Abstract
Involution is the process of post-lactational mammary gland regression to quiescence and it involves secretory epithelial cell death, stroma remodeling and gland repopulation by adipocytes. Though reportedly accompanying apoptosis, the role of autophagy in involution has not yet been determined. We now report that autophagy-related (ATG) proteins mediate dead cell clearance and suppress inflammation during mammary involution. In vivo, Becn1+/− and Atg7-deficient mammary epithelial cells (MECs) produced ‘competent’ apoptotic bodies, but were defective phagocytes in association with reduced expression of the MERTK and ITGB5 receptors, thus pointing to defective apoptotic body engulfment. Atg-deficient tissues exhibited higher levels of involution-associated inflammation, which could be indicative of a tumor-modulating microenvironment, and developed ductal ectasia, a manifestation of deregulated post-involution gland remodeling. In vitro, ATG (BECN1 or ATG7) knockdown compromised MEC-mediated apoptotic body clearance in association with decreased RAC1 activation, thus confirming that, in addition to the defective phagocytic processing reported by other studies, ATG protein defects also impair dead cell engulfment.
Using two different mouse models with mammary gland-associated Atg deficiencies, our studies shed light on the essential role of ATG proteins in MEC-mediated efferocytosis during mammary involution and provide novel insights into this important developmental process. This work also raises the possibility that a regulatory feedback loop exists, by which the efficacy of phagocytic cargo processing in turn regulates the rate of engulfment and ultimately determines the kinetics of phagocytosis and dead cell clearance.
Keywords: autophagy, mammary involution, efferocytosis, engulfment, inflammation, ductal ectasia, dead cell clearance, MERTK, ITGB5
Background
During development, the mammary gland undergoes well-orchestrated, hormone-dependent morphological changes to become a milk-producing machine during pregnancy and lactation, which subsequently regresses to a quiescent organ by involution.1,2 In early gestation, mammary epithelial cells (MECs) proliferate, whereas in late pregnancy and lactation, they differentiate for milk production. During involution, most secretory alveolar MECs undergo programmed cell death (PCD); remaining cells are remodeled into pre-pregnancy-like glandular structures, in a process consisting of two phases and completed within 10 d in mice.3 The first involution phase, lasting 48 h post-weaning, is regulated by local factors4,5 and is reversible, i.e., if pups are returned to mother, MEC death is halted and lactation resumes; the second phase is regulated by circulating hormones,3 is characterized by extensive tissue remodeling that is dependent on matrix metalloproteinases (MMPs),6 and is irreversible.
Within the first 72 h, residual milk and dead cells are primarily cleared from alveolar lumens by neighboring, living MECs in a highly conserved process resembling phagocytosis and uniquely termed efferocytosis (from the greek word ‘effero’, to carry to the grave).7,8 Professional phagocytes, such as macrophages, also infiltrate the gland, mostly after the third day of involution,8 and play a critical role in stroma remodeling,9 concomitantly creating a tumor-promoting microenvironment10,11 that potentially contributes to the poor prognosis of pregnancy-associated breast cancer (PABC).12
Leukemia inhibitory factor (LIF),13,14 death receptor ligands10,15 and transforming growth factor β 3 (TGFB3)16 are strongly upregulated during the first 12 h of involution, and the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway, induced by LIF-dependent STAT3 phosphorylation, is critical for apoptosis initiation and timely gland involution.13,14,17,18 CD14 is also upregulated during involution and exposed on MECs in a STAT3-dependent manner.10,19 Although PCD during mammary involution has been primarily attributed to apoptosis, a recent study showed that cell death during the first phase of involution is mediated by permeabilization of lysosomal membranes.20 It is, thus, important to note that the term “apoptosis” as used here also includes this newly described lysosome-mediated cell death.
Studies on bovine mammary glands have demonstrated that, in parallel to apoptosis, the self-catabolic process of autophagy21 is also activated during involution.22,23 In vitro, apoptosis and autophagy induction in bovine MECs is accompanied by TGFB1 and TGFBR2 upregulation,23 resulting in increased insulin growth factor-binding protein (IGFBP) levels and decreased AKT phosphorylation.24 Autophagy regulation in MECs is even more complex, as EGF also suppresses autophagy via MTOR activation, whereas 17β-estradiol and progesterone stimulate autophagy by suppressing MTOR.25
Despite evidence for autophagy induction during involution, its specific role during this important developmental process remains unclear. If autophagy is activated as a cell-survival mechanism in response to apoptosis-inducing signals triggered by cessation of lactation,26 autophagy defects may accelerate MEC apoptosis and gland remodeling; alternatively, autophagy may act as a cell death mechanism complementary to apoptosis22 and, thus, its impairment may decrease MEC death and delay involution; the autophagic machinery may also participate in dead cell removal.27 To date, there is no genetic evidence supporting a particular functional role for autophagy in mammary involution over another.
Using the Gfp-Lc3 mice,28 ubiquitously expressing a fluorescent GFP-fusion of the essential autophagy-related (ATG) protein LC3 (ortholog of ATG8), we determined that autophagy is induced early in involution. Monoallelic Becn1 loss or MEC-specific Atg7 deletion compromised autophagosome formation, but did not affect apoptosis during the first 48 h post-weaning; however, the second involution phase was prolonged in association with delayed alveolar collapse, dead cell accumulation in alveolar lumens and slower gland repopulation by adipocytes. Atg-defective mammary tissues exhibited persistent involution-associated macrophage infiltration, which possibly creates a tumor-modulating microenvironment, and developed ductal ectasia, that is indicative of long-term impairment in mammary remodeling. Our studies genetically confirm the critical role of MECs in dead cell clearance during involution and expand the accumulating evidence on the importance of ATG proteins in phagocytosis,29 innate immune response30,31 and efferocytosis32,33 by demonstrating that, in addition to the previously reported endocytic processing defect,29,32,34 impairment of autophagic machinery proteins compromises efferocytosis in vivo at the early step of engulfment.
Results
Atg-dependent autophagy induction in early mammary involution
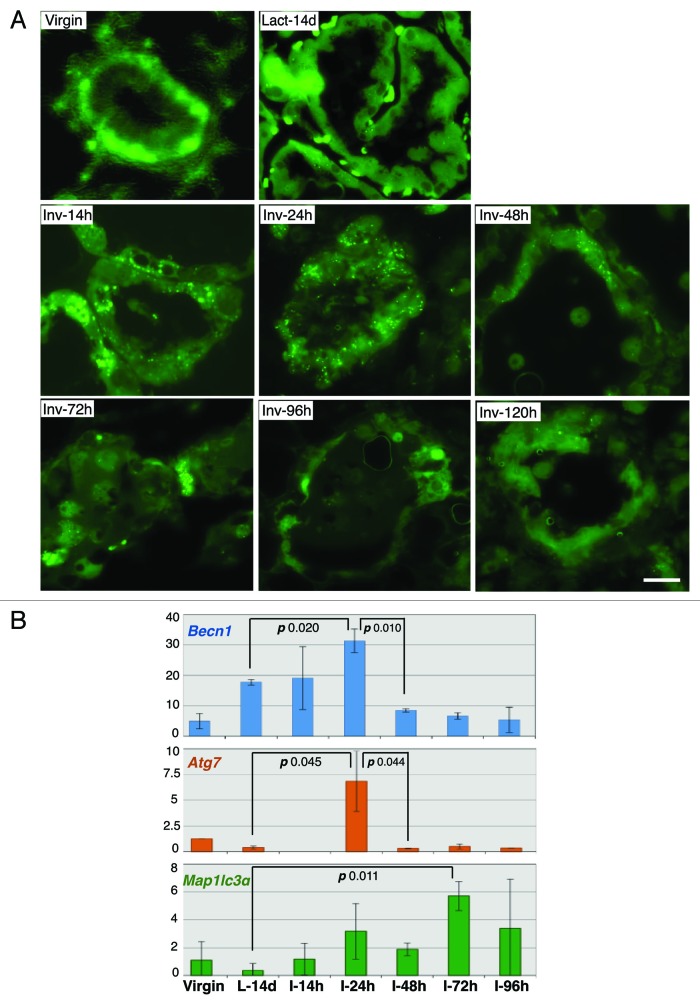
The functional status of autophagy at different stages of mammary gland development was investigated in the Gfp-Lc3 mice,28 ubiquitously expressing the ATG protein LC3 (ATG8) fused to GFP (Fig. 1A). The diffuse cytoplasmic LC3 distribution observed in virgin and lactating mammary glands changed to a perinuαclear punctate pattern, indicative of LC3 lipidation and translocation to the autophagosome membrane, within hours of forced pup-weaning, demonstrating autophagy induction very early in involution. Autophagosome formation peaked at 24 h post-weaning and returned to baseline after 72 h. Mammary gland Becn1 mRNA was induced during lactation and peaked at 24 h post-weaning, whereas Atg7 and Map1lc3α (Lc3) mRNA levels were induced only during involution (Fig. 1B).
Figure 1. Autophagy is induced in the early phase of mammary involution. (A) GFP fluorescence on frozen sections of mammary glands from GFP-LC3 mice. (B) RT-PCR quantification of Becn1, Atg7 and Map1lc3α mRNA in mammary glands from wild-type mice. L (Lact), lactation; I (Inv), involution; d, days; h, hours.
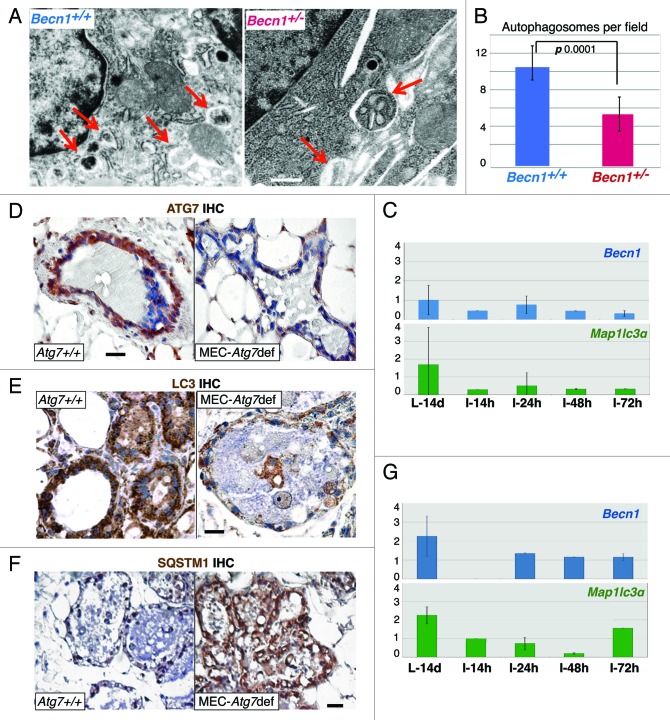
Allelic Becn1 loss or MEC-specific Atg7 deletion compromised involution-induced autophagy (Fig. 2A and B, and Fig. 2D–F, respectively) and was accompanied by severely blunted changes in Becn1 and Lc3 mRNA (Fig. 2C and G), thus indicating that upregulation of autophagy in the early phase of mammary involution is both Becn1- and Atg7-dependent, and defects in either ATG protein decrease postlactational autophagy induction. In contrast to the ubiquitous (in all cell types and tissues) monoallelic Becn1 deletion in Becn1+/− mice, in the mammary gland-specific Atg7-deficient mouse model described here, Atg7 was deleted only in secretory MECs after pregnancy and lactation,35,36 thus indicating that alveolar epithelial cells are primarily responsible for the autophagic response observed shortly after pup weaning. It is also of interest that both allelic Becn1 loss and MEC-specific Atg7 deletion compromised involution-associated LC3 upregulation, thus suggesting that LC3 induction is dependent on and/or coregulated with BECN1 and ATG7 expression. To our knowledge, this finding has not been reported before and is definitely worthy of further investigation.
Figure 2. Monoallelic Becn1 loss or MEC-specific Atg7 deletion compromises autophagy induction in mammary involution. (A) EM micrographs of Becn1+/+ and Becn1+/− mammary glands on day 2 post-weaning. Red arrows point to autophagosomes. (B) Quantification of autophagosomes in Becn1+/+ and Becn1+/− mammary glands on day 2 post-weaning by EM. (C) RT-PCR quantification of Becn1 and Map1lc3α mRNA in mammary glands from Becn1+/− mice. (D–F) ATG7, LC3 and SQSTM1 IHC respectively on mammary glands from parous wild-type and Atg7F/F;WAP-Cre mice on day 2 post-weaning. (G) RT-PCR quantification of Becn1 and Map1lc3α mRNA in mammary glands from parous wild-type and Atg7F/F;WAP-Cre mice type. L, lactation; I, involution; d, days; h, hours. Parous mice: after second pregnancy and lactation
Atg defects delay mammary involution in association with dead cell accumulation in alveolar lumens
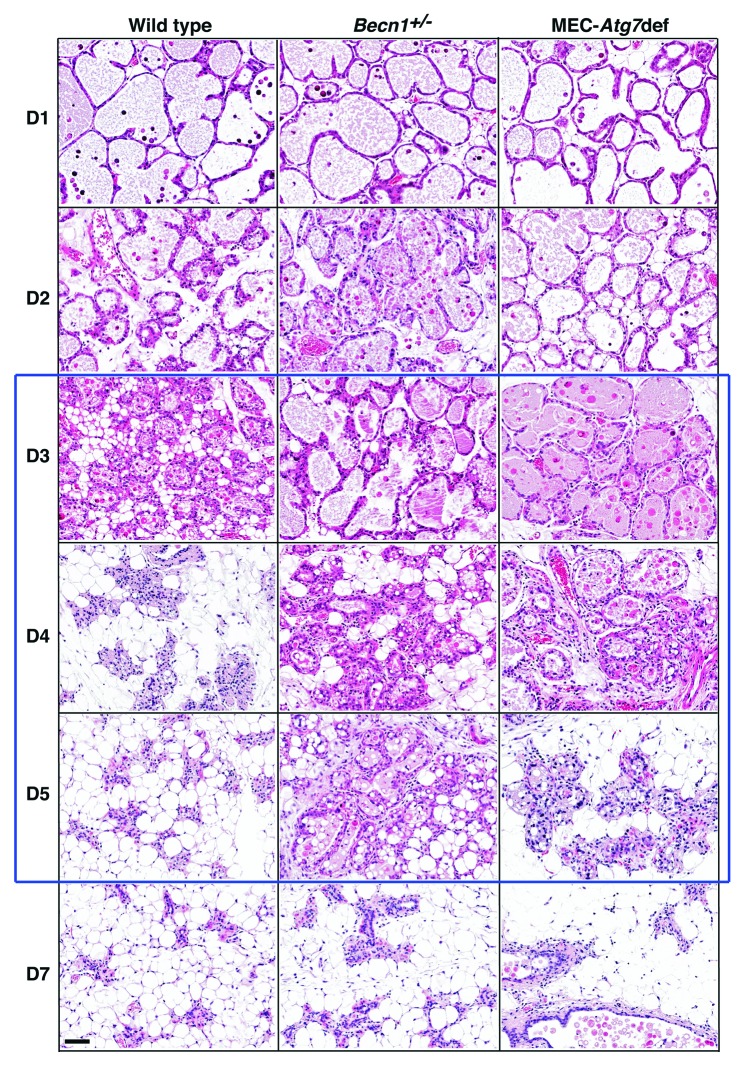
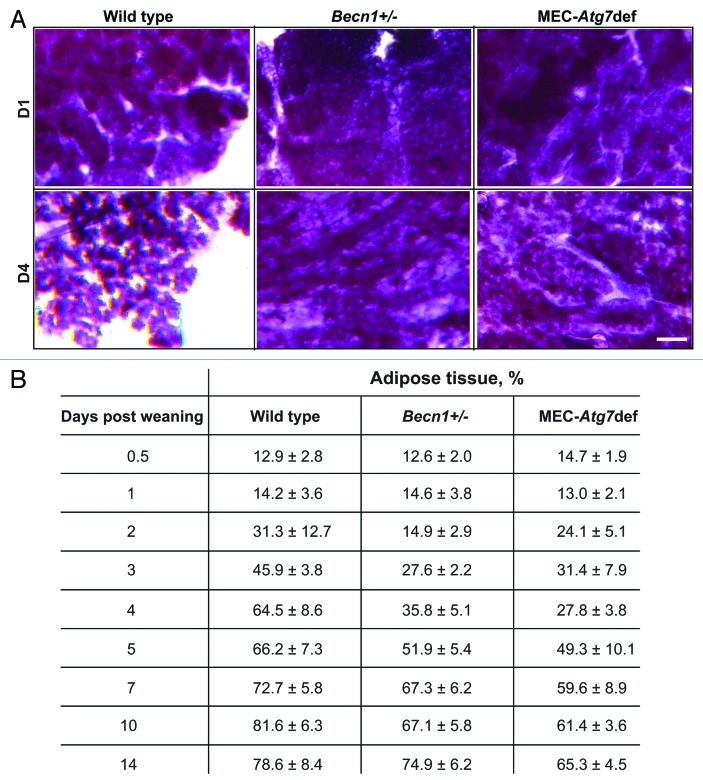
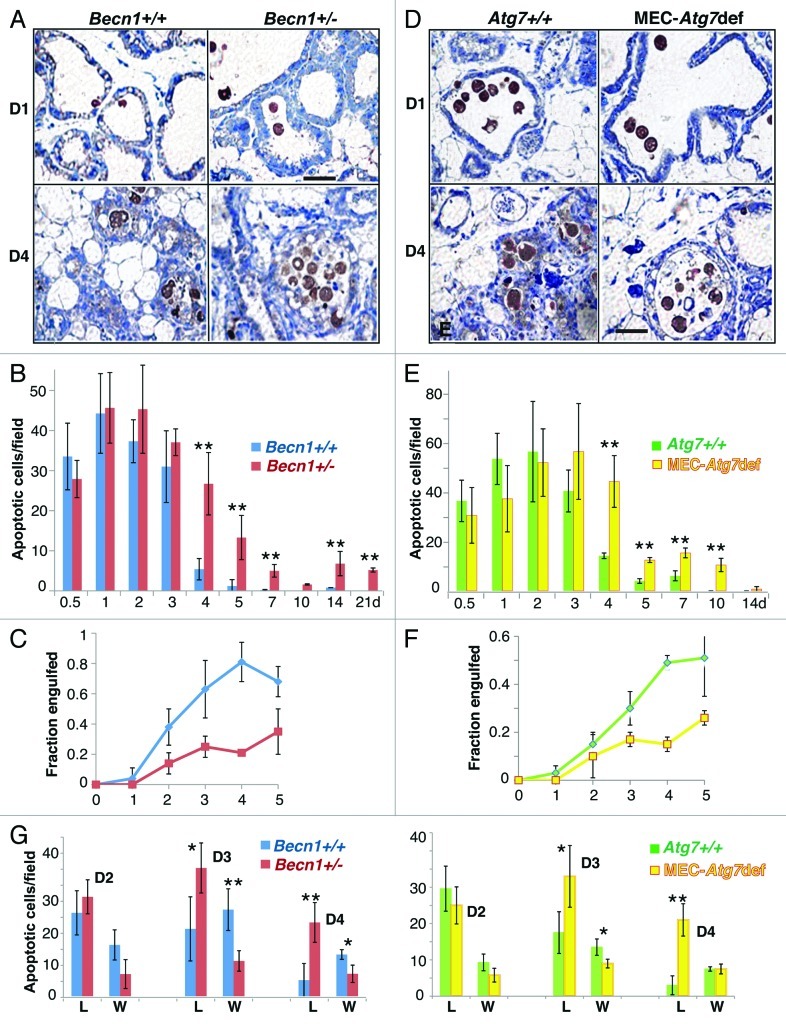
To investigate how Atg defects impact mammary involution, we examined wild-type, Becn1+/− and MEC-specific Atg7-deficient mammary glands at successive involution time points (Figs. 3 and 4). Transient involution delays, most apparent between days 3 and 5 post-weaning, were observed in both Atg-deficient genetic backgrounds. Involuting gland morphology appeared grossly unaffected by autophagy functional status within the first 48 h. However, Becn1+/− and MEC-specific Atg7-deficient mammary glands demonstrated abnormally distended milk- and dead cell-filled alveoli for at least 72 and 96 h post-weaning, respectively (Fig. 3) and an accompanying delay in stroma remodeling (Fig. 4). Becn1+/− and MEC-specific Atg7-deficient glands were repopulated by adipocytes to wild-type levels after 5 and 10 d, respectively (Fig. 4), indicating that MEC-specific Atg7 deficiency resulted in longer, though still temporary, gland remodeling delay than ubiquitous monoallelic Becn1 loss.
Figure 3. Monoallelic Becn1 loss or MEC-specific Atg7 deletion delays mammary involution. H&E-stained sections of formalin-fixed and paraffin-embedded mammary tissues from parous wild-type, Becn1+/− and Atg7F/F;WAP-Cre mice on days 1–7 post-weaning. Parous mice: after second pregnancy and lactation.
Figure 4. Monoallelic Becn1 loss or MEC-specific Atg7 deletion delays mammary involution. (A) Whole mammary gland mounts from parous wild-type, Becn1+/− and Atg7F/F;WAP-Cre mice on days 1–4 post-weaning. (B) Mammary gland repopulation by adipocytes (adipose tissue/total mammary tissue, %) during days 0.5–14 post-weaning.
To gain insight into the mechanism(s) by which defects in autophagic machinery proteins delay involution, MEC apoptosis was quantified (Fig. 5A, B, D and E). During the first 72 h post-weaning, MECs exhibited high apoptotic rates, independent of autophagic responsiveness (Fig. 5B and E). Although autophagy is regularly induced as a cytoprotective mechanism under stress,37-40 the observation that autophagy-defective MECs were not more susceptible than their wild-type counterparts to post-lactational cell death was not surprising, given that the prosurvival effects of autophagy become readily apparent only in an apoptosis-defective background.37-40 Quantification of apoptotic body (AB) clearance by neighboring, still living, MECs revealed that Atg-deficient MECs exhibited decreased engulfment compared with wild-type MECs after the first 24 h post-weaning, i.e., after the peak of autophagosome formation (Fig. 5C and F). Furthermore, engulfment difference between wild-type and Atg-deficient MECs continually increased between 48 and 96 h, remaining high at least until involution day 5 and probably accounting for the persistently high AB numbers in Becn1+/− and MEC-specific Atg7-deficient mammary glands beyond 72 h; abnormal intraluminal dead cell accumulation (Fig. 5G) and milk retention (Fig. 3) were in turn the most likely reasons behind delayed alveolar collapse in Atg-deficient mammary glands (Fig. 3).
Figure 5. Involution delay in Atg-deficient mammary glands is associated with dead cell accumulation in alveolar lumens. (A) Cleaved CASP3 IHC on mammary glands from wild-type and Becn1+/− mice on involution days 1 and 4. (B) Quantification of post-lactational apoptotic bodies in mammary glands from wild-type and Becn1+/− mice during involution days 0.5–21. (C) Fraction of apoptotic bodies engulfed by living MECs in mammary glands from wild-type (blue line) and Becn1+/− (red line) mice during involution days 1–5. (D) Cleaved CASP3 IHC on mammary glands from wild-type and MEC-specific Atg7-deficient mice on involution days 1 and 4. (E) Quantification of post-lactational apoptotic bodies in mammary glands from wild-type and MEC-specific Atg7-deficient mice during involution days 0.5–21. (F) Fraction of apoptotic bodies engulfed by living MECs in mammary glands from wild-type (green line) and MEC-specific Atg7-deficient (yellow line) mice during involution days 1–5. (G) Apoptotic body distribution between non-engulfed (i.e., in alveolar lumens, L) and engulfed (i.e., inside MECs in alveolar wall, W) fractions in mammary glands from wild-type (blue and green bars), Becn1+/− (red bars) and MEC-specific Atg7-deficient (yellow bars) mice during days 2–4 post-weaning. **p 0.001, *p 0.01.
Atg defects compromise MEC phagocytic ability in vivo without impairing PS exposure or MFGE8 levels, but in association with decreased expression of apoptotic body receptors on phagocytes
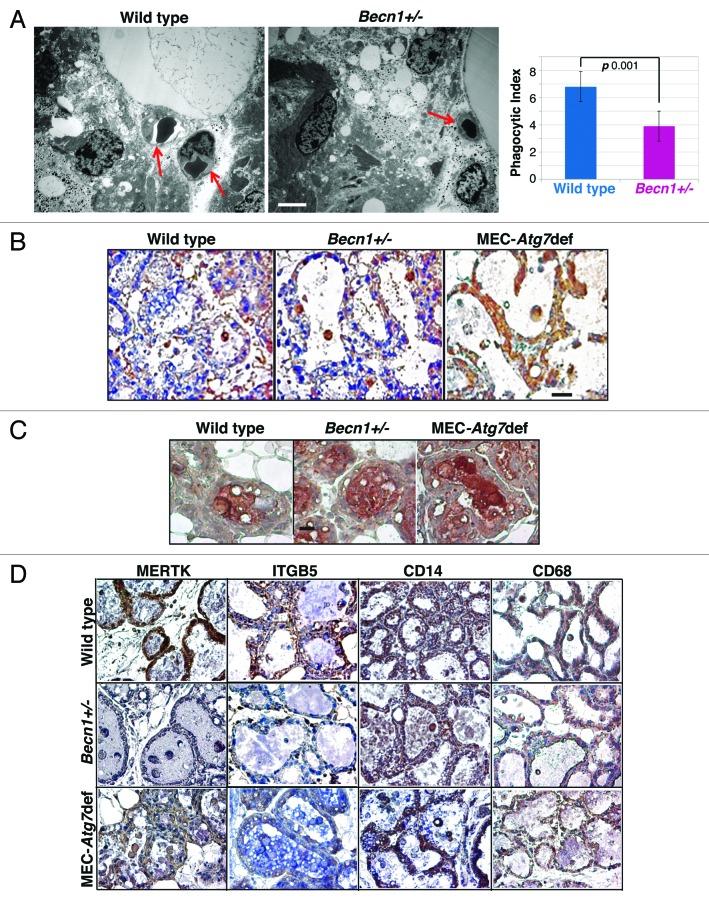
The in vivo phagocytic ability of wild-type and Atg-defective MECs during involution was quantified and a significantly decreased phagocytic index for Becn1+/− MECs was observed (Fig. 6A). Phagocytic index quantification by EM was performed only for Becn1+/− glands, as in this model all MECs were Becn1-heterozygous, whereas in the ATG7 model, despite the well-documented and efficient transgene deletion in the secretory epithelial cells,35,36 individual MECs scored for phagocytic ability could be wild type and might have negatively impacted the accuracy of phagocytic index determination.
Figure 6.Atg-deficiency impairs efferocytosis during mammary involution in association with decreased MERTK and ITGB5 levels, but without compromising PS exposure on apoptotic bodies or MFGE8 expression. (A) (Left panel) EM micrographs of Becn1+/+ and Becn1+/− mammary glands on day 2 post-weaning. Red arrows point to autophagosomes (engulfed by MECs in wild type, non-engulfed in Becn1+/− gland); (Right panel) Phagocytic index, i.e., fraction of engulfed over total number of autophagosomes in HPF. (B) Annexin V IHC on mammary glands from wild-type, Becn1+/− and MEC-specific Atg7-deficient mice on day 2 post-weaning. (C) MFGE8 IHC on mammary glands from wild-type, Becn1+/− and MEC-specific Atg7-deficient mice on day 3 post-weaning. (D) MERTK, ITGB5, CD14 and CD68 IHC on mammary glands from parous wild-type, Becn1+/− and MEC-specific Atg7-deficient mice at 48 h post-weaning.
To elucidate the mechanism underlying the dead cell clearance defect in Atg-deficient involuting mammary glands, we serially investigated the functionality of different efferocytosis players,41 starting with the apoptotic cells, which are known to ‘actively’ participate in their own removal by exposing ‘eat-me’ receptors (PS) on their cell membranes, thus providing recognition signals to neighboring phagocytes. Wild-type, Becn1+/− and MEC-specific Atg7-deficient involuting mammary glands showed similar PS exposure on dead cells (Fig. 6B), indicating that Atg-defective MECs give rise to “phagocytosis-competent” ABs in vivo.
Milk fat globule-EGF-factor 8 (MFGE8) is a glycoprotein at the apical plasma membrane of secretory MECs and on the milk fat globule membrane (MFGM) around secreted milk fat that acts as an opsonin and facilitates AB clearance; MFGE8 deficiency results in dead cell accumulation during involution,42 similar to that described above for Atg-deficient mammary glands (Fig. 5A, D and G). MFGE8 protein levels in wild-type and Atg-deficient mammary tissues were similar (Fig. 6C), thus excluding altered MFGE8 expression as contributing to AB accumulation in Atg-defective mammary glands.
We next examined the expression of AB receptors on phagocytes, concentrating on those implicated in MEC-mediated efferocytosis,10,27,43 and found that MERTK and integrin β5 (ITGB5) exhibited decreased levels in Atg-deficient mammary glands, whereas other receptors, such as CD14, which is upregulated during involution and becomes exposed on the surface of MECs in a STAT3-dependent manner,10,19 and CD68 showed genotype-independent expression (Fig. 6D).
Persistence of inflammation and ductal ectasia in Atg-defective mammary glands
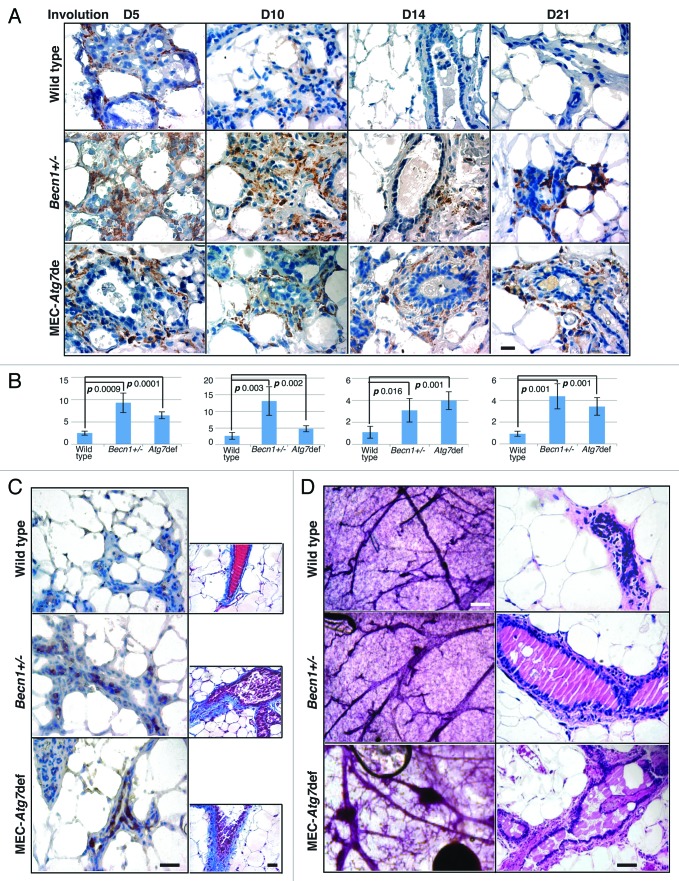
Defective dead cell clearance commonly results in inflammation, as secondary necrosis and highly immunogenic intracellular antigen exposure occur in parallel with decreased anti-inflammatory cytokine secretion by phagocytes.44 To investigate whether dead cell accumulation in Atg-defective mammary glands altered the inflammatory response normally associated with involution,27 macrophage infiltration was assessed (Fig. 7A and B). In wild-type glands, macrophages were clearly present until involution day 10, but depleted in fully involuted glands, whereas in Atg-deficient mammary tissues, macrophages persisted past involution completion (Fig. 7A and B), were of the alternatively activated M2 subtype45 (Fig. 7C, left column), and were accompanied by increased collagen deposition (Fig. 7C, right column). Atg-deficient mammary glands from older multiparous mice demonstrated prominent ductal ectasia (Fig. 7D), which—though not a preneoplastic condition itself—is indicative of abnormal ductal remodeling in association with defective dead cell clearance during mammary involution.
Figure 7. Involution delay due to Atg-deficiency is accompanied by persistence of inflammation, M2-macrophage infiltration, increased collagen deposition and ductal ectasia. (A) F4/80 IHC on mammary glands from wild-type, Becn1+/− and MEC-specific Atg7-deficient mice on days 5–21 post-weaning. (B) Macrophage quantification in mammary glands from wild-type, Becn1+/− and MEC-specific Atg7-deficient mice on days 5–21 post-weaning. (C) ARG1 IHC (left column); Mason trichrome staining (right column) on mammary glands from wild-type, Becn1+/− and MEC-specific Atg7-deficient mice on day 21 post-weaning. (D) Whole gland mount (left column) and H&E-stained sections (right column) of mammary glands from parous wild-type, Becn1+/− and MEC-specific Atg7-deficient mice.
ATG protein knockdown compromises iMMEC phagocytic ability in vitro in association with decreased apoptotic body engulfment and impaired Rac activation
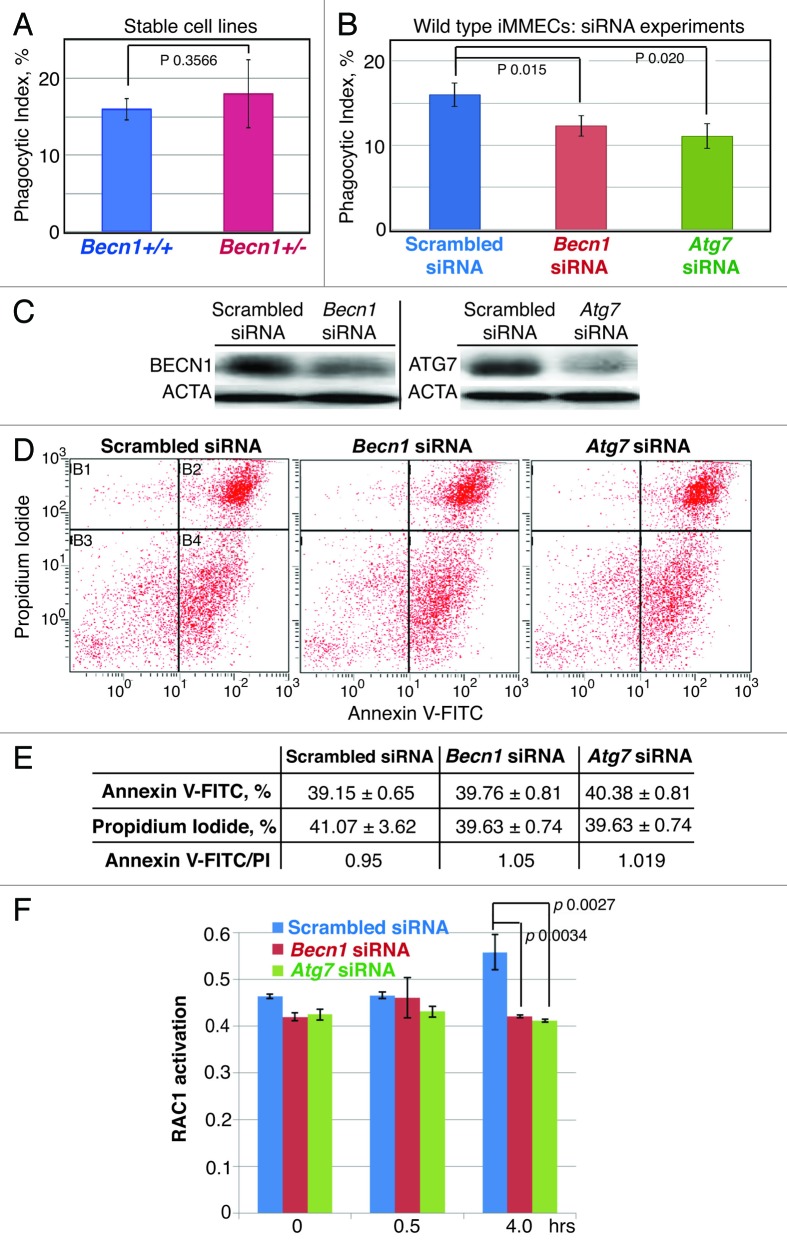
To further investigate the mechanism(s) underlying the in vivo efferocytosis defect of Atg-defective MECs, we initially used iMMEC cell lines derived from Becn1+/+ and Becn1+/− mice.39 To our surprise, Becn1+/− iMMECs, which are partially defective in autophagy induction under metabolic stress,39 did not exhibit any significant phagocytosis impairment (Fig. 8A). This result, though unexpected, has a precedent in earlier work,46 where the PS exposure defect documented in Becn1- and Atg5-null ES cells in a 3D-model of embryoid body cavitation was not observed in single layer ES cell culture treated with apoptosis-inducing agents, thus indicating that in vivo properties of Atg-defective cells may be altered in regular 2D-culture.
Figure 8. BECN1 or ATG7 knockdown in wild-type iMMECs compromises efferocytosis in vitro without affecting PS exposure on apoptotic bodies, but in association with decreased engulfment-related RAC1 activation. (A) Phagocytic index for stable Becn1+/− and Becn1+/− iMMEC lines in vitro. (B) Phagocytic index for wild-type iMMECs treated with scrambled or Atg (Becn1 or Atg7) siRNA. (C) Quantification of BECN1 and ATG7 knockdown in wild-type iMMECs. (D and E) FACS-based quantification of PS exposure on apoptotic bodies derived from wild-type iMMECs treated with scrambled or Atg (Becn1 or Atg7) siRNA. (F) Efferocytosis-induced RAC1 activation in wild-type iMMECs treated with scrambled or Atg (Becn1 or Atg7) siRNA.
In contrast, BECN1 or ATG7 knockdown (KD) by siRNA decreased the phagocytic index of wild-type iMMECs (Fig. 8B and C). Furthermore, similar to the in vivo results (Fig. 6B), BECN1 or ATG7 KD did not affect PS exposure on resultant ABs (Fig. 8D and E). Given decreased MERTK and ITGB5 levels in Atg-defective mammary glands during involution in vivo (Fig. 7E) and, thus, predicted impairment of their downstream effector RAC1, which mediates actin reorganization and phagocytic cup formation for effective dead cell engulfment,47 we investigated whether BECN1 or ATG7 KD affected RAC1 activation during efferocytosis in vitro and found that in both cases RAC1 upregulation was significantly compromised (Fig. 8F), again indicating that impaired efferocytosis due to Atg defects can be attributed, at least partially, to defective engulfment.
Discussion
Using the Gfp-Lc3 autophagy-reporter mouse model,28 we found that autophagy is induced shortly after weaning and is accompanied by transcriptional upregulation of ATG proteins. Our findings are in agreement with earlier DNA microarray studies,10,48 which included data on induction of ATG proteins and their regulators during early involution in parallel with downregulation of autophagy suppressors, such as AKT and MTOR. MEC apoptosis is maximal and autophagy status-independent during the first 72 h post-weaning (Fig. 5B and E). In wild-type mammary glands, apoptotic cell number drops acutely by involution day 4 and nears zero by day 5, suggesting that most dead cells are efficiently cleared between 72 and 96 h, i.e., after the peak in autophagosome formation and prior to significant macrophage infiltration from external sites.8 In contrast, in Becn1+/− or MEC-specific Atg7-deficient mammary glands, dead cells abnormally accumulated at involution days 4 and 5, indicating that Atg-deficient mammary tissues exhibited defective efferocytosis. It is of great interest that a recent study demonstrates the essential role of macrophages during the first phase of involution;49 our results complement this study, as well as earlier work,8,43 in showing that both MECs and macrophages have critical contributions to proper execution of this important developmental process.
Accelerated apoptosis cannot be ruled out as a reason behind dead cell accumulation in Atg-defective glands; however, the finding that Becn1+/− and Atg7-deficient MECs exhibit decreased dead cell engulfment even at 48 and 72 h post-weaning (Fig. 5C and F), when the total number of ABs is Atg-independent (Fig. 5B and E), clearly indicates a defect in dead cell clearance in Atg-deficient mammary glands.
An earlier study reports that Atg defects result in cell corpse persistence due to failed AB clearance during embryonic cavitation.46 In that case, dying cells in Atg5- or Becn1-null embryoid bodies (EBs) fail to generate energy-dependent engulfment signals, such as the ‘eat-me’ (PS exposure) and the ‘come-get-me’ (LPC secretion) signals, and the efferocytosis defect is attributed to the production of ‘defective’ ABs by Atg-defective cells. However, in the afore-mentioned study, other mechanisms potentially impairing AB clearance were not investigated, leaving open the possibility of defects in other dead cell clearance effectors, such as phagocytes.
Our results demonstrate proper PS exposure on Atg-deficient ABs in vivo (Fig. 6B) and in vitro (Fig. 8B) and, together with Atg-independent expression of the opsonin MFGE8 (Fig. 6C and D), indicate that defective phagocytes account for the efferocytosis impairment. This finding is in close agreement with studies on dead cell34 and pathogen29 phagocytosis by macrophages, apoptotic corpse clearance during C. elegans development,32,33 and the live-cell-engulfment process of entosis.32 The novel contribution of our work is in reporting for the first time that Atg defects in MECs impair efferocytosis during mammary involution in vivo at the early step of engulfment. Our results are greatly supported by a completely independent study that has recently reported involvement of BECN1 in engulfment of apoptotic cells by macrophages and fibroblasts in vitro.50 Thus, inefficient efferocytosis—and by extension, phagocytosis—likely involves more aspects of the phagocytic process than the previously documented endocytic processing. Although we did not investigate processing in Atg-deficient MECs, it is quite likely defective, given the emerging literature mentioned above. The combination of engulfment and processing defects in Atg-deficient MECs is very intriguing, especially in light of recent work indicating that engulfment depends on apoptotic material processing.51 In support of this hypothesis, we never detected Becn1+/− MECs “stuffed” with ABs; instead in all cases examined, multiple apoptotic corpses were near or on the phagocyte, but only one was ingested, as if engulfment rate were strictly regulated. Whether the engulfment defect in Atg-deficient MECs is primary or secondary to a feedback loop controlled by endocytic processing will be the subject of future studies. Elucidation of the nature of the engulfment defect in Atg-deficient MECs and its etiologic connection to decreased MERTK and ITGB5 expression will also be investigated.
Autophagy protein defects delay mammary involution
Autophagy induction was previously documented in involuting bovine mammary glands,52 but our studies are the first to demonstrate the functional role of ATG proteins in mammary involution using transgenic mouse models. Participation of the autophagic machinery in involution process is not surprising given accumulating evidence for the critical role of autophagy in diverse developmental events,53 such as spore formation in yeast,54,55 dauer formation in C. elegans,56 oocyte-to-embryo57 and embryo-to-neonate58-61 transitions in mammals. Autophagy induction during developmental tissue remodeling often accompanies apoptosis, as exemplified by germline P granule degradation in somatic cells in C. elegans62,63 and by salivary gland63 and midgut64 degradation in Drosophila; furthermore, autophagy is essential for midgut cell death even when apoptosis is inhibited.64 The latter studies implicate autophagy as an active PCD mechanism in Drosophila, which is a an intriguing finding given the more commonly observed cytoprotective roles of autophagy.65 In our studies, autophagy and apoptosis are induced in parallel during mammary involution and whole organ or MEC-specific Atg defects transiently delay mammary gland remodeling due to impaired dead cell clearance after the peak of autophagosome formation and without affecting apoptotic rates. Thus, although autophagy as a process of self-eating is not essential for MEC death, ATG proteins are required for AB clearance and, thus, act as late PCD and heterophagy effectors.66 In this respect, our studies shed light into the function of ATG proteins as mediators of efferocytosis which, though related to and sharing machinery with, is a process distinct from autophagy, as it does not involve self-cannibalism. Quite interestingly, whole mounts of Atg-deficient involuting mammary glands (Fig. 4) show persistent mammary tissue compared with wild-type glands, macroscopically reminiscent of the developmentally nondegraded atg-mutant Drosophila salivary glands.63 Given the more recently reported requirement for the engulfment receptor Draper in activation of autophagy regulators and degradation of Drosophila salivary glands67 and our results of impaired efferocytosis in Atg-deficient mammary glands in association with decreased MERTK and ITGB5 expression, it is conceivable that Drosophila salivary gland degradation and mammary involution may be very similar in their utilization of ATG proteins for efficient cell clearance during development. Further investigation will provide critical insight into the similarities and differences between these two developmentally important PCD processes.
Mammary epithelial cells as nonprofessional phagocytes
In the mammary gland, epithelial cell death followed by corpse clearance is a frequent event, first observed in puberty, when solid epithelial cords are canalized to form ductal outgrowths,68 and then recurring through the entire female reproductive life with each menstrual/estrous cycle, during which the mammary epithelium proliferates moderately and either continues to do so if pregnancy occurs or undergoes apoptosis.69 The most extreme form of this cell proliferation-followed-by-cell death process happens during pregnancy, lactation and subsequent involution, at which time AB clearance is essential for proper stroma remodeling and mammary gland regression to a “quiescent” state.1 It is of great interest that the cells responsible for dead body removal are living MECs acting as nonprofessional phagocytes.8,27 Recent work using mice deficient for different receptor tyrosine kinases that regulate efferocytosis in macrophages showed that MERTK, but not AXL or TYRO3, is critical for MEC-mediated efferocytosis in the postpartum mammary gland,43 a finding that is in agreement with our observation that MERTK and ITGB5 expression was suppressed in Atg-deficient involuting mammary glands. Indeed the Mertk-null mice exhibited the same post-lactational involution phenotype as our Atg-deficient models: AB accumulation and stagnant milk in alveolar lumens by involution day 3, followed by eventual dead cell and debri clearance in association with epithelial and stromal alterations. In the MERTK and BECN1 models, both MECs and macrophages were mertk−/− and Becn1+/−, respectively, raising the possibility that defective professional phagocytes were at least partially responsible for impaired efferocytosis. Transplantation of mertk−/− mammary epithelium into wild-type mammary fat pads resulted in a similar dead-cell-accumulation phenotype, suggesting that MECs are the primary efferocytes during involution. Our ATG7 mammary model combines phagocytosis-defective (atg7−/−) MECs and wild-type stroma in the same transgenic mouse for the first time and, thus, proves beyond a doubt that MECs are the primary efferocytes during involution.
Involution-associated inflammation
Transient inflammation is an undisputable characteristic of mammary involution, as macrophages infiltrate the mammary gland by day 4 post-weaning and orchestrate stroma remodeling. Increased immune mediator expression during involution is consistent with an immune cascade resembling an early acute-phase response and a wound healing process.10,15,48 In breast cancer, macrophage infiltration and PABC are both associated with poor clinical outcomes, an observation that led to the involution-hypothesis stating that the wound healing microenvironment of the involuting mammary gland is tumor-promotional and implicated in the high rate of associated distant metastasis.11,12 Examination of rodent and human mammary tissues revealed that involution macrophages and their associated stroma exhibit characteristics possibly driving breast cancer progression, such as an alternatively activated M2 phenotype, IL4 and IL13 proinflammatory cytokine production, fibrillar collagen accumulation and high cyclooxygenase 2 (PTGS2) expression.45,70 Our studies revealed that ineffective AB clearance due to Atg deficiencies result in inflammation that persists beyond full mammary gland regression (Fig. 7A–C), indicating that ATG protein defects may alter the tumor-modulating properties of the mammary microenvironment during involution and, thus, affect tumor progression risk. Atg-deficient mammary glands from retired breeders exhibit ductal ectasia, which, although not considered a preneoplastic change, is a manifestation of abnormal mammary gland remodeling. At this point, the etiologic origin of the ductal ectasia phenotype cannot be precisely defined, as more than one process can be contributing to it, including the intraductal persistence of ABs and its resultant proinflammatory response and/or the chronic inflammation due to defective autophagy-induced cell susceptibility to metabolic and tissue stressors. Along the same lines, not only inflammation may follow decreased AB clearance, but also defective autophagy-associated inflammation may impair the phagocytic ability of MECs, leading to a self-sustaining proinflammatory microenvironment. Whether impaired efferocytosis and persistent inflammation due to Atg deficiencies are mechanistically connected to lower BECN1 expression in human breast cancer71 remains to be investigated.
Materials and Methods
Mice and induction of involution
Becn1+/−,72Atg7F/F;WAP-Cre73 and Gfp-Lc374 mice were previously described.
PCR primers for genotyping were as following:
Becn1:
NeoESPCR2: 5′- CGC CTT CTA TCG CCT TCT TGA CG-3′
BCNKOT-1C: 5′-TGG AGG GCA GTC CAT ACC CTG-3′
BCNKOT-2R: 5′-GAG CTG GCT CCT GTG AGT ATG-3′
Atg7F:
Hind-Fw: 5′-TGG CTG CTA CTT CTG CAA TGA TGT-3′
Pst-Rv: 5′-CAG GAC AGA GAC CAT CAG CTC CAC-3′
Cre:
C001: 5′-ACC AGC CAG CTA TCA ACT CG-3′
C002: 5′-TTA CAT TGG TCC AGC CAC C-3′
Gfp-Lc3:
GFP(LC3)1: 5′-TCC TGC TGG AGT TCG TGA CCG-3′
LC3*rc3: 5′-TTG CGA ATT CTC AGC CGT CTT CAT CTC TCT CGC-3′
Mice were housed in a pathogen-free facility at the Child Health Institute of New Jersey. Animal protocols were IACUC-approved. Atg7F/F;WAP-Cre mice were used after two pregnancy-lactation cycles to ensure maximum CRE-recombination.35,36 For involution studies, litter size was normalized to 6–8 pups. After 14 d of nursing,8,42,45 dams were separated from their pups. Thoracic and abdominal mammary glands were harvested at 0–21 d time points after forced weaning, 3–5 mice per time point. Mammary glands were also harvested from nulliparous and multiparous mice at older ages.
qRT-PCR
Protein and RNA were isolated from nulliparous, lactating and involuting Becn1+/− and MEC-specific Atg7-deficient mammary glands using the PARIS kit (Applied Biosystems). Brilliant II SYBR Green 2-step QRT-PCR with AffinityScript Master Mix (Agilent) was used to convert 100 ng RNA to cDNA using a Veriti Thermal Cycler (Applied Biosystems). Relative mRNA expression was analyzed on a Stratagene MX3005P (Agilent), normalized to a housekeeping gene (Acta1) and compared with mRNA from virgin mammary glands using the ΔΔCt method performed by the Comparative module on the MxPro software (Agilent).
Primers (Integrated DNA Technologies) used were as following: Acta1 Forward: 5′-GGTATCGCT GACCGCATGCAGA-3′, Reverse: 5′-CCTGAG GAGAGAGAGCGCGAACG-3′; Becn1 Forward: 5′-CTCAAGTGGGGTCTTGCCTGGG T-3′ Reverse: 5′-GAAAGGATGCATGGGGCT GAGTC-3′; Map1lc3α Forward: 5′-TAATCAGACGG CGCTTGCAGCT-3′ Reverse: 5′-TGTGGGTGG TGAGCCGTCCA-3′
Cell culture
Becn1+/+ and Becn1+/− mouse mammary epithelial cells (MMECs) were isolated and immortalized (iMMECs) as previously described.39 Wild-type iMMECs were electroporated with scrambled, Becn1 or Atg7 mouse siRNAs (Thermo Scientific, D-001050-01-20; M-055895-01-0005; M-049953-02-0020) for ATG regulator knockdown in vitro.
Cells were seeded on 100-mm plastic dishes, 6-well plates, coverslips or chamber slides depending on specific experiment and grown in F12 medium supplemented with 10% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 5 ng/ml EGF, 100 U/ml penicillin and streptomycin at 37°C in a humidified atmosphere containing 5% CO2. To induce apoptosis, cells were incubated in glucose/serum-free Dulbecco’s modified Eagle’s medium (DMEM) supplemented with hormones and antibiotics for 14 h.
Mammary gland processing and analysis
Whole mount: Freshly dissected abdominal mammary glands were spread between glass slides and fixed in 4% paraformaldehyde (PFA) at room temperature (RT) for 2 h. After acetone defatting and alcohol rehydration, tissues were stained overnight in carmine alum solution (Carmine, Sigma C-1022; aluminum potassium sulfate, Sigma A-7167) followed by dehydration and clearing in xylene.
For H&E and IHC, glands were fixed in 4% PFA, embedded in paraffin and sectioned. H&E staining followed by digital scanning was performed at the Mouse Pathology Laboratory, University of California, Davis.
For frozen sections, tissues were incubated in sucrose gradient prior to processing. Sections were fixed in acetone and mounted with ProLong Gold antifade reagent (Invitrogen, P36930). EGFP-LC3 fluorescence was monitored by epifluorescence microscopy.
For IHC, sections were deparaffinized in xylene, rehydrated with alcohol and blocked with 3% peroxide, followed by 1 h incubation with 5% goat serum or 5% BSA, depending on primary antibody source. Vector M.O.M. Immunodetection kit (Vector Laboratories, BMK-2202) was used for rat primary antibody.
Tissue sections were incubated at RT for 1 h or at 4°C overnight with the following primary antibodies: rabbit polyclonal anti-ATG7 (Sigma, A2856), rabbit polyclonal anti-BECN1 (Santa Cruz, sc-11427), rabbit polyclonal anti-LC3 (Novus, NB100-2331), rabbit polyclonal anti-cleaved caspase-3 (Cell Signaling, 9661), rat monoclonal anti-F4/80 (AbD Serotec, A3-1, MCA497), rabbit polyclonal anti-iNOS/NOS Type II (BD Transduction Laboratories, 610333), goat polyclonal anti-Arginase-1 (Santa Cruz, sc-18351), rabbit polyclonal anti-MERTK (FabGenuix, MKT-121AP), rabbit polyclonal anti-ITGB5 (Santa Cruz, sc-14010), rabbit polyclonal anti-CD14 (Abcam, ab106285), rat monoclonal anti-CD68 (AbD Serotec, FA-11, MCA-1957GA), rabbit monoclonal anti-MFGE8 (kindly provided by Dr. Kamran Atabai, University of California, San Francisco). Sections were further incubated at RT for 1 h with the following biotinylated antibodies: goat polyclonal anti-rabbit (Novus, NB 730-B) or bovine anti-goat (Jackson ImmunoResearch, 711-065-152). Antibodies were diluted in 5% goat serum, 5% BSA or M.O.M. diluent, depending on specific antibody source. Vectastain Elite ABC Reagent (Vector Laboratories, PK-7100) and DAB+ Substrate Chromogen Kit (Dako, K3468) were used to visualize biotin signal.
For collagen staining, deparaffinized and rehydrated sections were incubated overnight at RT in Bouin’s solution (Sigma, HT10132-1L) prior to Masson trichrome staining (Sigma Accustain Trichrome Kit HT15).
For phosphatidylserine (PS) detection in vivo, glands were incubated in Annexin-V-Biotin in Ca-binding buffer (TACS Annexin V-Biotin kit, Trevigen 4835-01-K) according to the manufacturer’s instructions prior to fixation with 4% PFA, paraffin-embedding and sectioning. Dewaxed and rehydrated sections were incubated with 3% peroxide, rinsed and blocked with 5% goat serum, incubated at 4°C overnight with Vectastain Elite ABC Reagent, rinsed with 0.5% Tween-20 in PBS and developed in DAB+ Substrate.
For all staining described above, sections were mounted with Permount medium (Fisher, SP15-100).
Electron microscopy
1-mm3 specimens were excised from 3 randomly selected areas of abdominal mammary glands, fixed in 2.5% glutaraldehyde, 4% PFA, 8 mM CaCl2 in 0.1 M cacodylate buffer pH 7.4, and processed for conventional electron microscopy at UMDNJ Electron Microscopy Core facility (JEOL 1200EX electron microscope).
Phosphatidylserine analysis in vitro
Wild-type iMMECS transfected with scrambled, Becn1 or Atg7 siRNAs were grown in 6-well plates in regular growth medium for 48 h, then in serum/glucose-free DMEM to initiate apoptosis. Apoptotic cells were harvested 14 h later and stained (unfixed) with FACS Annexin V-FITC kit (Trevigen, 4830-01-K). FITC /PI cell ratio was determined within 1 h using Beckman-Coulter Cytomics FC500 Flow Cytometer. To visualize PS exposure, cells were grown on coverslips, starved overnight, stained with Annexin V-FITC/PI, and mounted with ProLong Gold antifade medium.
Macrophage quantification
Macrophage infiltration was quantified by using the measurement option of the ProgRes CapturePro 2.5 camera software. Basically, F4/80-stained areas for macrophages and hematoxylin-stained areas for mammary tissues were measured on paraffin-embedded specimen sections (images taken at 400x magnification). Ten fields/section and one section from 3 different mice per genotype per involution time point were analyzed. Results are presented as macrophage (F4/80 staining) to tissue (hematoxylin staining) ratios.
Phagocytosis assays
FACS-based assay: iMMECs transfected with scrambled, Becn1 or Atg7 siRNA recovered for 48 h post-transfection. Wild-type iMMECs labeled with red fluorescent cell linker PKH26 (Sigma, MINI26) were maintained in glucose/serum-free DMEM for 36 h for apoptosis induction. Cell death was verified by Annexin V-FITC/PI staining. PKH26-labeled apoptotic cells were added in 1:10 ratio to siRNA-transfected cells growing in regular growth medium and co-cultured for 14 h. Phagocyte monolayer was washed with PBS to remove bound, nonengulfed, apoptotic cells, stained with CellTracker Green BODIPY dye (Invitrogen, C2102), harvested by trypsinization and fixed with 1% PFA for 20 min on ice. Samples were acquired on an Amnis Imagestream 100 and analyzed by IDEAS 4.0 software. Internalization was determined by first gating for nuclear stain (Draq5) to identify intact cells. These were then analyzed for BODIPY Green-positive cells (phagocytes) by gating for Intensity BODIPY Green. Single cells were selected to remove large aggregates and doublets and then analyzed for percent PKH26-positive cells by gating for Intensity PKH26 vs. Area Draq5 to exclude conjugates and aggregates. PKH26-positive events were gated off of BODIPY Green cells alone as a control.
RAC1 activation assay
Cells were grown and siRNA transfections were performed as described above. Wild-type apoptotic cells were added in 1:10 ratio; phagocytosis was terminated by ice-cold PBS wash. Phagocyte monolayer was washed with PBS to remove bound, nonengulfed apoptotic cells and harvested by scraping at 0, 0.5 and 5 h time points after apoptotic cell addition. Assay was performed with G-LISA RAC Activation Assay Biochem (absorbance-based kit, Cytoskeleton BK125) according to manufacturer’s manual, except that cells were grown in serum-containing medium to avoid starvation-induced effect on autophagy status. Coverslips were stained for actin with Rhodamine Phalloidin (Cytoskeleton, PHDR1).
Statistical analysis
Student’s t-test was used for statistical analysis.
Acknowledgments
I.T., F.L., S.S., B.R. and V.K. designed and analyzed experiments; I.T., F.L., S.S. and S.P. performed experiments; N.B. and R.D.C. reviewed mouse mammary gland histology; V.K. supervised project; V.K. wrote manuscript; all authors reviewed manuscript.
We thank N. Mizushima (Tokyo Medical and Dental University; Gfp-Lc3 mice), Z. Yue and S. Jin (Mount Sinai School of Medicine and University of Medicine and Dentistry of New Jersey; Becn1 mice), M. Komatsu (Tokyo Metropolitan Institute of Medical Science; Atg7F/F mice), K. Atabai (University of California, San Francisco; MFGF-8 antibody). We also thank R. Patel for assistance with electron microscopy, N. Goldsmith-Kane for assistance with confocal microscopy, K. Bell at the Center for Comparative Medicine, University of California at Davis for assistance with mammary tissue processing and fixation, the Cancer Institute of New Jersey Tissue Analytical Services and the Flow Cytometry and Immunology Core Laboratory at University of Medicine and Dentistry of New Jersey-New Jersey Medical School. We are grateful for financial support from a New Jersey Commission on Cancer Research Predoctoral Fellowship (F.L.), NIH grant R00 (V.K.) and a Damon Runyon Clinical Investigator Award (V.K.).
Glossary
Abbreviations:
- AB
apoptotic body
- AKT
v-akt murine thymoma viral oncogene homolog/PKB
- ATG
autophagy-related
- ARG1
arginase 1
- AXL
AXL receptor tyrosine kinase
- BECN1
Beclin 1
- BSA
bovine serum albumin
- CaCl2
calcium chloride
- CASP3
caspase 3
- CD
cluster of differentiation
- DAB
3, 3′-diaminobenzidine
- DAPI
4’,6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle's medium
- EGF
epidermal growth factor
- EM
electron microscopy
- ES
embryonic stem
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GFP
green fluorescent protein
- H&E
hematoxylin and eosin
- HPF
high power field
- IGFBP
insulin growth factor-binding protein
- IHC
immunohistochemistry
- IL
interleukin
- iMMEC
immortalized mouse mammary epithelial cell
- iNOS
inducible nitric oxide synthase
- I (Inv)
involution
- ITGB5
integrin beta 5
- JAK
janus kinase
- L (Lact)
lactation
- LC3
microtubule-associated protein 1 light chain 3
- LIF
leukemia inhibitory factor
- LPC
lysophosphatidylcholine
- MEC
mammary epithelial cell
- MERTK
c-mer proto-oncogene tyrosine kinase
- MFGE8
milk fat globule-EGF-factor 8
- MFGM
milk fat globule membrane
- MMP
matrix metalloproteinase
- M.O.M.
mouse on mouse
- MTOR
mechanistic target of rapamycin
- RAC1
ras-related C3 botulinum toxin substrate 1
- PABC
pregnancy-associated breast cancer
- PCD
programmed cell death
- PFA
paraformaldehyde
- PI
propidium iodide
- PS
phosphatidylserine
- PTGS2
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- RT
room temperature
- SQSTM1
sequestosome-1/p62
- STAT3
signal transducer and activator of transcription 3
- TGFB
transforming growth factor beta
- TGFBR2
transforming growth factor beta receptor 2
- TYRO3
TYRO3 protein tyrosine kinase
- WAP
whey acidic protein
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23164
References
- 1.Watson CJ. Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Breast Cancer Res. 2006;8:203. doi: 10.1186/bcr1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 3.Lund LR, Rømer J, Thomasset N, Solberg H, Pyke C, Bissell MJ, et al. Two distinct phases of apoptosis in mammary gland involution: proteinase-independent and -dependent pathways. Development. 1996;122:181–93. doi: 10.1242/dev.122.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marti A, Feng Z, Altermatt HJ, Jaggi R. Milk accumulation triggers apoptosis of mammary epithelial cells. Eur J Cell Biol. 1997;73:158–65. [PubMed] [Google Scholar]
- 5.Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, et al. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci U S A. 1997;94:3425–30. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green KA, Lund LR. ECM degrading proteases and tissue remodelling in the mammary gland. Bioessays. 2005;27:894–903. doi: 10.1002/bies.20281. [DOI] [PubMed] [Google Scholar]
- 7.deCathelineau AM, Henson PM. The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem. 2003;39:105–17. doi: 10.1042/bse0390105. [DOI] [PubMed] [Google Scholar]
- 8.Monks J, Smith-Steinhart C, Kruk ER, Fadok VA, Henson PM. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol Reprod. 2008;78:586–94. doi: 10.1095/biolreprod.107.065045. [DOI] [PubMed] [Google Scholar]
- 9.Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41:207–20. doi: 10.1002/mc.20058. [DOI] [PubMed] [Google Scholar]
- 10.Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:R75–91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schedin P, O’Brien J, Rudolph M, Stein T, Borges V. Microenvironment of the involuting mammary gland mediates mammary cancer progression. J Mammary Gland Biol Neoplasia. 2007;12:71–82. doi: 10.1007/s10911-007-9039-3. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien J, Schedin P. Macrophages in breast cancer: do involution macrophages account for the poor prognosis of pregnancy-associated breast cancer? J Mammary Gland Biol Neoplasia. 2009;14:145–57. doi: 10.1007/s10911-009-9118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW, et al. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development. 2003;130:3459–68. doi: 10.1242/dev.00578. [DOI] [PubMed] [Google Scholar]
- 14.Schere-Levy C, Buggiano V, Quaglino A, Gattelli A, Cirio MC, Piazzon I, et al. Leukemia inhibitory factor induces apoptosis of the mammary epithelial cells and participates in mouse mammary gland involution. Exp Cell Res. 2003;282:35–47. doi: 10.1006/excr.2002.5666. [DOI] [PubMed] [Google Scholar]
- 15.Clarkson RW, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6:R92–109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen AV, Pollard JW. Transforming growth factor beta3 induces cell death during the first stage of mammary gland involution. Development. 2000;127:3107–18. doi: 10.1242/dev.127.14.3107. [DOI] [PubMed] [Google Scholar]
- 17.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–16. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology. 2002;143:3641–50. doi: 10.1210/en.2002-220224. [DOI] [PubMed] [Google Scholar]
- 19.Hughes K, Wickenden JA, Allen JE, Watson CJ. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol. 2012;227:106–17. doi: 10.1002/path.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreuzaler PA, Staniszewska AD, Li W, Omidvar N, Kedjouar B, Turkson J, et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol. 2011;13:303–9. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajewska M, Gajkowska B, Motyl T. Apoptosis and autophagy induced by TGF-B1 in bovine mammary epithelial BME-UV1 cells. J Physiol Pharmacol. 2005;56(Suppl 3):143–57. [PubMed] [Google Scholar]
- 23.Zarzyńska J, Gajkowska B, Wojewódzka U, Dymnicki E, Motyl T. Apoptosis and autophagy in involuting bovine mammary gland is accompanied by up-regulation of TGF-beta1 and suppression of somatotropic pathway. Pol J Vet Sci. 2007;10:1–9. [PubMed] [Google Scholar]
- 24.Gajewska M, Motyl T. IGF-binding proteins mediate TGF-beta 1-induced apoptosis in bovine mammary epithelial BME-UV1 cells. Comp Biochem Physiol C Toxicol Pharmacol. 2004;139:65–75. doi: 10.1016/j.cca.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Sobolewska A, Gajewska M, Zarzyńska J, Gajkowska B, Motyl T. IGF-I, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. Eur J Cell Biol. 2009;88:117–30. doi: 10.1016/j.ejcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Motyl T, Gajewska M, Zarzynska J, Sobolewska A, Gajkowska B. Regulation of autophagy in bovine mammary epithelial cells. Autophagy. 2007;3:484–6. doi: 10.4161/auto.4491. [DOI] [PubMed] [Google Scholar]
- 27.Monks J, Rosner D, Geske FJ, Lehman L, Hanson L, Neville MC, et al. Epithelial cells as phagocytes: apoptotic epithelial cells are engulfed by mammary alveolar epithelial cells and repress inflammatory mediator release. Cell Death Differ. 2005;12:107–14. doi: 10.1038/sj.cdd.4401517. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Kuma A. Autophagosomes in GFP-LC3 Transgenic Mice. Methods Mol Biol. 2008;445:119–24. doi: 10.1007/978-1-59745-157-4_7. [DOI] [PubMed] [Google Scholar]
- 29.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 30.Deretic V. Autophagy, an immunologic magic bullet: Mycobacterium tuberculosis phagosome maturation block and how to bypass it. Future Microbiol. 2008;3:517–24. doi: 10.2217/17460913.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–35. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–43. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruck A, Attonito J, Garces KT, Núnez L, Palmisano NJ, Rubel Z, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400. doi: 10.4161/auto.7.4.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108:17396–401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–30. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bierie B, Gorska AE, Stover DG, Moses HL. TGF-beta promotes cell death and suppresses lactation during the second stage of mammary involution. J Cell Physiol. 2009;219:57–68. doi: 10.1002/jcp.21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 38.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erwig LP, Henson PM. Clearance of apoptotic cells by phagocytes. Cell Death Differ. 2008;15:243–50. doi: 10.1038/sj.cdd.4402184. [DOI] [PubMed] [Google Scholar]
- 42.Atabai K, Fernandez R, Huang X, Ueki I, Kline A, Li Y, et al. Mfge8 is critical for mammary gland remodeling during involution. Mol Biol Cell. 2005;16:5528–37. doi: 10.1091/mbc.E05-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandahl M, Hunter DM, Strunk KE, Earp HS, Cook RS. Epithelial cell-directed efferocytosis in the post-partum mammary gland is necessary for tissue homeostasis and future lactation. BMC Dev Biol. 2010;10:122. doi: 10.1186/1471-213X-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravichandran KS. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J Exp Med. 2010;207:1807–17. doi: 10.1084/jem.20101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Brien J, Lyons T, Monks J, Lucia MS, Wilson RS, Hines L, et al. Alternatively activated macrophages and collagen remodeling characterize the postpartum involuting mammary gland across species. Am J Pathol. 2010;176:1241–55. doi: 10.2353/ajpath.2010.090735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y, Singh S, Georgescu MM, Birge RB. A role for Mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J Cell Sci. 2005;118:539–53. doi: 10.1242/jcs.01632. [DOI] [PubMed] [Google Scholar]
- 48.Clarkson RW, Watson CJ. Microarray analysis of the involution switch. J Mammary Gland Biol Neoplasia. 2003;8:309–19. doi: 10.1023/B:JOMG.0000010031.53310.92. [DOI] [PubMed] [Google Scholar]
- 49.O’Brien J, Martinson H, Durand-Rougely C, Schedin P. Macrophages are crucial for epithelial cell death and adipocyte repopulation during mammary gland involution. Development. 2012;139:269–75. doi: 10.1242/dev.071696. [DOI] [PubMed] [Google Scholar]
- 50.Konishi A, Arakawa S, Yue Z, Shimizu S. Involvement of Beclin 1 in engulfment of apoptotic cells. J Biol Chem. 2012;287:13919–29. doi: 10.1074/jbc.M112.348375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, et al. Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature. 2011;477:220–4. doi: 10.1038/nature10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motyl T, Gajkowska B, Zarzyńska J, Gajewska M, Lamparska-Przybysz M. Apoptosis and autophagy in mammary gland remodeling and breast cancer chemotherapy. J Physiol Pharmacol. 2006;57(Suppl 7):17–32. [PubMed] [Google Scholar]
- 53.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 55.Mukaiyama H, Kajiwara S, Hosomi A, Giga-Hama Y, Tanaka N, Nakamura T, et al. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology. 2009;155:3816–26. doi: 10.1099/mic.0.034389-0. [DOI] [PubMed] [Google Scholar]
- 56.Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 57.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 58.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 59.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 61.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–8. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 62.Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–7. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 63.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–48. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–6. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Curr Opin Cell Biol. 2010;22:206–11. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.deCathelineau AM, Henson PM. The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem. 2003;39:105–17. doi: 10.1042/bse0390105. [DOI] [PubMed] [Google Scholar]
- 67.McPhee CK, Baehrecke EH. The engulfment receptor Draper is required for autophagy during cell death. Autophagy. 2010;6:1192–3. doi: 10.4161/auto.6.8.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Humphreys RC, Krajewska M, Krnacik S, Jaeger R, Weiher H, Krajewski S, et al. Apoptosis in the terminal endbud of the murine mammary gland: a mechanism of ductal morphogenesis. Development. 1996;122:4013–22. doi: 10.1242/dev.122.12.4013. [DOI] [PubMed] [Google Scholar]
- 69.Andres AC, Strange R. Apoptosis in the estrous and menstrual cycles. J Mammary Gland Biol Neoplasia. 1999;4:221–8. doi: 10.1023/A:1018737510695. [DOI] [PubMed] [Google Scholar]
- 70.Lyons TR, O’Brien J, Borges VF, Conklin MW, Keely PJ, Eliceiri KW, et al. Postpartum mammary gland involution drives progression of ductal carcinoma in situ through collagen and COX-2. Nat Med. 2011;17:1109–15. doi: 10.1038/nm.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 72.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kongara S, Kravchuk O, Teplova I, Lozy F, Schulte J, Moore D, et al. Autophagy regulates keratin 8 homeostasis in mammary epithelial cells and in breast tumors. Mol Cancer Res. 2010;8:873–84. doi: 10.1158/1541-7786.MCR-09-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]