Abstract
These days, when we talk about the origin of a protein, or even a pathway, we are typically referring to evolutionary lineages based on nucleotide sequences. For example, is a particular protein’s function conserved? How far back did it first appear? Are there homologs in higher eukaryotes? However, a simpler question (or perhaps I should say, a non-molecular biology question) is when was the process first detected in the paleontological record? Of course I assumed that macroautophagy was ancient, but a new finding (see p. 632 in this issue of the journal) provides an unexpected—and exciting—piece of information for our field. For the first time, scientists have discovered fossil evidence for an actual subcellular pathway—and it looks like it might actually be autophagy (I admit I am biased, but you can decide for yourself).
Keywords: autophagy, fossil, lysosome, stress, vacuole
A joint team of archeologists, molecular biologists and imaging specialists in China’s Jiangsu’s province excavated the fossilized remains of what appears to be yet another new species of dinosaur, tentatively being called Autoraptor wanke. A. wanke is most closely related to Velociraptor sp., which lived approximately 71- to 75-million years ago. What is striking in this case is the amazing degree of preservation. Based on the recovery of samples, radar imaging of the shape of the skeleton and computerized reconstructions, the team, led by Dr. Kai M. Ao of Peking University, conclude that the dinosaur died of starvation on 4/1 approximately 72 million years ago. The first true indication that this specimen was special came from the identification of fossilized organs within the body cavity. “It was the most intact set of remains I have ever seen,” said Dr. May-Ahn Jin, of Tsinghua University, “including an amazing preservation of skin and organ tissue.” Pairing up with researchers at Fudan University, the team used state-of-the-art technology, including a previously unused method unofficially dubbed 4D-tomography, to provide, for the first time, a look at what is being interpreted as a subcellular process in a fossilized specimen.
“You have to realize that this is subject to interpretation,” said Dr. Lou Xu of Wuhan University (head of the molecular imaging team) through an interpreter. “After all, we are of course looking at mineral deposits.” Another problem concerns the degradation and erosion of the samples; however, even a quick look at relatively low resolution suggests that the scientists have clearly identified cells (Fig. 1), of a quality unsurpassed for this type of fossilized material.1 Obtaining higher resolution images, however, turned out to be much more involved. A deconvolution process that is based on multiple cell samples is the only way to overcome the overall loss of definition that results from this type of severe aging. The method is somewhat similar to single-particle electron microscopy, where literally thousands (or tens of thousands) of images need to be aligned, superimposed and averaged, which in this case—due to the mineral nature of the samples—was extremely time consuming. Nonetheless, after months of work, both in the field and back at Peking University, the researchers began to piece together what appears to be an actual pathway from subcellular samples (Fig. 2). In collaboration with researchers at the University of Michigan led by Dr. Kay T. Parzykat, a schematic was generated, which (I think) strikingly resembles the process of macroautophagy (Fig. 3).
Figure 1. Low-resolution image from a fossilized sample indicating cell boundaries. Scale bar: 20 µm.
Figure 2. Images from the archeological dig in Jiangsu province. (A) A fossilized sample from Autoraptor wanke that resembles a phagophore. (B) A fossilized sample from a different cell displaying a structure that may represent a complete autophagosome. Scale bar: 1 × 10−8 cubits.
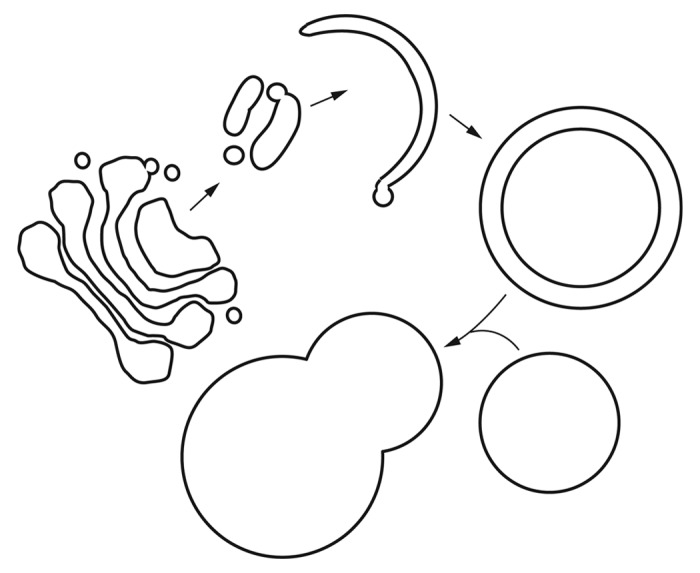
Figure 3. Reconstruction of a subcellular pathway from scientists at the University of Michigan, based on the 4D-tomography of the samples from Autoraptor wanke. Note that this schematic is only one possible interpretation, and the arrows were added by the editor-in-chief to aid the reader.
One key observation was the absence of any structure resembling an omegasome,2 the endoplasmic reticulum-associated structure that is thought to play a role in biogenesis of the phagophore. However, all of the researchers were quick to point out that this is a negative result. “We would not want to overinterpret these types of data” stated Dr. Ao. “Although we think we detected tubulovesicular structures, it would be quite a stretch for us to state that they were forming the phagophore.” (See Mari et al. for a description of these structures.3) The researchers are hopeful that the discovery of similarly well-preserved specimens will provide additional insight into the mechanistic details of macroautophagy.
Finally, I could not help asking Dr. Ao why he chose to submit his paper to Autophagy. “It is the top journal in the field” he said “and I wanted to be sure our paper would be reviewed by experts who could appreciate the nature of our studies.” I could not have put it better myself.4
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23907
References and Footnotes
- 1.Images from Wikipedia (http://en.wikipedia.org/wiki/Fossil and http://en.wikipedia.org/wiki/File:Petoskey_stone_unpolished_with_cm_scale.jpg).
- 2.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.“April 1. This is the day upon which we are reminded of what we are on the other three hundred and sixty-four.” Mark Twain, Pudd’nhead Wilson, 1894. [Google Scholar]


