Abstract
AMBRA1 is a positive regulator of the BECN1-dependent program of autophagy recently identified in mouse. In this study, we cloned the full-length cDNAs of ambra1a and ambra1b zebrafish paralogous genes. As in mouse, both Ambra1 proteins contain the characteristic WD40 repeat region. The transcripts of both genes are present as maternal RNAs in the eggs and display a gradual decline until 8 hpf, being replaced by zygotic mRNAs from 12 hpf onwards. After 24 hpf, the transcripts are mainly localized in the head, suggesting a possible role in brain development. To check their developmental roles, we adopted morpholino knockdown to block either translation (ATGMOs) or splicing (SPLICMOs). Treatment with ATGMOs causes severe embryonic malformations, as prelarvae could survive for only 3 and 4 days in ambra1a and b morphants, respectively. Treatment with SPLICMOs led to developmental defects only at a late stage, indicating the importance of maternally supplied ambra1 transcripts. Analysis of the levels of Lc3-II, an autophagosome-specific marker, in the presence of lysosome inhibitors evidenced a reduction in the rate of autophagosome formation in both MOs-injected embryos at 48 hpf, more pronounced in the case of ambra1a gene. Although some defects, such as body growth delay, curved shape and hemorrhagic pericardial cavity were present in both morphants, the occurrence of specific phenotypes, such as major abnormalities of brain development in ambra1a morphants, suggests the possible acquisition of specific functions by the two paralogous genes that are both required during development and do not compensate each other following knockdown.
Keywords: Ambra1, zebrafish, autophagy, development, morpholino
Introduction
Autophagy is an evolutionarily conserved catabolic process used to break down and recycle, through the lysosomal machinery, long-lived proteins and organelles in order to maintain a homeostatic environment within the cell. This process is tightly regulated and plays several important roles in embryo development, differentiation, normal physiology, survival during starvation, and aging.1,2 Three types of autophagy are known: macroautophagy (usually referred as “autophagy”), microautophagy, and chaperone-mediated autophagy, working with different cellular mechanisms and playing distinctive functions.3 Macroautophagy is responsible for the bulk degradation of macromolecules and damaged cytoplasmic organelles in a double-membrane-bound vesicle, the autophagosome.4 Failure to remove aggregates of mutated toxic proteins could explain the key pathogenic effect of impaired autophagy in neurodegenerative diseases, including Huntington disease, Parkinson disease, amyotrophic lateral sclerosis, and Alzheimer disease.5,6 Autophagy has also been identified as a critical process in oncogenesis and cancer progression.7,8 Moreover, in some congenital muscular dystrophies, defective activation of the autophagic machinery causes accumulation of dysfunctional mitochondria, leading to damage and the subsequent loss of muscle fibers.9
A great number of papers, reviewed by Di Bartolomeo and coworkers,10 suggest that autophagy, in eukaryotes, represents a crucial mechanism during the development of multicellular organisms. Several proteins, called AuTophaGy-related proteins (ATG), first identified in yeast,11 participate in the different steps of the autophagic process: vacuole initiation, elongation and fusion with the lysosomes. Autophagosome formation requires a BECN1-containing complex that recruits the class III phosphatidylinositol 3-kinase (PIK3C3) to produce phosphatidylinositol-3-phosphate. This activity is coordinated by the interaction of BECN1 with several other proteins, including the regulatory protein kinase PIK3R4/Vps15/p150, the UV radiation resistance-associated protein (UVRAG), SH3GLB1 (endophilin B1/Bif-1), AMBRA1 (autophagy/Beclin 1 regulator 1), BCL2 (B-cell CLL/lymphoma 2), ATG14 (autophagy-related 14), and KIAA0226 (RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein /RUBICON) to form different protein complexes that mediate distinct functions in membrane trafficking.12,13
A positive regulator of the BECN1-dependent program of autophagy has been recently identified in mouse as AMBRA1.14 AMBRA1 is normally docked in mammalian cells at a specific cytoskeletal site (the dynein light chain, DYNLL1), wherefrom it is unleashed upon autophagy induction to translocate to the autophagosome origin sites on the ER.15 AMBRA1 is primarily expressed, during mouse embryogenesis, in the neural plate of the CNS and it is crucial for the proper nervous system development. AMBRA1-deficiency in mouse embryos is lethal, due to neural tube developmental defects as well as abnormal neural cell proliferation.14
Currently, zebrafish is one of the leading models for studying not only early development, a phase of the life cycle in which zebrafish has a recognized superiority, but also cellular and physiological processes involved in human diseases. Zebrafish is more closely related to humans than invertebrate models and, at the same time, it offers numerous advantages, such as external fertilization, the rapid development of transparent embryos, and the large number of offspring, compared with nonfish vertebrate models. Within six days, the transparent embryos and larvae complete their development, allowing in vivo visualization and analysis of all developmental stages and organogenesis. Moreover, the zebrafish often contains duplicate copies of genes that are instead present as single copies in mammals. This is helpful to obtain additional insight into the multiple functions of the human counterpart.16 Herein, two zebrafish ambra1 genes were identified and analyzed for their function in embryonic development. Ambra1a and ambra1b genes appear to be paralogous and their expression was detected throughout embryonic development. Morpholino-based silencing of both genes resulted in developmental defects similar to those observed in the AMBRA1-deficient mouse model. Presence of paralog-specific alterations suggests the possibility of subfunctioning of the two genes and demonstrates that zebrafish is a viable model for the study of AMBRA1 functions.
Results
Isolation and characterization of the zebrafish ambra1 genes and deduced proteins
Two predicted sequences, XM_002667669 (coding for ambra1a, as named in this paper) and XR_084457 (coding for ambra1b), were identified by bioinformatics tools using the mouse AMBRA1 amino acid sequence (NP_766257) as query. The whole coding region of the two transcripts, as well as the 5′- and 3′-untranslated terminal regions (UTRs), were then sequenced using RNA extracted from 2- and 48-hpf (hours post-fertilization) embryos, according to the cloning strategy reported in Figure S1. Sequences obtained by RT-PCR and cloning display differences with the above-predicted sequences as well as the presence or absence of predicted exons. Data on the nucleotide features of the different transcript variants identified and sequenced in this work are described in Table 1.
Table 1. Summary of the principal features of the cloned transcripts for zebrafish ambra1 genes.
| 5′ UTR | 3′UTR | Coding region | Predicted ORF | GenBank Accession | 2 hpf | 48 hpf | |
|---|---|---|---|---|---|---|---|
|
ambra1a1 |
307 bp |
182 bp |
3948 bp |
1315 aa |
HE602022 |
x |
x |
| 4035 bp |
1344 aa |
- |
x |
x |
|||
|
ambra1a2 |
307 bp |
266 bp |
3297 bp |
1098 aa |
HE602023 |
x |
x |
| 3384 bp |
1127 aa |
- |
x |
x |
|||
|
ambra1a3 |
307 bp |
540 bp |
2367 bp |
788 aa |
FR846231 |
x |
x |
| 2454 bp |
817 aa |
- |
- |
x |
|||
|
ambra1a4 |
307 bp |
362 bp |
2175 bp |
724 aa |
HE602022 |
- |
x |
| 2262 bp |
753 aa |
- |
- |
- |
|||
| ambra1b | 350 bp | 1269 bp | 4083 bp | 1360 aa | FR846230 | x | x |
b= binding region
The genomic organization of the two genes, obtained by sequence analysis of the new cDNAs with the BLAT program, is described in Figure 1 and compared with that of the mouse Ambra1 gene. In Table S1, we report the sizes of each of the exons and introns. The canonical splice consensus sites “GT” and “AG” are present in all zebrafish ambra1 introns.
Figure 1. Genomic structure and organization of zebrafish and mouse ambra1 genes. Plain boxes indicate exons. The coding region is in color and numbered with Roman numerals. Boxes of exon 8 are dashed in transcripts where it may be absent. The positions of the WD40 domain, BECN1 and DYNLL1 binding regions are indicated. Introns, represented as lines, are not drawn to scale, but the corresponding lengths can be found in Table S1 together with the exon sizes. Exons in zebrafish ambra1a and b are similar in size to orthologous exons in mouse Ambra1.
The zebrafish gene ambra1a is alternatively spliced, generating four variant C-terminal sequences. The entire coding sequence of the four ambra1a transcripts was obtained by overlapping fragments of different lengths. Given the long and alternatively spliced transcripts, all four variants were also amplified with a set of primers localized before and after the start and stop codons (see Fig. S1). The fragments were then cloned and sequenced to verify the accuracy of the overlapping processes. All four variants cloned lacked exon 8, as it occurred in one of the two transcripts encoded by mouse Ambra1 gene (see Fig. 1, exon boxed with dashed line). However, PCR performed with a sense primer located on exon 7 and an antisense primer specific for each ambra1a form showed that exon 8 could be transcribed, but at very low levels with respect to the forms lacking this exon (data not shown). The presence of exon 8 as well as the expression of all the identified transcripts were checked at 2- and 48-hpf (see Table 1). No transcript containing exon 8 was found for the ambra1a4 variant at both 2- and 48-hpf.
Ambra1b gene contains 19 exons as ambra1a, with the same genomic organization. Though several 3′-RACE analyses using sense primers localized on different regions of its coding sequence were performed, only one transcript was identified for this gene. Moreover, no transcript variant lacking exon 8 was sequenced or identified by PCR analysis. The mouse Ambra1 gene (sequence used NP_766257), as the human one (not shown, sequence used Q9C0C7), presents the same genomic organization as the zebrafish genes, but it contains only 18 exons. Nevertheless, the mouse last exon (18th) contains a longer coding region with respect to the corresponding exon 18 of zebrafish ambra1 genes. This difference explains the low identity in the C-terminal regions of mammals and zebrafish full-size Ambra1 deduced proteins (Fig. 2). Cloning results show that, although there is no consistency with respect to intron size (Table S1), the overall gene architecture has been conserved across species.
Figure 2. Multiple sequence alignment of human (Hs, Homo sapiens, Q9C0C7), mouse (Mm, Mus musculus, NP_766257) and zebrafish (Dr, Danio rerio) Ambra1 proteins was originated with the program ClustalW. Zebrafish Ambra1 amino acid sequence is inferred from the coding sequences cloned in this study (Ambra1a1, HE602022; Ambra1a2, HE602023; Ambra1a3, FR846231; Ambra1a4, HE602024; Ambra1b, FR846230). In the alignment, identical residues in all sequences are indicated by ‘*’. Conservative and semi-conservative substitutions are indicated by ‘:’ and ‘.’, respectively. The WD40 repeats-region at the N-terminus is indicated in BOX 1, the region involved in the BECN1 interaction in BOX 2 and the binding site to the dynein light chain (DYNLL1) in BOX 3, with the TQT-domain in bold. Position of exon 8, inside BECN1 binding region, is underlined.
Alignment of the zebrafish Ambra1a1 and b amino acid sequences with those of human and mouse indicates that the primary structure of Ambra1 proteins is highly conserved among vertebrates (Fig. 2) and is represented by a WD40 protein of about 1,300 amino acids. Zebrafish Ambra1a1 and Ambra1b polypeptides exhibit 55% identity to each other, and 56% and 58% identity with human and mouse AMBRA1s, respectively (Table 2). The WD40 motif (amino acids 49–175 of human AMBRA1) shows very high identity among the human, mouse and zebrafish AMBRA1 polypeptides (Table 2). The region involved in the binding with the BECN1 protein presents a lower identity ranging from 45% to 52%, according to the presence or absence of exon 8. Binding sites to the dynein light chain (DYNLL1) are present only in Ambra1a1, Ambra1a2 and Ambra1b proteins, all three containing the double TQT domain (Table 2).
Table 2. Percentage of identity between Zebrafish Ambra1a1 and Ambra1b polypeptides and with human and mouse AMBRA1s.
| Dr-ambra1b | Mm-Ambra1 | Hs-AMBRA1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
Tot. identity |
WD40 |
Becn1 b |
DYNLL1 b |
Tot. identity |
WD40 |
BECN1 b |
DYNLL1 b |
Tot. identity |
WD40 |
BECN1 b |
DYNLL1 b |
| Dr-ambra1a |
55 |
88 |
45/50 |
69 |
56 |
90 |
46/51 |
61 |
56 |
90 |
47/52 |
66 |
| Dr-ambra1b |
- |
- |
- |
- |
58 |
90 |
49 |
74 |
58 |
90 |
50 |
79 |
| Mm-ambra1 | - | - | - | - | 95 | 100 | 94 | 92 | ||||
Conserved syntenies for ambra1 genes and phylogenetic analysis
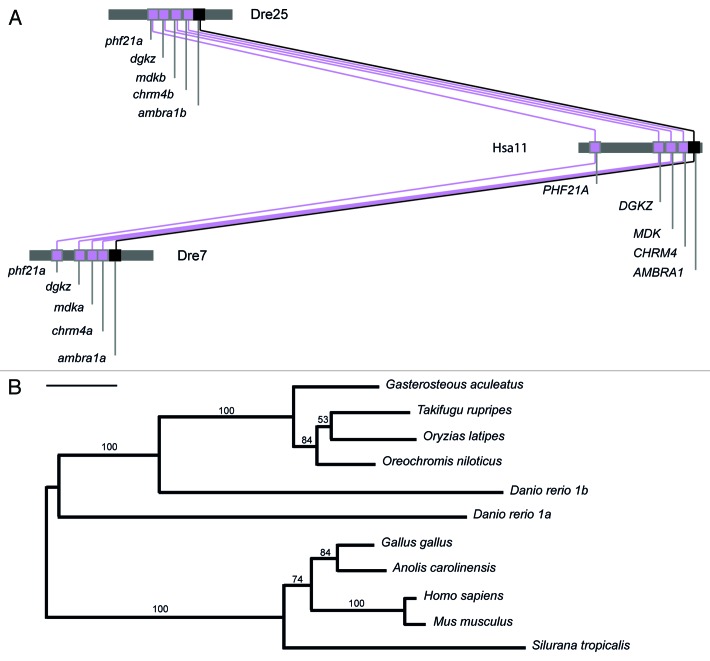
The duplicated ambra1 genes share strong synteny with the human AMBRA1. As shown in Figure 3A, zebrafish ambra1a and b lie on chromosomes 7 (Dre7) and 25 (Dre25), respectively. Analysis of conserved synteny between zebrafish and human genomes was performed using the Syntheny Database set up on the zebrafish genome assembly Zv9 and the Ensembl version 61.18 Their genomic neighborhoods contain 20 and 12 genes, respectively, with conserved synteny to the orthologous region in human chromosome 11 (Hsa11) in which the single AMBRA1 gene is located as expected if zebrafish ambra1 genes are co-orthologous to the human gene AMBRA1. Moreover, the Synteny Database analysis (sliding window size 25 genes) shows that the linkage fragments of chromosomes 7 and 25 containing ambra1a and b enclose other genes that are duplicated in zebrafish and present as a single gene in mammals, such as: midkine (mdk), diacylglycerol kinase-zeta (dgkz), M4 muscarinic cholinergic receptor (chrm4) and PDH finger protein 21A (phf21a). These findings suggest that the zebrafish ambra1a and ambra1b genes are paralogs, which most likely arose as a result of the genome-wide duplication that occurred at the base of the teleost radiation.19
Figure 3. Conserved synteny neighboring the AMBRA1 locus and evolutionary relationship of the known ambra1 genes. (A) Graphical representation of conserved synteny neighboring the AMBRA1 locus between D. rerio chromosomes 7 (Dre7) and 25 (Dre25) and H. sapiens chromosome 11 (Hsa11). The analysis was performed with the Synteny Database program (http://teleost.cs.uoregon.edu/acos/synteny_db/) with a sliding window size of 25 genes. Synteny analysis shows that the portion of zebrafish chromosome 7 (Dre7) that contains ambra1a and the portion of zebrafish chromosome 25 (Dre25) that contains ambra1b possess other four genes that are orthologous to genes in the portion of human chromosome 11 (Hsa11) that contains AMBRA1. Gene names are from Ensembl (www.ensemblgenomes.org/) or NCBI (www.ncbi.nlm.nih.gov/gene/). Genes are drawn as squares. The figure depicts the relative locations of genes, but is not drawn to physical scale. The positions of ambra1a, ambra1b and AMBRA1 are marked in black. Oblique lines connect presumed paralogs within chromosome groups. (B) Evolutionary relationship of the known ambra1 genes. The phylogenetic tree was calculated using the Maximum likelihood method with the RaxML 7.2.6 program and by applying the evolutionary model JTT+G.17 Bar represents 0.1 substitutions per site. Comparisons were made to the amino acid sequences of D. rerio Ambra1a (CCE04070), D. rerio Ambra1b (CCA61107), Takifugu rubripes (ENSTRUG00000013113), Gasterosteus aculeatus (ENSGACG00000007844), Orizyas latipes (ENSORLP00000017709), Silurana tropicalis (XP_002934144), Anolis carolinensis (XP_003214662), Gallus gallus (ENSGALP00000013594), Oreochromis niloticus (XP_003458340), Mus musculus (NP_766257), Homo sapiens (Q9C0C7). Numbers indicate the values supporting the branching pattern from 1000 bootstraps.
To determine whether the presence of two ambra1 genes is a general feature of teleost genomes, genomic database from Tetraodon nigroviridis, Takifugu rubripes, Gasterosteus aculeatus and Oryzias latipes were screened on the BLAT program (http://genome.ucsc.edu/cgi-bin/hgBlat?command=start) using Ambra1a and b sequences as queries. However, no duplicated ambra1 genes could be detected in these genomes, suggesting that, in these species, one of the duplicated genes was silenced and subsequently lost. Moreover, analysis of teleost fishes EST libraries did not suggest the presence of transcripts deriving from duplicated genes.
A phylogenetic analysis was performed to study the evolutionary relationships among the AMBRA1 genes using the amino acid sequence of zebrafish Ambra1a and b and other published Ambra1 sequences. The resulting tree (Fig. 3B) shows that tetrapods and teleostean fishes share a common ancestor for the AMBRA1 gene family. Moreover, the tree suggests that a duplication arose within the teleostean clade giving birth to the paralogous forms of ambra1 found in zebrafish. The separation of the two paralogous zebrafish genes is very well supported (bootstrap value: 1000/1000). Moreover, Danio rerio ambra1b gene is the orthologous form of the other teleostean ambra1s.
Expression patterns of ambra1a and b mRNAs during zebrafish development
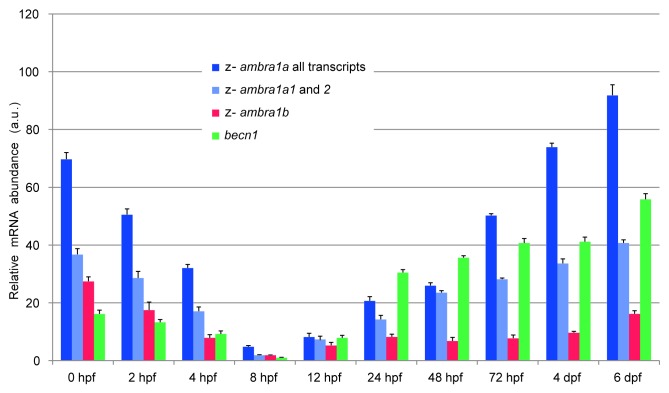
To investigate the temporal expression pattern of zebrafish ambra1 genes, together with becn1, we performed qPCR analysis on cDNA obtained from different developmental stages (Fig. 4). Gene-specific primer sets were designed to span known introns, thus revealing any contaminating genomic DNA as larger sized PCR fragments. For ambra1a, a set of primers was designed to simultaneously amplify the four transcript variants, whereas a second set could amplify only the two longer forms, ambra1a1 and ambra1a2 (see Fig. S1). Due to constraints in fragment length for qPCR amplification, it was not possible to select more specific primer sets for the shorter forms, ambra1a3 and ambra1a4.
Figure 4. Temporal expression patterns of the duplicated zebrafish ambra1 genes. The graph shows the relative mRNA transcript abundance of ambra1a (all transcript variants), ambra1a1 and 2 and ambra1b, as well as becn1 mRNAs in whole zebrafish embryos, from 0 to 6 dpf, as determined by qPCR. Error bars indicate SEM.
Simultaneous detection of arp and 18S rRNA gene expression was used to normalize the expression level of ambra1 variants. For the comparison of differences in gene expression over time, all values were adjusted to the stage with the lowest expression level, corresponding to becn1 expression at 8 hpf which was set at an arbitrary unit of 1. Similarly, all ambra1 variants present the lowest expression at this stage.
qPCR analysis revealed that ambra1a and b transcripts, as well as becn1, were maternally deposited into the embryo. These transcripts dramatically declined throughout the first 8 hpf, being replaced, thereafter, by the corresponding embryonic mRNAs up to a maximum at 6 dpf (days post-fertilization), the last stage examined. Transcripts of ambra1a present a higher expression level at 4 and 6 dpf with respect to initially deposited maternal mRNAs, whereas ambra1b expression was somewhat reduced. The contribution of the short forms (ambra1a3 and 4), deduced from the difference between all ambra1a transcripts and the two long forms, seems to increase during embryo and larval development. Similarly, embryonic and larval becn1 mRNA is much higher than the maternal messenger.
To further investigate the spatio-temporal expression of maternal and zygotic ambra1a and b mRNAs during zebrafish development, WMISH assays were performed on WT embryos and larvae from 0.2 hpf to 6 dpf using digoxigenin-labeled antisense ambra1a and ambra1b mRNA probes. In order to ensure specificity and to avoid cross-hybridizations, each probe covered fragments of the C-terminus and the 3′-UTR region.
Consistently with the qPCR results, ambra1a1 and b mRNAs could be detected in one-cell stage embryos and distributed evenly in the blastodisc (Fig. 5). No or a very low signal was found at 10 hpf (tail bud), in agreement with qPCR results showing a minimum concentration of both transcripts at 8 hpf during gastrulation. At 1 dpf, the maternal ambra1a mRNA was replaced by zygotic expression observed in the brain and otic vesicles. At 1 dpf, ambra1b mRNA was concentrated in the head and in the trunk, but its expression was less well defined with respect to ambra1a. At 4 and 6 dpf, both transcripts are localized in the otic vesicles, oral cavity, intestine, swim bladder and trunk. As shown by histological analysis, the signal for ambra1b is more evident in the apical border of the intestinal epithelium with respect to ambra1a.
Figure 5. Spatio-temporal expression of ambra1a1 and ambra1b mRNA during zebrafish development as evidenced by whole-mount in situ hybridization performed at the indicated stages. All embryos are lateral views with the animal pole up (1 cell and 10 hpf) and head pointing to the left (1, 4 and 6 dpf). 10× magnification of the embryo (dorsal view) on the left. Scale bar: 200 μm. (A and B) Transverse histological sections 6 μm thick of the 6 dpf ambra1a1 (A) and ambra1b (B) labeled embryos (the dashed lines in 6 dpf whole-mount embryos indicate the position of the section). Ov = otic vesicle, Sb = swim bladder, In = intestinal cavity.
Developmental consequences of Ambra1a and Ambra1b proteins depletion in zebrafish embryos and larvae
MOs effectiveness
To determine the functions of ambra1a and b during zebrafish development, gene-specific antisense MOs targeted to the ATG region (MO1-ambra1a and MO1-ambra1b) were injected into 1-cell embryos for translation knockdown. Five-nucleotide-mismatched MOs (MISMMOs) (MO1-ambra1a-5m and MO1-ambra1b-5m) were used as negative controls.
Due to the lack of specific antibodies validated to detect zebrafish Ambra1a or Ambra1b proteins, the efficacy of MO1-ambra1a and MO1-ambra1b in targeting and blocking protein translation for each paralogous gene was analyzed by an alternative approach. This was based on in vitro transcribed mRNAs encoding in-frame EGFP fusion constructs containing the MO-targeted sequence of each ambra1 paralog. Coinjection of ambra1a- or ambra1b-EGFP fusion construct mRNAs with the corresponding 5-bp MISMMO, resulted in bright ubiquitous EGFP protein expression, thus demonstrating that the relative knockdown depends precisely on the complementary MO sequence (Fig. S2A). Coinjection of the EGFP fusion mRNAs for each paralog with the corresponding morpholino resulted in a drastic reduction in the EGFP expression, thus indicating that both MOs are able to efficiently inhibit protein translation (Fig. S2A). To confirm the absence of cross-target effects, each MO was coinjected with the EGFP fusion mRNA for the other paralog gene. In this case, no interference with EGFP expression was detected, indicating that it is unlikely that cross-targeting of MOs is occurring (Fig. S2A).
To discriminate between the contribution of maternal and zygotic ambra1 transcripts, gene-specific antisense MOs targeted to the exon 3 - intron 3 junction of the predicted pre-mRNA of each gene were injected (MO2-ambra1a and MO2-ambra1b). These SPLICMOs caused the skipping of the exon 3 thus altering the translation reading frame of exon 4 with introduction of a premature stop codon and production of a truncated Ambra1a and Ambra1b proteins lacking all previously identified binding domains. The size of the mis-spliced product was consistent with the loss of the targeted exon 3, as confirmed by sequencing of both the mis-spliced RT-PCR products (Fig. S2B). Total RNAs extracted from 4, 6, 8, 12 and 24-hpf WT and MO2-ambra1a- and MO2-ambra1b-injected embryos were analyzed by RT-PCR with two sets of specific primers flanking the splice site of each paralogous gene. Zygotic ambra1b expression takes place already at 6 hpf, although at a very low level, as indicated by the predicted mis-spliced transcript detected as a lower band when embryos are injected with MO2-ambra1b. At 8 and 12 hpf, both ambra1 zygotic RNAs become predominant and almost completely abolished by SPLICMOs. At 24 hpf, MO2-ambra1a still efficiently abrogates ambra1a splicing, while the effect of MO2-ambra1b started to decrease as shown by the reappearance of normally spliced band.
Optimization of MO injections
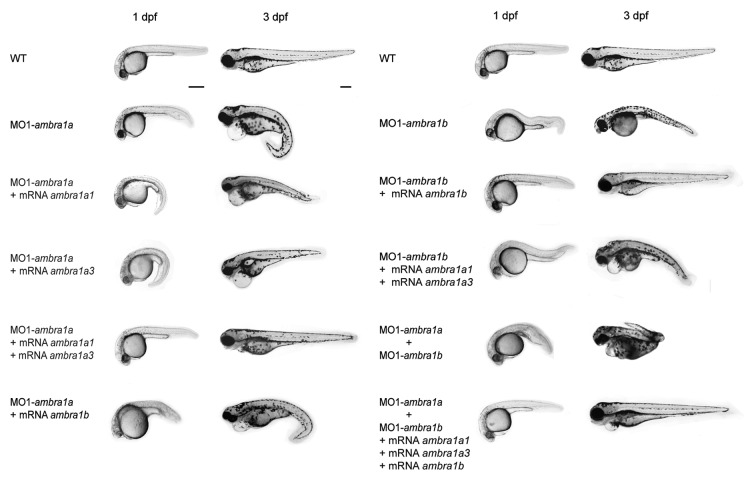
In order to optimize MO injections, 8.2, 10.3, 15.5, 20.6 and 25.8 ng of MO1-ambra1a or MO1-ambra1b, as well as of SPLICMOs (MO2-ambra1a or MO2-ambra1b) and MISMMOs (MO1-ambra1a-5m or MO1-ambra1b-5m) per embryo were injected in triplicates. Depending on the severity of defects compared with controls, morphant embryos from ATGMOs were classified into two graded phenotypes, less affected as class I and more affected as class II (Fig. 6). As expected, the highest numbers of dead embryos were associated with the highest dose of both ATGMOs (25.8 ng). On the other hand, the lowest dose of both ATGMOs (8.2 ng) showed the least number of abnormal phenotypes (Table S2). Accordingly, 15.5 ng of MO1-ambra1a and 20.6 ng of MO1-ambra1b were chosen as optimal dosages to knockdown translation. At the same dosages, MO1-ambra1a-5m and MO1-ambra1b-5m had no or very low effects on zebrafish embryogenesis.
Figure 6. Phenotypes of embryos or larvae at 1 and 3 dpf after treatment with the different ambra1 ATGMOs alone or together with tp53 MO or with SPLICMOs. Morphants phenotypes are compared with control groups (WT or MISMMOs). Phenotypes of tp53 MO morphants are also reported. Animals are presented as lateral view, anterior to the left. Scale bar: 200 μm.
In the MO-coinjection experiment, just 5 ng of MO1-ambra1a plus 4 ng of MO1-ambra1b were selected as the best dosages for the simultaneous knockdown of the two paralogous genes. In this case, morphant embryos were classified into three graded phenotypes, with class III corresponding to highly deformed morphants (Fig. 6).
Moreover, accordingly to the optimization analysis, 18.5 ng of MO2-ambra1a and 15.5 ng of MO2-ambra1b were chosen as optimal dosages to knock down ambra1 splicing (Table S2). Injected embryos were analyzed by light microscopy and their morphology was recorded.
To further prove the specificity of the observed Ambra1 knockdown phenotypes, each ambra1 ATGMO was coinjected with tp53/p53 MO. The rationale is based on a report that MOs can nonspecifically activate the tumor suppressor TP53-induced apoptosis, causing off-target phenotypic effects that are not caused by the specific MO used.20 However, the tp53 MO failed to eliminate the morphant phenotypes and showed that these were specifically target-related (Fig. 6).
Analysis of morphant phenotypes
This analysis showed that treatment with ambra1 ATGMOs causes severe embryonic malformations, because, at the selected MO dosages, ATGMOs morphants could survive for only 3 and 4 dpf, respectively.
During early development (cleavage period, 0–3 hpf), MO1-ambra1a- and MO1-ambra1b-injected embryos could not be morphologically distinguished from the controls (non-injected and MISMMO-embryos). However, by 4 hpf (sphere stage), both ATGMO-injected and coinjected embryos often exhibited a slight developmental delay, which was more evident in the MO1-ambra1a and coinjected morphants.
At 1 dpf, growth impairment was manifest, since both morphant embryos of class II had smaller head, reduced eyes and trunk, curved or twisted tail and delayed pigmentation compared with controls. Otoliths were also smaller in size. Coinjection of both ATGMOs produced more extensive morphological alterations with a complete derangement of the body plan in class III morphants (Fig. 6).
At 2 dpf morphants were analyzed by light microscopy analysis of morphology as well as by scanning electron microscopy (SEM) and toluidine blue-stained semithin sections. As shown in Figure 7, morphants showed abnormal head development involving smaller eyes and pronounced hydrocephalus, that is present in both midbrain and hindbrain ventricles in MO1-ambra1a morphants compared with wild-type and control morphants (Fig. 7B). This problem is instead present only in hindbrain ventricles in MO1-ambra1b morphants. In coinjected morphants hydrocephalus was present in both midbrain and hindbrain and was more severe, as shown by the corresponding semithin sections, where also the notochord appeared disorganized (Fig. 7E).
Figure 7. Two-dpf embryos treated with different ambra1 ATGMOs and compared with controls. (A) SEM (scanning electron microscope) images of dorsal view, anterior to the left. The white dashed lines indicate position of the section reported in (C). (B) Light microscope image showing hydrocephalus (dashed lines). Asterisks indicate midbrain-hindbrain boundary. Lateral view head to the left. (C) Toluidine blue staining of semithin transverse sections. (D) SEM images of lateral view tails showing the morphants deformities compared with controls. (E) Transversal semithin sections stained with toluidine blue displaying notochord malformations in different morphants. Scale bar: 100 μm.
At 3 dpf, these defects were further aggravated, as hatched MO1-ambra1b and in particular MO1-ambra1a larvae were very poorly developed (Fig. 6). Abnormalities were more severe in MO1-ambra1a larvae (Fig. 6), which displayed higher degrees of ventral curvature of the spine with misshapen tail (Fig. 6). MO1-ambra1a larvae of class I show a slight ventralizated phenotype that is more evident in ambra1a and ambra1b morphants of class II as reported in Table S6.
At 3 dpf, light microscopy analysis of morphology of less deformed morphants showed that otic vesicles were much smaller in MO1-ambra1a and coinjected morphants (dashed lines in Fig. 8). Eyes were smaller with both MOs, while MO1-ambra1b and coinjected morphants showed also mild cyclopia (close-set eyes, white arrowheads in Fig. 8). Both morphants presented pericardial edema (arrows in Fig. 8) and persistent voluminous and oedematous yolk sac.
Figure 8. Close-up lateral and ventral light microscopy views of 3 dpf wild-type and ATGMOs-injected embryos. The genes targeted by ATGMOs are indicated for each image. Otic vesicles are smaller in MO1-ambra1a and ATGMO-coinjected morphants (dashed lines). White arrowheads indicate cyclopia in ambra1b and coinjected morphants. All morphants presented pericardial edema (arrow). Scale bar: 200 μm
The network of subintestinal vessels at the yolk stalk, examined at 3 dpf, was devoid, in both morphants, of the 2 to 3 loop series observed in WT, following blood vessel visualization by endogenous alkaline phosphatase activity. The pattern of intersegmental vessels in the trunk and tail was also less defined (Fig. S3). The lack of a properly established circulation could explain the large yolk sac still present in 48 and 72 hpf morphant embryos.
Knockdown of ambra1a and/or ambra1b indicates that both genes act in a synergistic fashion during development, as the gravity and number of abnormalities when both ATGMOs were injected was greater than the sum of those obtained following injection of each ATGMO separately (Figs. 6–8). With both MISMMOs, no morphological abnormalities were observed up to 7 dpf.
Moreover, the analysis of morphant phenotypes showed that MO2-ambra1a- and MO2-ambra1b-injected embryos (SPLICMOs) are comparable to controls during the first developmental period (0–24 hpf). By 1 to 3 dpf morphant obtained by SPLICMOs seem only slightly deformed with a lightly curved tail shape (Fig. 6). SEM analyses, performed at 2 dpf, highlighted a pronounced hydrocephalus in the hindbrain area of both MO2-ambra1a and MO2-ambra1b morphants (Fig. 9).
Figure 9. Lateral SEM and light microscopy views of wild-type and SPLICMOs-injected embryos. Top panel: Two-dpf embryos treated with ambra1a and b SPLICMOs and compared with controls. SEM images of dorsal view showing the morphant deformities compared with control. Botton panel: Close-up lateral and ventral light microscopy views of 5 and 10 dpf WT and SPLICMOs-injected embryos. The genes targeted by SPLICMOs are indicated for each image. Otic vesicles are smaller in MO2-ambra1a (dashed lines). Both SPLICMO morphants presented pericardial edema (arrow) and reduced eyes (asterisks). Scale bar: 200 μm
Further abnormalities appeared later in development: by 5 dpf, MO2-ambra1a-injected embryos showed developmental defects, such as a pronounced pericardial edema, highly reduced eyes and smaller otic vesicles compared with controls. The same abnormalities were evident in MO2-ambra1b embryos by 10 dpf (Fig. 9) and were similar to those displayed by ATGMOs-injected embryos earlier in development (Fig. 8). MO2-ambra1a- and MO2-ambra1b-injected embryos died within 12 dpf.
Rescuing of MO1-ambra1b or MO1-ambra1a knockdown with synthetic ambra1a and b mRNAs
To rescue the knockdown phenotype, the MO1-ambra1a was coinjected with in vitro-transcribed mutated zebrafish ambra1a1 and 1a3 mRNAs encoding the full length of each transcript variant, but containing eight silent mutations in the region recognized by MO1-ambra1a. Similarly the MO1-ambra1b was coinjected with in vitro-transcribed mutated zebrafish ambra1b (Fig. 10; Table S3).
Figure 10. Phenotype comparison among ATGMOs treated embryos and uninjected controls (WT) after the different rescue experiments. The genes targeted by ATGMOs and the mRNAs injected are indicated for each image. All the embryos are lateral view, anterior to the left. Scale bar: 200 μm.
We found that coinjection, at the one-cell stage, of 15.5 ng MO1-ambra1a with either 200 pg of ambra1a1 mRNA or 150 pg of ambra1a3 mRNA was not able to fully rescue the ambra1a loss-of-function phenotype, proving that this phenotype derives from the loss of both long and short Ambra1a proteins. This was confirmed by coinjection of 15.5 ng of MO1-ambra1a with 200 pg of ambra1a1 and 150 pg of ambra1a3 mRNAs that led to highly efficient rescue of the loss-of-function phenotype. The same result was obtained with coinjection of 20.6 ng of MO1-ambra1b with 200 pg of ambra1b mRNA, verifying that this phenotype is a result of loss-of-function of ambra1b.
To test whether Ambra1a has equivalent functional capacities to Ambra1b, we attempted to rescue the ambra1a loss-of-function phenotype by coinjection with ambra1b mRNA and vice versa. In both cross-injection experiments (15.5 ng of MO1-ambra1a with 200 pg of ambra1b mRNA or 20.6 ng of MO1-ambra1b with 200 pg of ambra1a1 and 150 pg of ambra1a3 mRNAs) the phenotypes were not rescued. Moreover, in the MO1-ambra1a (4 ng) plus MO1-ambra1b (5 ng) loss of function, a complete rescue was obtained only with coinjection of ambra1a1 (200 pg), ambra1a3 (200 pg) and ambra1b (150 pg) mRNAs (Fig. 10; Table S3).
Inhibition of ambra1a and b expression results in reduced autophagy and increased apoptosis during embryogenesis
In order to elucidate whether Ambra1 expression is required for autophagy induction during zebrafish development, LC3-II levels were examined in ATGMOs- and SPLICMOs-injected embryos by immunoblotting analysis at 48 hpf. The analysis was also performed in the presence of the lysosomal inhibitor bafilomycin A1 (Baf A1) to prevent autophagosome degradation, thus making it possible to measure the whole amount of LC3-II produced at this stage. As shown in Figure 11, a high rate of autophagy was detected in control embryos (WT and MISMMOs) as revealed by the strong increase of LC3-II signal observed following bafilomycin A1 treatment. In contrast, the inhibition of either ambra1a or ambra1b expression led to a reduced levels of LC3-II in the same experimental settings. In particular, ambra1a ATGMOs-injected embryos show the lowest amount of LC3-II, indicating a main contribution of this gene product at this stage, possibly because of its higher expression level when compared with ambra1b and zygotic ambra1a expression (Fig. 4) or due to subfunction partitioning of the two ambra1 paralogous genes.
Figure 11. Analysis of autophagy and of BECN1 levels in ambra1-MOs injected embryos. (A and B) Analysis of autophagy in ambra1-MOs injected embryos. Protein extracts were prepared from WT and ambra1a (A) or ambra1b (B) MOs-injected embryos at 48 hpf and subjected to immunoblotting analysis using an anti-LC3 antibody. A parallel set of embryos were incubated with the lysosome inhibitor bafilomycin A1 for 6 h before lysis, in order to assess the rate of autophagic flux upon Ambra1 downregulation. A graph reporting data from three independent experiments is shown together a representative immunoblot image. Values represent the densitometric measurement of LC3-II band intensities normalized to the signals of the loading control actin. A.U.: arbitrary units. Please note that this LC3 antibody shows a stronger reactivity for the zebrafish type II form of LC3 than the type I, which is detected only at longer exposure times. However, to ensure that the detected LC3 isoform was LC3-II, protein extracts from HeLa cells were run as a reference marker. (C) Analysis of Becn1 levels in ambra1-MOs injected embryos. Protein extracts were prepared from WT and ambra1-MOs-injected embryos at 48 hpf and subjected to immunoblotting analysis using an anti-BECN1 antibody. Actin expression was monitored as a loading control.
Moreover, we analyzed whether the injection of MOs targeting the ambra1a or the ambra1b genes affects the expression of the proautophagic factor Becn1, a component of the class III phosphatidylinositol 3-kinase complex whose activity is regulated by its interaction with Ambra1.14 Interestingly, Ambra1 downregulation led to increased levels of Becn1 protein, particularly evident with morpholinos targeting both zygotic and maternal transcripts (MO1-ambra1a and ambra1b) (Fig. 11C), which may be interpreted as an attempt of the system to counterbalance the absence of a positive regulator of the autophagic process.
To examine whether Ambra1a and b protein deficiency causes apoptosis, whole-mount TUNEL-staining was used to detect apoptotic cells in WT and morphant embryos analyzed at 24 hpf. Minimal evidence of apoptosis was found in WT (Fig. 12), whereas a highly increased number of TUNEL-positive cells was detectable in the head region of both ATGMOs- and SPLICMOs-injected embryos (Fig. 12), suggesting that the developmental defects may be partially caused by an increase of apoptosis.
Figure 12. TUNEL analysis to detect apoptotic nuclei in WT embryos and ambra1 ATGMOs (MO1-ambra1a and MO1-ambra1b), SPLICMOs (MO2-ambra1a and MO2-ambra1b) and MISMMOs (MO1-ambra1a-5m and MO1-ambra1b-5m) embryos at 24 hpf. Minimal evidence of apoptosis was found in WT while a highly increased number of TUNEL-positive cells was detectable in the head region of ATGMOs- and SPLICMOs-injected embryos. Scale bar: 200 μm. Insert: differences in the TUNEL-positive cell number between both ATGMOs- and SPLICMOs-injected embryos compared with the control embryos (WT and MISMMOs). Values represent the mean ± SD (n = 5). “*” indicates that the difference in the expression levels are significantly different (p < 0.05; **p < 0.01; ***p < 0.001).
The t-test showed significant differences in the apoptotic cell number between both ATGMOs- and SPLICMOs-injected embryos compared with the control embryos (WT and MISMMOs) (graphic in Fig. 12).
Alteration of dorso-ventral patterning in Ambra1-deficient embryos
To further characterize the ATGMOs phenotypes, the expression of marker genes that are critical for dorso-ventral patterning and notochord formation was analyzed by WMISH in morphants and compared with WT or MISMMOs embryos during different developmental stages. For each marker and each developmental stage, 15 embryos from three different microinjection experiments were used (Fig. 13).
Figure 13. WMISH showing expression of the developmental markers, gsc, chd and shha, in ambra1-MOs-injected and control embryos. Dorsal views with the head pointing to the top in 1 dpf embryos. Scale bar: 200 μm.
The in situ hybridization assay showed that MO1-ambra1b and coinjected morphants displayed a decrease in the expression of gsc and a reduction of chd staining in the dorsal axis. The modifications are less evident in MO1-ambra1a morphants and absent with SPLICMOs.
WMISH analysis of sonic hedgehog (shha), a marker for notochordal and floor plate cells, showed undulating notochord in both ATGMOs-injected embryos compared with control ones. This was not evident with SPLICMOs-injected embryos. SPLICMOs do not seem to interfere with the early developmental stages likely due to the presence of the ambra1 maternal transcripts during the first developmental stages.
Discussion
In this study, we report the identification, through genome searches and targeted cloning, of two paralogous ambra1 genes in the zebrafish genome. The conserved synteny with human AMBRA1 gene located in chromosome 11, suggests that ambra1a and ambra1b genes, like other genes in chromosomes 7 and 25 they are linked to, arose as part of the fish-specific whole-genome duplication that occurred after divergence of the fish and tetrapod lineages.21 Analysis performed on other available teleostean genomes shows that only D. rerio retained both ambra1 paralogs, whereas the fate of the second ambra1 gene in the other teleosts examined so far was likely nonfunctionalization and loss. Phylogenetic analysis showed that the D. rerio ambra1b gene is the orthologous form of the other sequenced teleostean ambra1s. Retention of initially redundant genes in teleost genomes is expected to be caused by either neofunctionalization or subfunction partitioning of the multiple roles of their mammalian ortholog, as the maintenance of functional redundancy over long times is unlikely. Thus, the study of paralogous gene function in zebrafish can help in understanding the roles of the homologous human gene in health and disease.16
Zebrafish ambra1a and ambra1b genes share the same genomic organization of mammalian Ambra1 genes, but present some mutual differences: ambra1a is spliced as four alternative transcripts coding for proteins reduced in three ways in their C-terminal region, whereas only one transcript was identified for the ambra1b gene. Interestingly, Ambra1a3 and Ambra1a4 proteins lack the DYNLL1 binding domain, suggesting that these Ambra1 isoforms escape from the inhibitory interaction with the dynein complex. Moreover, the majority of ambra1a transcript variants lacked exon 8 that is instead always present in ambra1b mRNA. Exon 8 is located inside the region involved in the interaction with the BECN1 protein, suggesting a possible modulation of the BECN1 binding properties of AMBRA1 and of its autophagic potential. Although the exon 8 is located inside the region involved in the interaction with BECN1 protein, human AMBRA1 isoforms either containing or not exon 8 are able to bind BECN1,14,22 suggesting the sequence encoding by this small exon may regulate other functions of the protein.
During embryogenesis, at all time points analyzed through the first 6 d of development, the expression of ambra1a mRNA was greater than that of ambra1b messenger, suggesting that ambra1a may have more important functions during development. We find that the expression pattern of these two genes, analyzed by WMISH, is partially overlapping, but the ambra1a probe was more specifically localized in the anterior brain and otic vesicles until 4 dpf. At 6 dpf the signal of both genes was weak and hardly detectable in the brain, but evident in the alimentary canal and in the swim bladder. Although this overlapping expression might suggest a potential for at least partially functional redundancy, this was not supported by the rescue experiments.
We selectively depleted the Ambra1 proteins both separately and together by MO-mediated translation and splicing blockages. The present work demonstrates that both the maternal and zygotic ambra1a and b mRNAs are required for normal embryogenesis and larval development because the MO-induced deficiency of the corresponding proteins is associated with several developmental abnormalities and increased loss of viability at 3 to 4 dpf after fertilization in ATGMOs-injected embryos and at 12 dpf in SPLICMO-injected embryos. This conclusion is supported by the incorporation of several MO controls in the experimental design (see Results). However, the neural abnormalities observed in Ambra1a and b knockdowns are not due to activation of the TP53 pathway because they were still observed after coinjection with tp53 MO. Embryos coinjected with both ATGMOs display even more severe malformations with a complete derangement of the body plan in class III morphants. The increased mortality in zebrafish Ambra1’s targeted loss-of-function is consistent with previous data obtained in a mouse model.14 Rescue experiments show that the two genes could not compensate each other's deficiencies, at least morphologically. Moreover, in the Ambra1a knockdown experiments, the normal phenotype was restored only after coinjection of short and long transcripts, suggesting specific functions also for Ambra1a3 and Ambra1a4 proteins that lack the DYNLL1 binding domain.
The phenotype that we observed with Ambra1a and b silencing with both ATGMOs occurred as early as 24 hpf, suggesting that both paralog genes may play a role in the early development of zebrafish and potentially during gastrulation. AMBRA1 deficiency in mice also results in neural abnormalities, including exencephaly due to failure in neural tube closure.14
Moreover, the two SPLICMOs (MO2-ambra1a and MO2-ambra1b) that target only zygotic ambra1 transcripts, led to developmental defects only at a late stage, indicating the importance of maternally supplied ambra1 transcripts and fundamental functions of the corresponding proteins in early embryonic development.
At the molecular level, the downregulation of Ambra1 expression leads to autophagy impairment associated with increased cell death, confirming the crucial role of Ambra1 in the crosstalk among these processes during embryogenesis and gametogenesis.23,24 In accordance with what was observed in mice,14 an early increased rate of cell proliferation (manuscript in preparation) was observed in zebrafish, that is largely counterbalanced by an excess of cell death.
The comparative analysis of zebrafish morphant phenotypes of autophagy genes confirms a clear difference between the core members of the BECN1 complex and the genes involved at later stages of the autophagic pathway, such as ATG5 and ATG7,25,26 in the regulation of development, as previously reported in mammals. Indeed, the silencing of AMBRA1 results in early embryonic lethality both in fishes and mice with similar defects in neural tube formation. Conversely, zebrafish atg5 silencing causes a longer embryonic survival with respect to ambra1,27 similarly to mice in which death occurs only after birth.25 The earlier appearance of Ambra1a and b morphant phenotypes suggests that Ambra1 may play multiple roles in early development, not all directly related to its positive regulation of the autophagic process. This conclusion is also supported by the observation that conversion to the LC3-II form is observed in zebrafish embryos starting from 32 hpf28 (and data not shown), relatively late compared with the onset of the developmental defects.
Other interesting differences were also observed between Ambra1 and ATG5 morphant embryos when the expression profiles of patterning-regulating genes were analyzed. Indeed, Ambra1 morphant embryos show a ventralized phenotype with reduced or modified expression of the dorsal chd and gsc genes, while ATG5 downregulation causes embryonic over-dorsalized deformities.26 The chd and gsc are organizer-specific genes, expressed in the dorsal mesoderm, which induce the morphogenesis of dorsal structures, such as the notochord.29,30 Therefore, the pattern observed in the in situ hybridization experiment with ATGMOs may explain the ventralized phenotype of morphants. Also shha pattern, a marker of the notochord,31 was differently shaped in morphants resulting in detectable undulations in the notochord with respect to WT, as previously observed in mouse by Fimia and colleagues.14
Importantly, the inhibition of Ambra1 expression interferes with the regulation of cell proliferation and death during embryogenesis in both zebrafish and mice, while this effect was not detected for atg5 gene.27 This is probably due to the fact that the upstream regulators of the autophagic process play a more complex role by interacting and regulating the activity of members of other cell pathways.32 The functional link between proliferation, apoptosis and autophagy suggests that these processes are coordinately regulated in early development in order to limit uncontrolled cell demise and/or cell growth during the profound remodeling occurring in the course of organogenesis.
Collectively, these findings confirm an important developmental role for Ambra1 in early vertebrate embryogenesis and provide compelling evidence for subfunctionalization of ambra1 gene paralogs in zebrafish after ancestral duplication.
Materials and Methods
Fish maintenance and embryo collection
Zebrafish of wild-type (WT) strain, purchased from local pet shops, were maintained according to Westerfield and Lawrence.33,34 Embryos were obtained by natural spawning and cultured in zebrafish Fish Water solution at 28.5°C with a photoperiod of 14 h light/10 h dark. The developmental stages were determined according to the hpfs and morphological features.35
RNA extraction, reverse transcription and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from pools of 20 to 60 embryos at the desired stages of development using TRIzol reagent (Invitrogen, 15596-018) according to the manufacturer’s instructions. The number of embryos was chosen according to the embryo size at each time point. The RNA samples were kept at −80°C until use.
For qPCR, 1 µg of total RNA obtained from three different pools of embryos at each developmental stage was used for cDNA synthesis, employing iScript cDNA Synthesis Kit (Bio-Rad) and following the manufacturer’s protocol. PCRs were performed with SYBR green method in a iQ5 iCycler thermal cycler (Bio-Rad, 179-8891). All samples were analyzed in triplicate in 20 μl volumes. Each reaction mixture consisted of 2 μl of diluted (1/10) cDNA, 10 μl of 2× concentrated iQ TM SYBR Green Supermix (Bio-Rad, 170-8882), containing SYBR Green as a fluorescent intercalating agent, 0.3 μM of forward primer and 0.3 μM of reverse primer. Real-time PCR conditions were optimized after trying various times and temperatures for each cycling step. The reaction conditions were as follows: enzyme activation at 95°C for 3 min followed by 45 cycles of denaturation (30 sec at 95°C), annealing (30 sec at 60°C for arginine rich protein, arp, 30 sec at 54°C for ambra1a and ambra1b, 30 sec at 55°C for 18S, and 30 sec at 60°C for becn1, and extension (20 sec at 72°C). Fluorescence monitoring occurred at the end of each cycle. The extension phase of the last cycle was prolonged by 10 min. Primer specificity and the absence of primer-dimer formation during real-time PCR analysis was indicated in each sample by the presence of a single peak in the dissociation (melt) curve at the end of the amplification program. The specificity of each primer set was also analyzed by sequencing of the obtained fragment. 18S and arp mRNAs were used as normalizers in each sample in order to standardize the results by eliminating variations in mRNA and cDNA quantity and quality. The data obtained were analyzed using the iQ5 optical system software version 2.0 (Bio-Rad) including GeneEx Macro iQ5 Conversion and genex Macro iQ5 files. Modification of gene expression is represented with respect to the control sampled at the same time of the treatment. The primer sequences are reported in Table S4.
Cloning of zebrafish ambra1a and ambra1b cDNAs
A BLASTN search for zebrafish ambra1 cDNA sequence was performed in the NCBI database (www.ncbi.nlm.nih.gov) using the amino acid sequence of mouse AMBRA1 (NP_766257) as query. Two Danio rerio cDNA sequences were obtained: XM_002667669 (coding for ambra1a, as named in this paper) and XR_084457 (coding for ambra1b). ESTs databases were also analyzed but, due to the length of the ambra1 cDNA, no full-length clones were found, but only fragments of both sequences.
Based on the predicted sequences, primers covering overlapping fragments were designed for RT-PCR and to obtain the 5′- and 3′-UTR regions according to the cloning strategy described in Figure S1. RNA ligase-mediated rapid amplification of cDNA 5′-ends (RLM-5′-RACE) and 3′-RACE were performed using the FirstChoice RLM-RACE kit (AM1700M, Ambion), following the manufacturer’s instructions, with the primers reported in Figure S1. These analyses were accomplished using RNA extracted from 2- and 48-hpf-old embryos. The full-length open reading frame (ORF) of zebrafish ambra1a (accession number: XM_002667669) and ambra1b (accession number: XR_084457) were amplified from 2-hpf zebrafish embryo total cDNA with pair of primers located outside of the start and stop codon regions (see Fig. S1) and cloned into pCR®-Blunt vector. Both strands were sequenced for each genes.
Synteny and phylogenetic analyses
Genomic analysis of conserved syntenies between human and zebrafish ambra1 genes were examined using the Synteny database with zebrafish as the source genome (release Sanger Zv9) and the human genome (release GRCh37.p2) as the outgroup. The sliding window size was settled at 25 genes.18 This program is designed for analyzing genomes that have undergone complete duplication during evolution. It identifies pairwise clusters of orthologs and paralogs simultaneously.
Phylogenetic relationships of zebrafish ambra1 genes were derived by aligning their deduced amino acidic sequences together with AMBRA1 sequences reported for other vertebrate species using the Clustal W program.36 All alignment positions were included in the analysis. The tree was generated by the Maximum likelihood method,37 as implemented in the RaxML 7.2.6 program.38 Insertions and deletions were not taken into account. Bootstrap resamplings were also performed to test the robustness of the trees and 1000 replicates were done.39 Program setting is detailed in the figure legend.
Morpholinos
All the MOs were obtained from Gene Tools (Philomath, OR, USA). The antisense MOs used for the silencing of zebrafish ambra1a and ambra1b were designed against the ATG translation initiation site according to the following sequences: MO1-ambra1a (5-CTC CAA ACA CTC TTC CTC ACT CCC T -3′) and MO1-ambra1b (5′-TTT TCC TCT TTA GTG CTC CAC GGC C-3′).
The antisense splice variant MOs were designed for both genes on the sequence at the exon 3-intron 3 junction: MO2-ambra1a: 5′-TGT AAT CAA AGT GGT CTT ACC TGT C-3′ and MO2-ambra1b: 5′- TGA AAT TGA TTG TTA CCT ATC TGG A-3′ (the exon complementary sequence is underlined).
As controls, we used five-nucleotide-mismatched control MOs: MO1-ambra1a-5m (5′- CTC gAA AgA CTg TTC CTg AgT CCC T-3′) and MO1-ambra1b-5m (5′-TTa TgC TgT TTA GTc CTC CAC cGC C-3′) (lowercase bold letters indicate the mismatch changes). Finally, we used an MO against the ATG translation initiation site of tp53 mRNA, tp53 MO: 5′-GCG CCA TTG CTT TGC AAG AAT TG-3′ (MO4-tp53).20
All the MOs were reconstituted at 1 mM in nuclease-free water. Different MOs concentrations were tested in a range between 1 and 2.5 µg/µl. MOs and mRNAs (see below) were injected into the yolk mass of 1- or 2-cell embryos. These were then incubated in 1× Fish Water (50×: 25 g Istant Ocean, 39.25 g CaSO4, and 5 g NaHCO3 for 1 l) at 28.5°C. MOs- and/or mRNAs-injected embryos were raised to the desired stages for observations or collected for further analysis.
In order to verify the knockdown effectiveness of MO1-ambra1a and MO1-ambra1b, the 5′-UTR region of both paralogs, containing the MOs target sequence, was cloned in pCR®-BluntII-TOPO vector (Invitrogen, K280002), in-frame upstream of the fluorescent protein EGFP coding region. The nucleotide portion including MOs target sequence and EGFP was XbaI and EcoRI digested and subcloned into pCS2+ expression vector. The resulting expression vectors were verified by sequencing. Linearized plasmids (1 μg) were used as templates for the synthesis of capped synthetic mRNAs with the mMESSAGE mMACHINE® SP6 Kit (Ambion, AM1340). Synthetic mRNAs were precipitated by centrifugation in LiCl and their integrity confirmed by formaldehyde-agarose electrophoresis. Stock of synthetic mRNAs solutions were quantified by absorbance and diluted to desired concentrations in 1× Danieau’s buffer for microinjection.
The effectiveness of SPLICMOs for exon skipping was confirmed by the presence of a significant amount of shorter RNA products detected by RT-PCR with two set of specific primers flanking the splice site (Am1a-F5 with Am1a-R6 for ambra1a and Am1b-F5 with Am1b-R4 for ambra1b, see Table S4).
In vitro transcription
For rescue experiments, since the injected synthetic mRNAs should not contain the MOs target sequences, modified cDNAs were amplified for ambra1a1, ambra1a3 and ambra1b with forward primers containing silent mutations and the restriction site of ClaI and reverse primers containing the restriction site of XhoI (see Fig. S1 and Table S4 for details). The resulting PCR products were cloned into the pCR®-Blunt vector and, after complete sequencing to check for lack of nucleotide changes that could alter the amino acidic sequence or interrupt the translation reading frame, subcloned into pCS2+ expression vector using ClaI and XhoI restriction enzymes.
Linearized plasmids (1 µg) were used as templates for the synthesis of capped synthetic mRNAs, which were purified as described above.
The pGEM plasmids containing the last 280 and 267 nucleotides plus the 3′-UTR regions of ambra1a1 and ambra1b, respectively, were used to synthesize digoxigenin (DIG)-labeled riboprobes for whole-mount in situ hybridization (WMISH). The PCR fragments were then cloned into pGEM-T Easy vector. DIG-labeled riboprobes were synthesized by in vitro transcription with SP6 and T7 RNA polymerases, respectively (Sp6 RNA polymerase, 10 810 274 001; T7 RNA polymerase, Roche, 10 881 767 001), following the manufacturer’s instructions.
Whole-mount in situ hybridization and microscopy analyses
WMISH was performed, on normal and treated embryos, essentially as reported by Thisse and Thisse.40 Embryos were fixed overnight in 4% paraformaldehyde (Sigma, PFA, P6148) in phosphate-buffered saline (PBS) at the desired stage of development. For embryos older than 24 hpf, pigmentation was suppressed by raising embryos in 0.03% PTU (1-phenyl-2-thiourea, Sigma, P7629) in Fish Water. Afterwards, the embryos were washed in PBT (PBS plus 0.1% Tween 20, P1379, Sigma) and dechorionated with forceps. They were treated with methanol and stored at −20°C. Methanol-stored embryos were rehydrated in methanol/PBS series, permeabilized by proteinase K (10 µg/ml), prehybridized, and then hybridized overnight at 65°C with 1 ng/µl of the appropriate riboprobe in the Hybridization Mix (HM: 50% formamide, 5 × SSC, 0.1% Tween 20, 50 µg/ml heparin and 500 µg/ml tRNA). After HM/SSC and SSC/PBT washing series, embryos were preincubated in blocking solution (2% sheep serum, and 2 mg/ml BSA in PBT) and then incubated overnight at 4°C with alkaline phosphatase (AP)-conjugated anti-DIG antibodies (Roche, 11 093 274 910) diluted 1:3,000 in blocking solution. After PBT washing, embryos were pre-soaked in staining buffer and then incubated in NBT/BCIP (5-nitro-blue tetrazolium chloride/bromo-4-chloro-3′-indolyphosphate p-toluidine salt Stock solution, Roche, 11 681 451 001) for blue staining. WMISH-stained embryos were mounted in 80% glycerol in PBT or cleared and mounted in 2:1 benzyl benzoate/benzyl alcohol, observed under a Leica DMR microscope (Leica Microsystems Srl) and photographed with a Leica DC500 digital camera (Leica Microsystems Srl)
All riboprobes for WMISHs are listed in Table S5.
For histology analysis, zebrafish WMISH-stained larvae at 6 dpf were fixed in 4% paraformaldehyde at RT for 2 h and processed into an alcohol–xylene series followed by paraffin embedding.
Blood vessels were visualized by endogenous alkaline phosphatase activity at 3 dpf following the protocol of Serbedzija et al.41
For scanning electron microscopy (SEM) material was fixed in 6% glutaraldehyde in 0.1 M cacodylate buffer (pH 6.9) overnight at 4°C. The post-fixed samples were dehydrated in a graded ethanol series, dried at the critical point and coated with gold. The samples were observed with a Stereoscan 260 (Cambridge Instruments, Ltd.) scanning electron microscope, operating at 12 kV. For semithin section, material was fixed in 6% glutaraldehyde in 0.1 M cacodylate buffer (pH 6.9) overnight at 4°C. After washing in cacodylate buffer, the specimens were post-fixed in 1% OsO4 in the same buffer for two h and dehydrated in a graded ethanol series followed by propylene oxide. The specimens were stained with uranyl acetate while undergoing dehydration in 75% ethanol. The samples were embedded in Araldite resin. Thin sections were cut with an Ultracut S, Reichert ultramicrotome, stained with toluidine blue and examined with a light microscope.
Immunoblotting analysis
Embryos were homogenized in CelLytic Mammalian Tissue extraction reagent (Sigma-Aldrich, C3228). Twenty micrograms of protein extracts were separated on a 13.5% SDS-polyacrylamide gel and electroblotted onto PVDF (Millipore, IPVH20200) membranes. Blots were incubated with a rabbit anti-LC3 antibody, (Cell Signaling, 2775) and a rabbit anti-BECN1 antibody (Santa Cruz, H-300 11427) in 5% nonfat dry milk in TBS plus 0,1% Tween20 overnight at 4°C. Detection was achieved using horseradish peroxidase-conjugate anti mouse IgG antibody (Jackson Immunoresearch, 711-036-152) and visualized with ECL PRIME (GE Healthcare, RPN2232) using ECL-Hyperfilm (GE Healthcare, 28-9068-7). Rabbit anti-Actin antibody (Sigma Aldrich, A2066), was used to monitor equal protein loading. The antibody is made against the SGPSIVHRKCF peptide that is 100% identical in β and gamma Actin and 91% in α Actin.
To assess the rate of autophagic flux upon Ambra1 downregulation, Lc3-II levels were analyzed in 48 hpf embryos incubated with the lysosome inhibitor bafilomycin A1 (1 μM) (Sigma Aldrich, B1793) for 6 h and then fixed for immunoblotting analysis.
Quantification of chemiluminescent signals was performed with the Gel Doc densitometer (BioRad,) using the Quantity One imaging software, version 4.4.0. All samples were run in triplicate and averaged.
Apoptotic assays
Apoptotic cells in the embryos were detected by the TdT-mediated fluorescein-dUTP nick-end labeling (TUNEL) assay. The TUNEL assay was performed using alkali stable digoxigenin-dUTP and TdT (terminal deoxynucleotidyl transferase) (Roche, 03 333 574 001). Embryos were fixed in 4% PFA (overnight, 4°C), treated with methanol and stored at −20°C. Methanol-stored embryos were rehydrated in methanol/PBS series and permeabilized by proteinase K (10 µg/ml). Afterwards, the embryos were washed in PBT (5 × 5 min, RT) and in ethanol/acetic acid (2:1) (20 min RT). After incubation with TUNEL buffer (30 min, RT), embryos were incubated in 100 µl TUNEL reaction mixture (overnight, RT). The reaction was stopped by washing samples with PBT/EDTA 1mM (2 × 1 h, RT). Samples were then processed as for the whole-mount in situ hybridization and stained with the AP substrate Fast Blue [Fast Blue BB 4-benzoylamino-2,5-diethoxybenzenediazonium chloride hemi (zinc chloride) salt, Sigma, F3378] plus NAMP (3-hydroxy-2-naphthoic acid 2,4-dimethylanilide phosphate, Sigma, N5000). Stained embryos were mounted in 80% glycerol in PBT and examined with Leica SP5 confocal microscope.
Apoptotic cells were estimated within the same area in five embryos of each experimental condition. All data were represented as the mean ± SD. Statistical significance was tested by Student's t-test. Groups were considered significantly different if p < 0.05.
Nucleotide sequencing
Sequencing was performed on double-stranded DNA using the ABI PRISM Dye Terminator Cycle Sequencing Core Kit (Applied Biosystems). Electrophoresis of sequencing reactions was completed on the ABI PRISM model 377, version 2.1.1 automated sequencer.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. E. Negrisolo for help in phylogenetic analysis and comments and L. Pivotti for the valuable assistance in the fish facility. Research was supported by 60% funding from the Ministry of the University and Scientific and Technological Research of Italy (L.D.V. Padova, O.C. Ancona and M.P. Rome), FIRB “Accordi di programma” (M.P.), the Italian Ministry of Health (“Ricerca Corrente” M.P. and G.M.F.), “Ricerca Finalizzata” (RF08MDPINK M.P.).
Glossary
Abbreviations:
- AMBRA1
autophagy/Beclin 1 regulator 1
- ATG
autophagy-related proteins
- ATG5
autophagy-related 5
- ATG7
autophagy-related 7
- ATG14
autophagy-related 14
- ATGMOs
translation blocking morpholinos
- Baf A1
bafilomycin A1
- BCL2
B-cell CLL/lymphoma 2
- BECN1
Beclin 1, autophagy-related
- cDNA
complementary DNA
- chd
chordin
- chrm4
cholinergic receptor, muscarinic 4
- CNS
central nervous system
- DIG
digoxigenin
- dgkz
diacylglycerol kinase, zeta
- DYNLL1
dynein, light chain, LC8-type 1
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- EST
expressed sequence tags
- gsc
goosecoid homeobox
- KIAA0226
RUBICON/RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein
- MAP1LC3/LC3
microtubule-associated protein 1A/1B-light chain 3
- mdk
midkine (neurite growth-promoting factor 2)
- MISMMOs
five-nucleotide-mismatched MOs
- MOs
morpholinos
- mRNA
messenger RNA
- NBT/BCIP
5-nitro-blue tetrazolium chloride/bromo-4-chloro-3′-indolyl phosphate p-toluidine salt
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PFA
paraformaldehyde
- PIK3C3
phosphatidylinositol 3-kinase, catalytic subunit type 3
- PIK3R4/Vps15/p150
phosphoinositide-3-kinase, regulatory subunit 4
- phf21a
PHD finger protein 21A
- qPCR
real time-polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcriptase-polymerase chain reaction
- SEM
scanning electron microscopy
- S.E.M.
standard error of the mean
- shha
sonic hedgehog a
- SH3GLB1
SH3-domain GRB2-like endophilin B1
- SPLICMOs
splicing blocking morpholinos
- TP53
tumor protein p53
- TUNEL
terminal deoxynucleotidyl transferase-mediated fluorescein-dUTP nick-end labeling assay
- UVRAG
UV radiation resistance-associated
- WMISH
whole mount in situ hybridization
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/23278
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23278
References
- 1.Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, et al. Mammalian macroautophagy at a glance. J Cell Sci. 2009;122:1707–11. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp ZD. Aging and TOR: interwoven in the fabric of life. Cell Mol Life Sci. 2011;68:587–97. doi: 10.1007/s00018-010-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21:691–8. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–12. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 6.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 7.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–20. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 10.Di Bartolomeo S, Nazio F, Cecconi F. The role of autophagy during development in higher eukaryotes. Traffic. 2010;11:1280–9. doi: 10.1111/j.1600-0854.2010.01103.x. [DOI] [PubMed] [Google Scholar]
- 11.Huang WP, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct. 2002;27:409–20. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- 12.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Y, Wang QJ, Yue Z. Atg14L and Rubicon: yin and yang of Beclin 1-mediated autophagy control. Autophagy. 2009;5:890–1. doi: 10.4161/auto.9162. [DOI] [PubMed] [Google Scholar]
- 14.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 15.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–90. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 18.Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volff JN. Genome evolution and biodiversity in teleost fish. Heredity (Edinb) 2005;94:280–94. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- 20.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–45. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- 22.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012;19:1495–504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gioacchini G, Dalla Valle L, Benato F, Fimia GM, Nardacci R, Ciccosanti F, et al. Autophagy and apoptosis interplay in Danio rerio follicles development: role of probiotic. Reprod Fertil Dev. 2012 doi: 10.1071/RD12187. In press. [DOI] [PubMed] [Google Scholar]
- 25.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011;7:1514–27. doi: 10.4161/auto.7.12.18040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009;5:520–6. doi: 10.4161/auto.5.4.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller-Bertoglio VE, Fisher S, Sánchez A, Mullins MC, Halpern ME. Differential regulation of chordin expression domains in mutant zebrafish. Dev Biol. 1997;192:537–50. doi: 10.1006/dbio.1997.8788. [DOI] [PubMed] [Google Scholar]
- 30.Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho KW, De Robertis EM, Nüsslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–52. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- 31.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–44. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 32.Fimia GM, Piacentini M. Regulation of autophagy in mammals and its interplay with apoptosis. Cell Mol Life Sci. 2010;67:1581–8. doi: 10.1007/s00018-010-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 3rd Edition. Eugene, OR, University of Oregon Press 1995. [Google Scholar]
- 34.Lawrence C. The husbandry of zebrafish (Danio rerio): a review. Aquaculture. 2007;296:1–20. doi: 10.1016/j.aquaculture.2007.04.077. [DOI] [Google Scholar]
- 35.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsenstein J. Inferring phylogenies. Sunderland (MA): Sinauer Associates. 2004. [Google Scholar]
- 38.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Confidence limits on phylogenies: an approach using bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 40.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 41.Serbedzija GN, Flynn E, Willett CE. Zebrafish angiogenesis: a new model for drug screening. Angiogenesis. 1999;3:353–9. doi: 10.1023/A:1026598300052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.