complete annotated 3′ UTR of EFHD2 is 1.6 kb in length (pink box). In contrast to testes the 3′ UTR of this RNA in sperm is truncated to ∼100 nt due to alternative polyadenylation. (B) An isoform of PKM2 specific to sperm contains exons (pink ovals) not observed together in other tissues. Examination of paired reads from these regions indicates splicing between these neighbouring exons and that these regions are transcribed at levels comparable with the remainder of the transcript. Previously reported isoforms containing either the M1 or M2 exon are presented below.
As determined by comparison of the T-libraries, several novel isoforms appear to be unique to sperm. These include the multi-functional pyruvate kinase PKM2 that is closely associated with the regulation of cellular energy pathways (48). Of note, conservation of this sperm transcript extends to Arabidopsis thaliana sperm homologue AT4G26390 (49) suggesting significance. In human sperm, this mRNA possesses two additional exons, which are detected at levels comparable with the remainder of the transcript (Figure 2B; boxes). Interestingly, each exon has been observed in other isoforms in testis and amygdala. Individual mapping of paired-end reads indicates that both exons are present in novel sperm-specific isoforms not observed in testes, the UHR RNAs or in unfertilized human oocytes (50). The sperm PKM2 isoforms also exhibit mutually exclusive use of exons 9 or 10. The exon 9 isoform is linked with aerobic respiration oxidative phosphorylation and plays a role in ATP production in sperm flagella (51). The M2 exon 10 isoform is a member of the aerobic glycolytic pathway often observed in growing cells (52). Delivery of these unique sperm transcripts may provide a paternal regulator which silences OCT4 maintaining the pluripotent state (53) as required post-fertilization.
Sperm/testes-associated transcripts
Transcripts that are abundant in both testes and sperm (RPKM > 25, SPIA-samples), but are absent or at very low levels in the UHR sample (RPKM < 1) were also apparent. This set of 102 transcripts (Supplementary Table S8) comprises those that are present throughout spermatogenesis and retained through the final stages of maturation. This group was independently validated in a recent testes microarray data set (41). Within this set of 102 RNA-seq sperm transcripts, 90 transcripts were interrogated by microarray probes, of which 42 spermatid transcripts were concordant. These RNAs are likely critical to gametogenesis, e.g., structural components—flagellum (six genes, P = 4.2 e-09), acrosomal vesicle (five genes, P = 1.1 e-06) and nucleosome (three genes, P = 2.2 e-03). Of particular significance are the large number of genes (22 genes, P = 1.6 e-11) in this group that are associated with infertility. Mouse knockout data from MGI (54) shows that 13 are associated with a male factor contribution to infertility (Supplementary Table S8). This includes PCYT2, which is only found in testes and sperm. It is the primary regulator of the CDP–ethanolamine pathway, which when deleted, confers complete embryonic lethality (55). One can pose that like PCYT2 those that await characterization may similarly contribute other essential male factors.
Abundant sperm transcripts
Of the 22 302 unique transcripts surveyed, 726 displayed RPKM levels greater than 50 in the aggregate of T-sperm libraries and were thus considered as representative of the highly abundant sperm transcript class (Supplementary Table S9). Of these transcripts, 565 were identified by ontological analysis as linked to testes (P = 6.5 e-48), 54 were spermatid related (P = 3.2 e-19), 44 were associated with spermatogenesis (P = 3.0 e-15) and 76 were linked to infertility (P = 7.8 e-09). This is consistent with these RNAs providing a critical male function and suggests that they may provide a resource of diagnostic markers of fitness (56). Enrichment of genes associated with sperm motility (six genes, P = 4.6 e-05), acrosomal vesicle (eight genes, P = 1.4 e-04), components of the RAN pathway (four genes, P = 4.4 e-04) associated with regulation of centrosomal activity during mitotic division of spermatogonia (57,58), and structural components of sperm flagella such as cytoskeleton (84 genes, P = 1.3 e-06) and microtubule (29 genes, P = 1.9 e-07), clearly reflect transcripts coding for critical functions. On one hand, it is likely that some abundant sperm transcripts including the protamine genes that are required earlier in spermatogenesis and RNAs associated with nuclear organization (10 genes, P = 5.1 e-08) and the chromosome (33 genes, P = 6.3e-04) are no longer required in fully differentiated sperm and are simply residual. On the other hand, some abundant sperm transcripts may also function post-fertilization. For example, RANBP2, one of the intact sperm RNAs, is known to play an important role in nucleocytoplasmic transport and mitosis, reduction of which directly effects aneuploidy (59). This raises the intriguing possibility that RANBP2 may be an example of a sperm-delivered transcript encoding a factor specifically involved in restructuring the paternal chromatin following fertilization. Consistent with a post-fertilization function, its knockout confers embryonic lethality (60,61).
Sperm-enhanced transcripts
Sperm transcripts appear to encompass a wide spectrum of RNAs that encode proteins spanning past, current and future roles. Comparison of the sperm and testes RNAs (SPIA-libraries) permits delineation of the transcripts and transcript groups that appear significant in sperm by their conspicuous abundance relative to testes. As transcription is believed to halt in the latter stages of spermiogenesis, the majority of RNAs which can be classified as present in sperm also appear in testes. Of the 2200 RNAs in the top decile of sperm-abundant transcripts, >85% are also present in testes at levels that are at least 50% of that observed in sperm. Most notable, however, are a number of sperm RNAs that appear to increase markedly in the latter stages of spermatogenesis, based on comparison of relative abundance of relatively intact transcripts in parallel sperm and testes libraries. This group likely represents a final burst of transcriptional activity during spermatid differentiation responsible for producing those RNAs with functions specific to the final stages of spermiogenesis or function subsequent to delivery. For example, among this group are keratins KRT33A, KRT5 and JUP, which are necessary structural components of sperm cytoskeleton and microtubules (62,63). Another RNA in this class is SIX3, a member of the SIX family of transcription factors, which includes the newly identified sperm-associated SIX5, critical to spermiogenesis (64) that is associated with DNA binding (65). Also in this group and identified by multiple-mapped reads, is the RNA encoded by the Y-chromosome gene HSFY 1/2 that is deleted in some cases of severe male infertility (66). Potential post-fertilization effector roles also include signalling pathway members like SOCS1 that is encoded immediately downstream of the protamine locus (67). It is both a regulator of the JAK/STAT signal-transduction pathway (68) and acts to block insulin action (69), perhaps regulating early embryonic growth. Similarly, NRARP, a coordinator of Notch and Lef1-dependent Wnt signalling (70) and EGR3, an early growth response regulator of genes controlling biological rhythm, were also identified as members of this sperm-enhanced class.
Non-coding RNAs
The classes of small non-coding sperm RNAs have been previously described (10). Several classes of larger non-coding transcripts are also present at significantly higher levels within the SPIA-sperm libraries relative to testes. This includes members of the small nuclear RNU family of RNAs. Together with protein factors, these transcripts comprise both the major (nuclear) and minor (cytosolic) spliceosomes. In addition to their role as components of spliceosome, these elements have also been found to be integrally associated with polyadenylation (71). Sperm RNU transcripts above the 80th percentile of abundance yet essentially absent in SPIA-Testes include RNU11, RNU4-1, RNU4-2, RNU5A, RNU5E and RNU6ATAC. As sperm are considered to be transcriptionally and translationally silent, there is presumably no active role for a spliceosome in the mature spermatozoon even though as independently shown (e.g., RNU11, Supplementary Figure S3) they appear intact. The abundance of these transcripts in sperm despite the absence of spliceosome activity suggests that they may serve a role on delivery to the oocyte. In late metaphase II (MII), prior to fertilization and until zygotic genome activation, the human oocyte is transcriptionally silent (72,73). Coincident with embryonic genome activation, dormant ribosomal and spliceosomal machinery must be reactivated (74). Perhaps the sperm RNU transcripts activate dormant zygotic post-transcriptional RNA-processing pathways.
Several snaR (small NF90-associated RNAs) transcripts including snaR-C3, -C4, -E, -F, -G1 and -I are also abundant in sperm compared with testes. These novel small non-coding RNAs are unique to human and chimpanzee and are embedded in genomic regions that contain many genes important to reproduction, development and regulation of male fertility (75). Of particular note is the observation that all snaR elements also overlap hypomethylated regions (HMRs) in sperm. These regions (34) occupy ∼4% of the total genome, and transcripts in the vicinity of these regions are abundant in sperm (discussed below). The observation that these regions overlap snaR elements suggests that local chromatin is specifically organized with respect to these transcripts. Even though their precise role remains to be determined, the following two observations require consideration. First, snaR-G1 is both intact as shown in Supplementary Figure S3, and one of the most abundant RNAs within the sperm-enhanced class. Second, the sequence encoding this RNA directly overlaps the predicted promoter region for the beta 1 subunit (CGB1) of chorionic gonadotropin glycoprotein hormone (CGβ), a regulator of early placental development and implantation (76).
A number of other novel classes of ncRNAs (non-coding) were additionally noted to be uniquely abundant in sperm as compared with testes and observed at relatively similar levels in all sperm samples examined. Included among these are at least 250 RNAs that appear to be a specific intron of transcripts expressed in testes, but for which the spliced mature form is no longer present in sperm (Supplementary Table S12). Recent studies have suggested that intron-encoded RNAs may function as precursors of miRNAs or as regulatory factors (77,78). Some sperm RNAs appear as relatively short regions overlapping the UTRs or individual exons of several low abundance sperm RNAs (Supplementary Table S13). Local sequence analysis suggests that these regions are expressed independently of the transcripts in which they are located and that some may possess a poly(A) consensus site (AATAAA) indicative of antisense transcription (79–81). Sense-antisense regulation is a pervasive feature of transcriptomes and contributes to gene silencing, selective transcript editing, promoter inactivation and epigenetic modification (82–86). Previous studies in other tissues have shown that such transcripts are often independently regulated and only co-expressed with a corresponding sense transcript target at specific stages of development. Additionally, many RNAs present in sperm overlap regions identified by Gencode annotation (preV13) as either lncRNAs or as antisense elements (Supplementary Table S14) or isoforms. A final class of RNAs for which there is evidence in sperm is that of chromatin-associated RNAs (CARs). These RNAs are transcribed from intergenic or intronic regions of the genome, are found to bind to DNA and may play a role in regulation of DNA architecture or transcriptional regulation (87). Several previously identified CAR regions of the genome are transcribed at significant levels in sperm (Supplementary Table S15). Further work is being done to more fully characterize and assess the functional importance of each of these ncRNA classes in sperm.
Pri-mir-RNAs
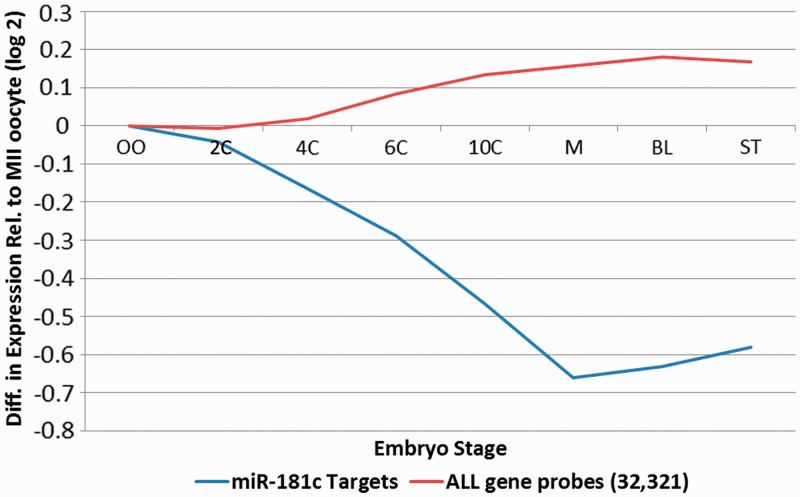
In addition to the previously characterized population of mature sperm micro-RNAs (10), the paternal gamete also harbours a number of ∼200–300 nt micro-RNA precursors (pri-miRNAs) that are essentially absent in testes. Examples of pri-mirRNAs present at high levels in sperm include pri-mir-1181, -miR-3648, -miR-3687, -mir-663 and -mir-181c. The most abundant of these, pri-mir-181c, is not observed in its shorter mature form in sperm (10) nor in either form in testes (Supplementary Figure S4). This strongly suggests that this transcript is delivered to the oocyte as a precursor mir-RNA requiring post-fertilization activation. The ability of fertilized mouse oocytes to process sperm-borne pri-mi-RNAs into mature forms capable of modulating target RNAs has been demonstrated (16). To evaluate the potential post-fertilization role of mir-181c, 27 high-confidence targets were computationally identified. Several of the putative targets including FIGN, ONECUT2, ZNF197, POU2F1, ROD1, ACRV2A, ACRV2B, TCERG1 and FOXP1 are associated with differentiation (77), while TNRC6B is linked to the regulation of miRNA pathway (78). Their average relative level as a function of the 2-, 4-, 6-, 10-post-fertilized human oocyte (30) is illustrated in Figure 3 and detailed in Supplementary Table S10. Greater than 70% of the targets decreased in expression from the oocyte to morula stage, with nine of these decreasing by a factor of 2 or more. Downregulation of targets from oocyte to morula stage is significantly greater than all genes (P < 0.0001) compared with the average expression of all 32 321 gene probes interrogated. This is consistent with the view that the sperm-delivered pri-miR-181c is processed to regulate genes of developmental consequence following delivery to the oocyte. Recent work has demonstrated that mature miR-181c modulates the expression levels of CARM1, a key regulator of critical pluripotency factors in human and mouse embryonic stem cells and blastomeres (79–81).
Figure 3.
Average level of miR-181C targets as a function of preimplantation embryonic development. The average level of 27 high-confidence predicted miR-181C targets through preimplantation embryonic development OO: Oocyte, M:Morula, BL:blastocyst, ST:Stem cell is shown in blue. This is compared to the relative level of non-targeted genes indicated in red.
Sperm RNA integrity
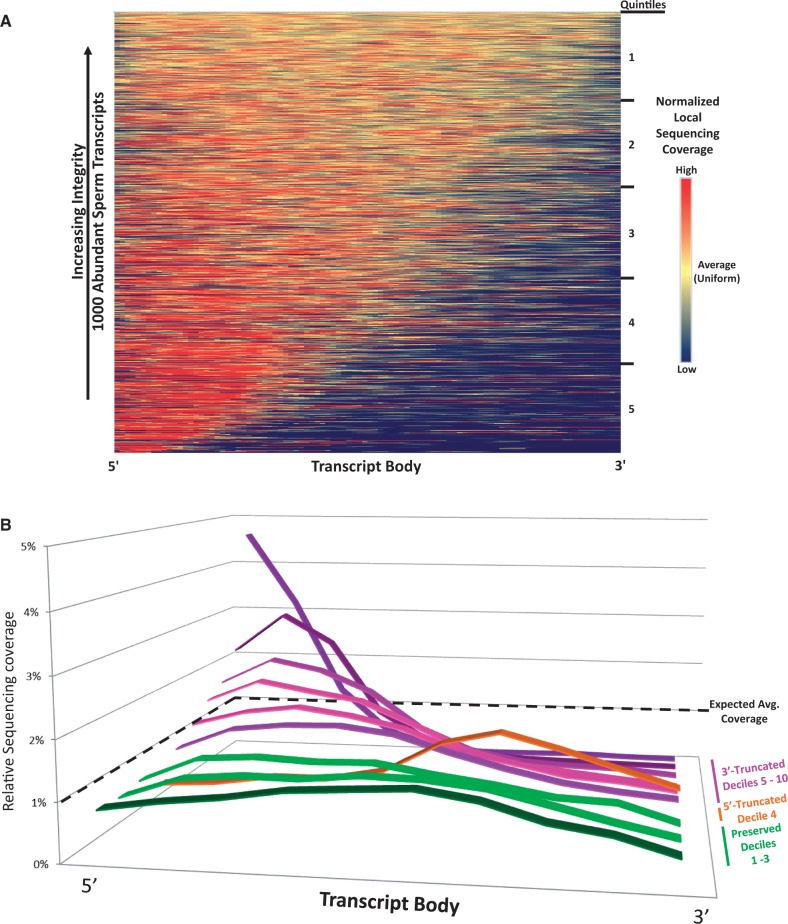
A computational approach was developed to globally identify the population of intact RNAs amongst the various fragmented forms. The RNA-seq profiles from the 1000 most abundant T-sperm RNAs (Sample Sp_D62[Tt]) were used to establish a means of measuring individual transcript integrity. This analysis was predicated on the assumption that sequencing coverage would vary less across the length of an intact transcript relative to one which was fragmented. To determine the uniformity of coverage for each, RNA transcripts were divided into 100 bins, and a 5-bin moving average was used to calculate localized variations in sequencing coverage. The squared deviation from expected coverage for each bin was summed and used as an intactness score to rank the 1000 RNAs according to their stability (Figure 4A). This highlighted a subpopulation of intact RNAs (top, yellow) while accentuating the biased representation of the 5′-end of the transcripts and variable depth of sequencing coverage (bottom left, red). It is important to note that shortened 3′ UTRs due to APA (see Alternative Polyadenylation and Isoforms) may contribute to decreased 3′ representation of some transcripts. Three mRNAs of interest were selected from the top 200 most intact profiles to test in silico predictions of stability by RT-PCR using transcript spanning primers (Supplementary Table S1 and Figure S3). These included NDUFA13, IZUMO4 and CIB1, which respectively encode a component of complex I of the mitochondrial respiratory chain, a protein involved in sperm-egg fusion and a calcium-binding protein essential to spermatogenesis (82–84). As described, three non-coding RNAs, snaR-G1, RNU11 and lnc-ERGIC1-1, which were not considered in the above integrity analysis due to their reduced length were also evaluated. As shown in Supplementary Figure S3, full length cDNA products corresponding to these transcripts were detected in all samples. The observed differential fragmentation and preservation of select RNAs may reflect the diverse roles of these transcripts during spermiogenesis or following fertilization. To determine if RNA stability was functionally correlated, the ranked transcripts were divided into quintiles and subjected to ontological analysis (1 most intact, 5 least intact; Supplementary Table S11). Transcripts in the first quintile were associated with mature spermatids (24 genes, P = 2.8 e-16) and male infertility (26 genes, P = 3.8 e-8). Subsequent quintiles generally highlighted ontological classes that included posttranscriptional regulation of gene expression (12 genes, P = 4.9 e-6) and RNA binding (20 genes, P = 9.4 e-5). The number of RNAs found within mature spermatids and male infertility ontologies decreased with each subsequent quintile, with not one spermatid or infertility transcript being observed in the 5th and most deteriorated.
Figure 4.
The most abundant sperm RNAs exhibit differential levels of stability. (A) Normalized RNA-seq coverage across the coding regions and UTRs of the 1000 most abundant sperm transcripts at a 100-bin resolution was used to identify intact RNAs. Transcripts are ranked in descending order of intactness and were divided into quintiles prior to ontological analysis. Profiles corresponding to stable RNAs exhibit uniform sequence coverage across the transcript (top, yellow). Profiles exhibiting increasing heterogeneity of coverage (bottom, red) are indicative of truncation. (B) A scaling function was applied to the above integrity scores to rank transcripts by their directionality and degree of fragmentation. The average sequencing coverage profiles for decile bins are presented. The expected level of coverage for an ideal transcript is shown as a dashed line. To highlight their likely functional importance, the flattest transcript deciles are prioritized to the foreground.
A scaling function was then developed to highlight directionality and degree of fragmentation. As shown in Figure 4B, the average sequencing coverage profile for each ranked and ordered decile highlighted three general categories of RNA stability: flat/intact (green; deciles 1–3), 5′ truncated (orange; decile 4) and 3′ truncated (purple; deciles 5–10). The presence of a pronounced curve at either end of a profile was observed for those sets of RNAs with a low intactness score. Interestingly, this analysis uncovered a group of 5′ truncated RNAs that were not readily observed in Figure 4A. These compromised sets were only weakly, if at all, associated with the ontological categories shared between the flat intact RNA groups. Though it is unlikely that truncated sperm RNAs serve any functional role following fertilization, some may serve as clinical biomarkers of successful spermatogenesis alongside the subpopulation of full-length transcripts identified above.
In contrast, the profiles that exhibited reduced variance in Relative Sequence Coverage were comprised mainly of transcripts with a high intactness score in the prior ranking and were enriched in similar ontological classes as the top intact quintile. These RNAs appear preserved and therefore are the most likely group to contain candidates for function following fertilization. This set of intact transcripts likely remains incomplete, reflecting the computational and technical (26) challenge of identifying full-length RNAs amongst a larger pool of transcriptional remnants. However, included in this preliminary set of flat and intact sperm RNAs is INTS1. This transcript encodes a subunit of the integrator complex that is required to process components of spliceosome. Inst1 knockout mice arrest at the early blastocyst stage and exhibit a concomitant accumulation and reduction of precursor and mature U2 snRNA, respectively (85). The level of this transcript increases (fold-change 1.5) in human two-cell embryos relative to the unfertilized oocyte, suggesting a paternal contribution (30).
Epigenetic modification of sperm transcript-associated genes
During spermatogenesis, the majority of the nucleosomes are replaced with protamines, compacting chromatin to a near crystalline state (86,87). This transition to a protamine-bound genome is evident from the significant underrepresentation in sperm of the >100 transcripts encoding both canonical and variant histones present in testes and/or somatic cells. Confirming previous results, some histone transcripts, including HIST1H1T, HIST1H2AA, HIST1H2BA (TSH2B) and H1FNT, revealed by RNA-seq are more abundant in testes than in somatic cells (UHR; 88). Additionally, histone variants H1FNT, H2AFJ, H3F3C and HILS1 are retained at a fairly high level in mature sperm. While H1FNT (H1T2) plays a role in spermatid elongation and DNA condensation (89,90) and HILS1 is similarly noted to aid in chromatin remodelling during spermiogenesis (91), the specific function of the other sperm-retained histone RNAs is not yet known. They may represent the last histones used prior to replacement by the transition proteins and protamines, perhaps facilitating chromatin compaction or essential for paternal imprinting.
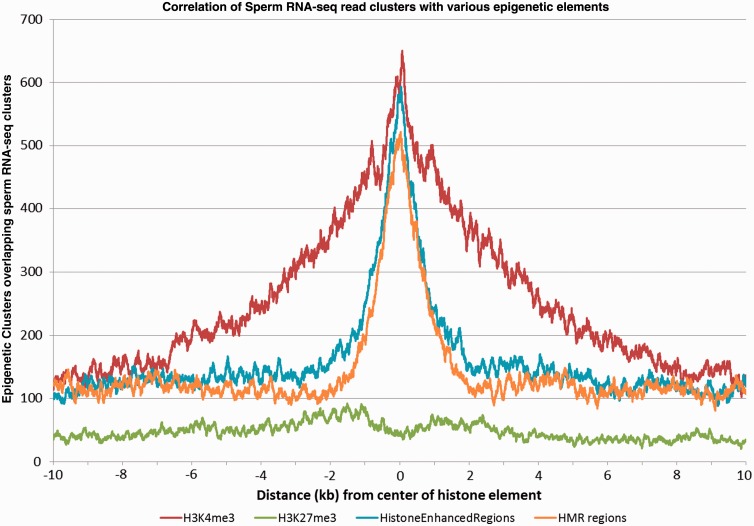
A comparatively small fraction of the paternal genome (5–15%) escapes protamination (92). Perhaps these regions encode transcripts required during compaction, encompass regions important early in development, or ensure protamine replacement with histones upon fertilization (8,93,94). These nucleosome-bound regions may be further differentiated based on presence or absence of specific histone modifications, DNA methylation and other genomic marks. The relationship between retained or specifically modified histones as a function of the associated sperm RNAs is illustrated in Figure 5. Sperm RNAs are associated with HMRs, H3K4me3 marks and histone-retained regions but not H3K27me3 enrichment. The high level of H3K4me3 observed in sperm in association with developmental factors EVX1, EVX2, ID1, STAT3, KLF5, FGF9, SOX7 and SOX9 has previously been suggested to mark sites poised for transcription following fertilization (33). Although these genes are marked by histone modifications correlated with active transcription when delivered to the oocyte (95–97), the coincident occurrence of their corresponding transcripts in sperm must be considered. Perhaps the H3K4me3 mark correlated with the high levels of EVX2, KLF5, SOX9 and EVX1 RNAs retained by the mature male gamete represents a transcriptional ghost. In contrast, the developmental regulatory HOX genes are not expressed in spermatogenesis and yet they are hypomethylated and enriched in nucleosomes, some of which contain the active H3K4me3 and repressive H3K27me2 modified histones (Supplementary Figure S4A and C; 33). Accordingly, the transcriptionally silent yet epigenetically poised bipotential state of the HOX clusters in sperm infers that these genes are packaged for early embryonic use. These domains also harbour EVX1 and EVX2, two regulators of early embryonic development, which are evolutionarily related to the HOX genes (98,99). Examination within the vicinity of EVX1 and EVX2 revealed significant retention of an RNA encoded by the last two exons of EVX2 in addition to transcripts corresponding to sequences proximal to the 3′-ends of both genes (Supplementary Figure S5B and D). Reads from these regions appear unique to sperm compared with testes, and may be indicative of novel non-coding, potentially regulatory transcripts.
Figure 5.
Sperm RNAs are correlated with the genomic positioning of histones and epigenetic elements. A total of 19 521 highly expressed RNA-seq clusters were identified in the promoter and exonic regions of sperm sample Sp_D62[Tt]. Each distance correlation is centered on the genomic coordinates of elements within one of the four following classes, H3K4me3 and H3K27me3 enriched regions (n = 34 912 and 38 337, respectively; 95), histone-enriched regions (n = 25 114; 33) and hypomethylated DNA regions (n = 79 124; 34).
The population of RNA in sperm as revealed by RNA-seq provides a window into the developmental history, functional viability and potential elements present by sperm that may serve a role in the final stages of spermiogenesis or at fertilization. These include a number of coding and non-coding RNAs, the primary role of which in either sperm or the oocyte can now be delineated. Correlation of RNA abundance in sperm with epigenetic marks has permitted a more complete picture of how these marks may act to regulate both the pre- and post-fertilization genome. The observation that an experimentally derived mouse gynogenote may not strictly require a paternal contribution (100) remains to be reconciled. It is likely that in this case, the confrontation–consolidation pathway [reviewed in (101)], in which the paternal RNAs may participate in genome compatibility recognition, was sidestepped. Further, thought must be given to the documented role of the paternally derived mir-34c mediating first cell division (102), the most abundant human sperm micro RNA (10) or the function of other RNAs as paternal epigenetic modifiers [reviewed in (103)]. The identification of transcripts based on traits including relative abundance, integrity, knock-out phenotypes and presence or absence in the unfertilized oocyte, provides the essential foundation to elucidate the role of male factors critically important to the fertilization, birth and long-term health of a child.
ACCESSION NUMBERS
Sequencing data is deposited in GEO as:
| GSM721696 | AS062 (Sp_D62[Tt]) |
| GSM721697 | AS064 (Sp_D64[Tt]) |
| GSM721698 | AS066 (Sp_D66[Tt]) |
| GSM721699 | AS062PLUS (Sp_D62[A+]) |
| GSM721700 | Testes Ambion (Te_PAm[A+]) |
| GSM721701 | Testes Clonetech (Te_PCl[A+]) |
| GSM1037852 | AS062 (Sp_D62[SP]) |
| GSM1037853 | AS064 (Sp_D64[SP]) |
| GSM1037854 | AS066 (Sp_D66[SP]) |
| GSM1037855 | Pool2 (Sp_P2[SP]) |
| GSM1037856 | Testes Ambion (Te_PAm[SP]) |
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–15, Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
GENI pilot grant from Harvard School of Public Health (to S.A.K. and R.H.); National Institute of Environmental Health Sciences [ES017285 to R.H.]; Charlotte B. Failing Professorship (to S.A.K.); Michigan Core Technology grant from the State of Michigan’s 21st Century Fund Program (to the Wayne State University Applied Genomics Technology Center). Funding for open access charge: Charlotte B. Failing Professorship (to S.A.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ester Anton Martorell and Meritxell Jodar Bifet for review and comments to the manuscript and Leah Sendler for her visual review and identification of many transcript features. Samples were also prepared for sequencing by Claudia Lalancette. Support by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Contract 25PM6 in collaboration with the LIFE Study Working Group, Division of Epidemiology, Statistics and Prevention Research, which provided some of the semen samples for analysis, is appreciated. Conceived and designed the experiments: M.P.D., R.H., S.A.K.; Performed the experiments: E.S., G.D.J., R.J.G.; Analyzed the data: E.S., G.D.J., S.M., S.A.K.; Contributed reagents/materials/analysis tools: M.P.D., R.H.; Wrote the manuscript: E.S., G.D.J., S.A.K.; Reviewed the manuscript: E.S., G.D.J., S.M., R.J.G., M.P.D., R.H., S.A.K.
REFERENCES
- 1.Krawetz SA. Paternal contribution: new insights and future challenges. Nat. Rev. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 2.Pessot CA, Brito M, Figueroa J, Concha II, Yanez A, Burzio LO. Presence of RNA in the sperm nucleus. Biochem. Biophys. Res. Commun. 1989;158:272–278. doi: 10.1016/s0006-291x(89)80208-6. [DOI] [PubMed] [Google Scholar]
- 3.Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 4.Miller D. RNA in the ejaculate spermatozoon: a window into molecular events in spermatogenesis and a record of the unusual requirements of haploid gene expression and post-meiotic equilibration. Mol. Hum. Reprod. 1997;3:669–676. doi: 10.1093/molehr/3.8.669. [DOI] [PubMed] [Google Scholar]
- 5.Miller D, Ostermeier GC. Towards a better understanding of RNA carriage by ejaculate spermatozoa. Hum. Reprod. Update. 2006;12:757–767. doi: 10.1093/humupd/dml037. [DOI] [PubMed] [Google Scholar]
- 6.Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–777. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- 7.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J. Biol. Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 8.Wykes SM, Visscher DW, Krawetz SA. Haploid transcripts persist in mature human spermatozoa. Mol. Hum. Reprod. 1997;3:15–19. doi: 10.1093/molehr/3.1.15. [DOI] [PubMed] [Google Scholar]
- 9.Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. A suite of novel human spermatozoal RNAs. J. Androl. 2005;26:70–74. [PubMed] [Google Scholar]
- 10.Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP. A survey of small RNAs in human sperm. Hum. Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, Lamb DJ, Matzuk MM. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum. Mol. Genet. 2006;15:3411–3419. doi: 10.1093/hmg/ddl417. [DOI] [PubMed] [Google Scholar]
- 12.Platts AE, Dix DJ, Chemes HE, Thompson KE, Goodrich R, Rockett JC, Rawe VY, Quintana S, Diamond MP, Strader LF, et al. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum. Mol. Genet. 2007;16:763–773. doi: 10.1093/hmg/ddm012. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C, Huo R, Wang FQ, Lin M, Zhou ZM, Sha JH. Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil. Steril. 2007;87:436–438. doi: 10.1016/j.fertnstert.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 14.Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- 15.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 16.Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl Acad. Sci. USA. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambard S, Galeraud-Denis I, Martin G, Levy R, Chocat A, Carreau S. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Mol. Hum. Reprod. 2004;10:535–541. doi: 10.1093/molehr/gah064. [DOI] [PubMed] [Google Scholar]
- 18.Miller D. Analysis and significance of messenger RNA in human ejaculated spermatozoa. Mol. Reprod. Dev. 2000;56:259–264. doi: 10.1002/(SICI)1098-2795(200006)56:2+<259::AID-MRD10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19.Waclawska A, Kurpisz M. Key functional genes of spermatogenesis identified by microarray analysis. Syst. Biol. Reprod. Med. 2012;58:229–235. doi: 10.3109/19396368.2012.693148. [DOI] [PubMed] [Google Scholar]
- 20.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson GD, Sendler E, Lalancette C, Hauser R, Diamond MP, Krawetz SA. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol. Hum. Reprod. 2011;17:721–726. doi: 10.1093/molehr/gar054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Syst. Biol. Reprod. Med. 2007;53:161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- 28.Johnson GD, Platts AE, Lalancette C, Goodrich R, Heng HH, Krawetz SA. Interrogating the transgenic genome: development of an interspecies tiling array. Syst. Biol. Reprod. Med. 2011;57:54–62. doi: 10.3109/19396368.2010.506000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, Belmonte JC. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138:3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert I, Bissonnette N, Boissonneault G, Vallee M, Robert C. A molecular analysis of the population of mRNA in bovine spermatozoa. Reproduction. 2007;133:1073–1086. doi: 10.1530/REP-06-0292. [DOI] [PubMed] [Google Scholar]
- 36.Goodrich RJ, Anton E, Krawetz SA. Isolating mRNA and small noncoding RNAs from human sperm. Methods Mol. Biol. 2013;927:385–396. doi: 10.1007/978-1-62703-038-0_33. [DOI] [PubMed] [Google Scholar]
- 37.Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozov R, Halperin E, Shamir R. MGMR: leveraging RNA-Seq population data to optimize expression estimation. BMC Bioinformatics. 2012;13(Suppl. 6):S2. doi: 10.1186/1471-2105-13-S6-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salemi M, Calogero AE, Di Benedetto D, Cosentino A, Barone N, Rappazzo G, Vicari E. Expression of SPANX proteins in human-ejaculated spermatozoa and sperm precursors. Int. J. Androl. 2004;27:134–139. doi: 10.1111/j.1365-2605.2004.00461.x. [DOI] [PubMed] [Google Scholar]
- 41.Chalmel F, Lardenois A, Evrard B, Mathieu R, Feig C, Demougin P, Gattiker A, Schulze W, Jegou B, Kirchhoff C, et al. Global human tissue profiling and protein network analysis reveals distinct levels of transcriptional germline-specificity and identifies target genes for male infertility. Hum. Reprod. 2012;27:3233–3248. doi: 10.1093/humrep/des301. [DOI] [PubMed] [Google Scholar]
- 42.Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol. Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legendre M, Ritchie W, Lopez F, Gautheret D. Differential repression of alternative transcripts: a screen for miRNA targets. PLoS Comput. Biol. 2006;2:e43. doi: 10.1371/journal.pcbi.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji X, Kong J, Liebhaber SA. An RNA-protein complex links enhanced nuclear 3′ processing with cytoplasmic mRNA stabilization. EMBO J. 2011;30:2622–2633. doi: 10.1038/emboj.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta V, Bamezai RN. Human pyruvate kinase M2: a multifunctional protein. Protein Sci. 2010;19:2031–2044. doi: 10.1002/pro.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, Becker JD. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 2008;148:1168–1181. doi: 10.1104/pp.108.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich A, Klatsky P, Carson S, Wessel G. The transcriptome of a human polar body accurately reflects its sibling oocyte. J. Biol. Chem. 2011;286:40743–40749. doi: 10.1074/jbc.M111.289868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krisfalusi M, Miki K, Magyar PL, O'Brien DA. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- 52.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 2012;40:D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fullerton MD, Hakimuddin F, Bakovic M. Developmental and metabolic effects of disruption of the mouse CTP:phosphoethanolamine cytidylyltransferase gene (Pcyt2) Mol. Cell Biol. 2007;27:3327–3336. doi: 10.1128/MCB.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lima-Souza A, Anton E, Mao S, Ho WJ, Krawetz SA. A platform for evaluating sperm RNA biomarkers: dysplasia of the fibrous sheath–testing the concept. Fertil. Steril. 2012;97:1061–1066 e1061–1063. doi: 10.1016/j.fertnstert.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 58.Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- 59.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, Shuai K, Grosschedl R, van Deursen JM. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, Joseph J, Kehlenbach RH, van Deursen JM. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J. Cell Biol. 2011;194:597–612. doi: 10.1083/jcb.201102018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeGregori J, Russ A, von Melchner H, Rayburn H, Priyaranjan P, Jenkins NA, Copeland NG, Ruley HE. A murine homolog of the yeast RNA1 gene is required for postimplantation development. Genes Dev. 1994;8:265–276. doi: 10.1101/gad.8.3.265. [DOI] [PubMed] [Google Scholar]
- 62.Calvin HI, Hwang FH, Wohlrab H. Localization of zinc in a dense fiber-connecting piece fraction of rat sperm tails analogous chemically to hair keratin. Biol. Reprod. 1975;13:228–239. doi: 10.1095/biolreprod13.2.228. [DOI] [PubMed] [Google Scholar]
- 63.Cowin P, Kapprell HP, Franke WW, Tamkun J, Hynes RO. Plakoglobin: a protein common to different kinds of intercellular adhering junctions. Cell. 1986;46:1063–1073. doi: 10.1016/0092-8674(86)90706-3. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar PS, Paul S, Han J, Reddy S. Six5 is required for spermatogenic cell survival and spermiogenesis. Hum. Mol. Genet. 2004;13:1421–1431. doi: 10.1093/hmg/ddh161. [DOI] [PubMed] [Google Scholar]
- 65.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 66.Tessari A, Salata E, Ferlin A, Bartoloni L, Slongo ML, Foresta C. Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol. Hum. Reprod. 2004;10:253–258. doi: 10.1093/molehr/gah036. [DOI] [PubMed] [Google Scholar]
- 67.Kramer JA, Adams MD, Singh GB, Doggett NA, Krawetz SA. A matrix associated region localizes the human SOCS-1 gene to chromosome 16p13.13. Somat. Cell Mol. Genet. 1998;24:131–133. doi: 10.1023/b:scam.0000007115.58601.87. [DOI] [PubMed] [Google Scholar]
- 68.Komazaki T, Nagai H, Emi M, Terada Y, Yabe A, Jin E, Kawanami O, Konishi N, Moriyama Y, Naka T, et al. Hypermethylation-associated inactivation of the SOCS-1 gene, a JAK/STAT inhibitor, in human pancreatic cancers. Jpn. J. Clin. Oncol. 2004;34:191–194. doi: 10.1093/jjco/hyh035. [DOI] [PubMed] [Google Scholar]
- 69.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 70.Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, et al. Nrarp coordinates endothelial Notch and Wnt signaling to control vessel density in angiogenesis. Dev. Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Awasthi S, Alwine JC. Association of polyadenylation cleavage factor I with U1 snRNP. RNA. 2003;9:1400–1409. doi: 10.1261/rna.5104603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 73.Prioleau MN, Huet J, Sentenac A, Mechali M. Competition between chromatin and transcription complex assembly regulates gene-expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 74.Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35:6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hallast P, Rull K, Laan M. The evolution and genomic landscape of CGB1 and CGB2 genes. Mol. Cell Endocrinol. 2007;260–262:2–11. doi: 10.1016/j.mce.2005.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cox GA, Mahaffey CL, Nystuen A, Letts VA, Frankel WN. The mouse fidgetin gene defines a new role for AAA family proteins in mammalian development. Nat. Genet. 2000;26:198–202. doi: 10.1038/79923. [DOI] [PubMed] [Google Scholar]
- 78.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu Z, Jiang J, Xu C, Wang Y, Sun L, Guo X, Liu H. MicroRNA-181 regulates CARM1 and histone aginine methylation to promote differentiation of human embryonic stem cells. PLoS One. 2013;8:e53146. doi: 10.1371/journal.pone.0053146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, Langford CF, Glover DM, Zernicka-Goetz M. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–2645. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan W, Leisner TM, McFadden AW, Clark S, Hiller S, Maeda N, O'Brien DA, Parise LV. CIB1 is essential for mouse spermatogenesis. Mol. Cell Biol. 2006;26:8507–8514. doi: 10.1128/MCB.01488-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang G, Lu H, Hao A, Ng DC, Ponniah S, Guo K, Lufei C, Zeng Q, Cao X. GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I. Mol. Cell Biol. 2004;24:8447–8456. doi: 10.1128/MCB.24.19.8447-8456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellerman DA, Pei J, Gupta S, Snell WJ, Myles D, Primakoff P. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 2009;76:1188–1199. doi: 10.1002/mrd.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hata T, Nakayama M. Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim. Biophys. Acta. 2007;1773:1039–1051. doi: 10.1016/j.bbamcr.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 86.Oliva R, Ballesca JL. Altered histone retention and epigenetic modifications in the sperm of infertile men. Asian J. Androl. 2012;14:239–240. doi: 10.1038/aja.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, Khochbin S. Establishment of male-specific epigenetic information. Gene. 2005;345:139–153. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 88.Govin J, Caron C, Rousseaux S, Khochbin S. Testis-specific histone H3 expression in somatic cells. Trends Biochem. Sci. 2005;30:357–359. doi: 10.1016/j.tibs.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 89.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl Acad. Sci. USA. 2005;102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singleton S, Zalensky A, Doncel GF, Morshedi M, Zalenskaya IA. Testis/sperm-specific histone 2B in the sperm of donors and subfertile patients: variability and relation to chromatin packaging. Hum. Reprod. 2007;22:743–750. doi: 10.1093/humrep/del439. [DOI] [PubMed] [Google Scholar]
- 91.Yan W, Ma L, Burns KH, Matzuk MM. HILS1 is a spermatid-specific linker histone H1-like protein implicated in chromatin remodeling during mammalian spermiogenesis. Proc. Natl Acad. Sci. USA. 2003;100:10546–10551. doi: 10.1073/pnas.1837812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction. 2011;141:21–36. doi: 10.1530/REP-10-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balhorn R. A model for the structure of chromatin in mammalian sperm. J. Cell Biol. 1982;93:298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y, Lalancette C, Miller D, Krawetz SA. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian J. Androl. 2008;10:535–541. doi: 10.1111/j.1745-7262.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 96.Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011;26:2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vavouri T, Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat. Rev. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 99.Kalisz M, Winzi M, Bisgaard HC, Serup P. EVEN-SKIPPED HOMEOBOX 1 controls human ES cell differentiation by directly repressing GOOSECOID expression. Dev. Biol. 2012;362:94–103. doi: 10.1016/j.ydbio.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 100.Kawahara M, Wu Q, Takahashi N, Morita S, Yamada K, Ito M, Ferguson-Smith AC, Kono T. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat. Biotechnol. 2007;25:1045–1050. doi: 10.1038/nbt1331. [DOI] [PubMed] [Google Scholar]
- 101.Bourc'his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- 102.Liu WM, Pang RT, Chiu PC, Wong BP, Lao K, Lee KF, Yeung WS. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl Acad. Sci. USA. 2012;109:490–494. doi: 10.1073/pnas.1110368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.