Abstract
Autophagy is a conserved degradation process, which plays important pathophysiological roles. The lack of effective inhibitors of autophagy has been an obstacle in both basic research and understanding the physiological role of autophagy in disease manifestation. The most widely used inhibitor, 3-methyladenine (3-MA), is poorly soluble at room temperature and is effective only at high concentrations. In this study, we synthesized a library of small compounds by chemically modifying 3-MA and screened this library for autophagy inhibitors. Three 3-MA derivatives generated through this approach showed improved solubility and effectiveness in inhibiting autophagy. We demonstrated that chemical modification of an existing autophagy inhibitor is an effective method to generate improved autophagy inhibitors.
Keywords: autophagy, inhibitor, 3-MA, synthesis, derivatives
Introduction
Autophagy is an evolutionarily conserved degradation process. During autophagy, cellular contents are engulfed by double-membrane vesicles named autophagosomes and subsequently delivered to lysosomes for degradation.1 Autophagy plays important physiological roles and various diseases have been linked to autophagy dysfunction. There are strong genetic and animal data supporting a direct connection between autophagy and cancers, neurodegenerative diseases, and chronic inflammation. For example, mice lacking Becn1, an essential autophagy gene, are more prone to tumor formation.2,3 Mice carrying a neuron-specific knockout of another autophagy gene, Atg7, develop age-dependent neuron degeneration.4 A third essential autophagy gene, ATG16L1, has recently been identified to be a Crohn disease susceptibility gene in humans.5 Although the exact roles of autophagy in each of these diseases remain to be determined, altering the autophagy level has become a potential target for therapeutic approaches to various diseases.
The lack of effective inhibitors of autophagy has been an obstacle to both basic research and to proper understanding of the physiological role of autophagy in disease manifestation. Only a few drugs are currently available for inhibiting autophagy activity.6-9 The most widely used is 3-methyladenine (3-MA), a class III phosphatidylinositol 3-kinase (PtdIns3K) inhibitor. Despite its wide usage, 3-MA is a difficult compound to work with: it is effective only when a very high concentration is used, and its solubility at room temperature is poor. Finally, a recent study indicated that 3-MA also inhibits the class I PI3K (phosphoinositide 3-kinase).10 Thus, development of more soluble, effective, and specific autophagy inhibitors is essential for developing autophagy-based therapeutic approaches for various diseases.
To develop autophagy inhibitors with both high solubility and high inhibitory activity, we synthesized a library of 3-MA derivatives. Through imaging-based screening, we identified three molecules, which inhibit autophagy and show improved specificity, solubility and effectiveness compared with 3-MA.
Results
Synthesis of 3-MA derivatives
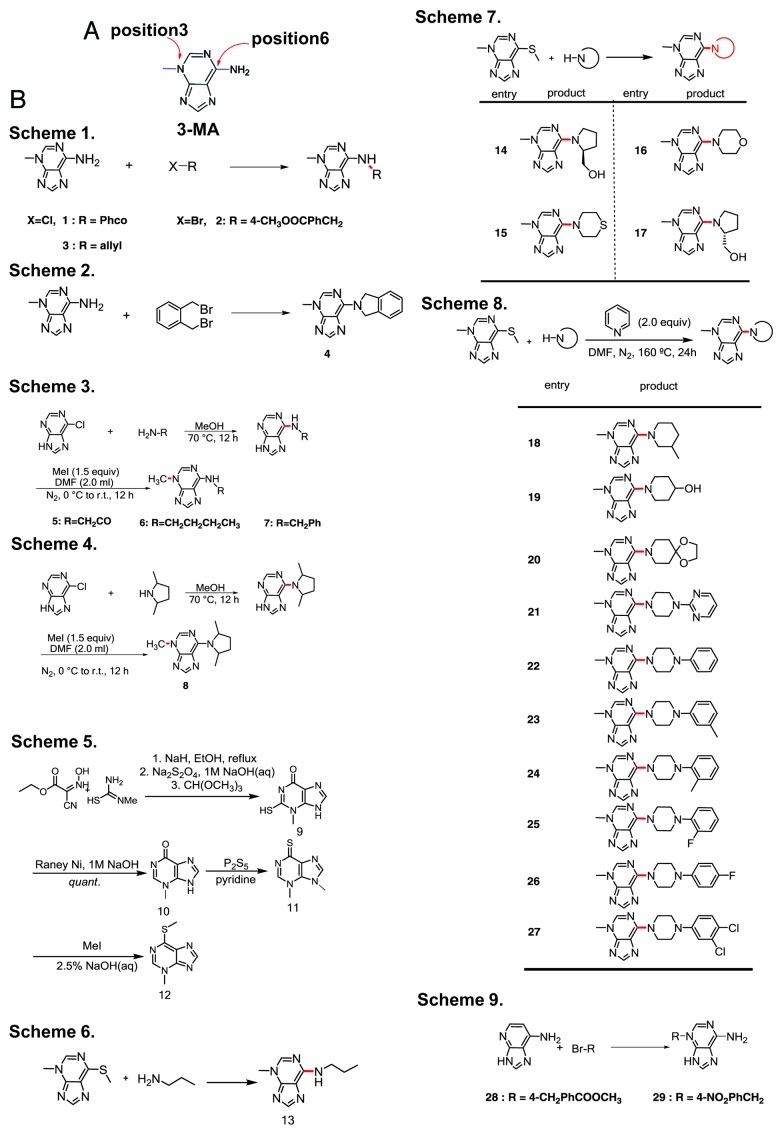
To develop drugs that are potentially better autophagy inhibitors than 3-MA, we designed a small-molecule library based on 3-MA with various modifications and substitutions. The NH2 group at the C6 position of 3-MA lends itself to the introduction of polar functionalities that may modify the solubility and bioaffinity of 3-MA. Although 3-MA is an ambident nucleophile, alkylations and acylations tend to occur primarily at the exocyclic amino group.
We used several different strategies to prepare 29 derivatives of 3-MA (Fig. 1). Direct modification of 3-MA with electrophiles generated N-substituted or N,N-disubstituted products (Fig. 1B, Schemes 1–2). In a different approach, we treated 6-chloro-adenine with several primary and secondary amines, then methylated the products to give N-methyl-adenines that are differentially substituted at the 6-amino position (Fig. 1B, Schemes 3–4). A complementary approach relies on nucleophilic substitution to displace a 6-methylthio group with various amines (Fig. 1B, Schemes 5–8). For benzyl halides with an electron-withdrawing group, alkylation at N3 is feasible (Fig. 1B, Scheme 9).
Figure 1. Preparation of analogs of 3-MA. (A) The structure of 3-MA. (B) Schemes 1 to 2: 3-MA was directly modified with various electrophiles to get N-substituted or N,N-disubstituted products (compounds 1–4). Schemes 3 to 4: 6-chloro-adenine was reacted with several primary and secondary amines, then the products were methylated to afford N-methyl-adenines that are differentially substituted at the 6-amino position (compounds 5–8). Schemes 5 to 8: A thioether precursor was first synthesized as previously described.11-13 N-substituted 3-MAs (compounds 9–13 and 14–27) were obtained by using primary and secondary amines to substitute the methylthio group. Scheme 9: 3-MA alkylation reactions (compounds 28 and 29).
Screening autophagy inhibitors from the 3-MA derivative library
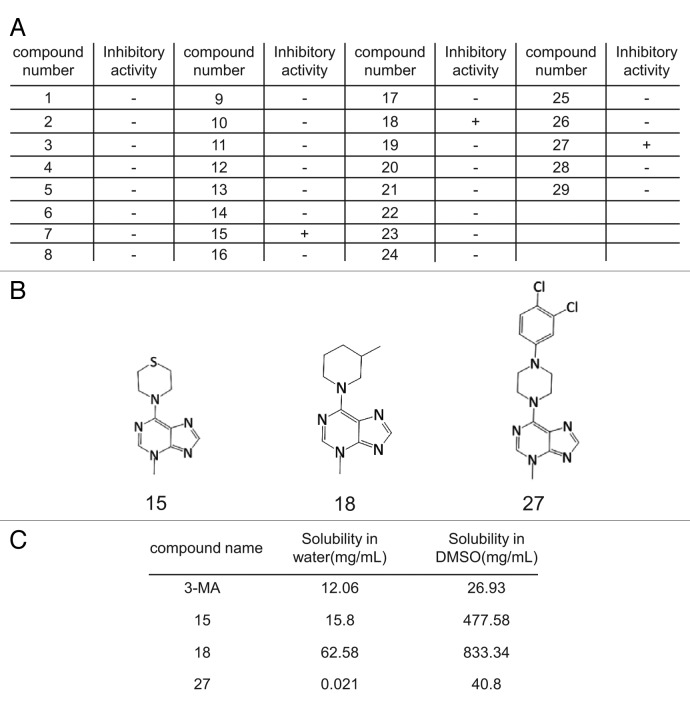
Autophagy can be monitored by imaging autophagosome formation. After induction of autophagy, the autophagy marker LC3, which is normally dispersed in the cytosol is cleaved and lipidated to generate the LC3-II form, and translocates to the autophagosome membrane, where it can be visualized by microscopy as puncta. Thus, autophagosomes can easily be monitored by following the localization of GFP-LC3.14,15 Using a normal rat kidney (NRK) cell line in which GFP-LC3 is stably expressed, we screened the 3-MA derivative library for autophagy inhibitors (Fig. 2A). Among 29 molecules tested, we found 3 compounds that inhibit autophagy (Fig. 2B), namely compounds 15 [4-(3-methyl-3H-purin-6-yl)thiomorpholine], 18 [3-methyl-6-(3-methylpiperidin-1-yl)-3H-purine] and 27 [6-(4-(3,4-dichlorophenyl)piperazin-1-yl)-3-methyl-3H-purine]. The solubility of these three active compounds was determined in water and dimethyl sulfoxide (DMSO) and compared with 3-MA (Fig. 2C). All three were more soluble than 3-MA in DMSO, and 15 and 18 were more soluble than 3-MA in water.
Figure 2. Identification of autophagy inhibitors by imaging-based screening. (A) GFP-LC3-NRK cells were starved for 4 h with or without 3-MA derivatives at 0.1 mM, 1 mM or 10 mM and at least three replicates were performed. Cells were observed by confocal microscopy and formation of GFP-LC3 puncta was assessed by visual screening. Activity of the compounds was scored as “+” if the formation of starvation-induced puncta was inhibited by the addition of the respective compound (at least with the two higher doses) and “-” if no inhibition of puncta formation was observed. (B) The structures of the three autophagy inhibitors identified by screening. (C) The solubility of 3-MA and the three active derivatives in DMSO or water at 37°C.
Derivatives 15, 18 and 27 show improved effectiveness in autophagy inhibition
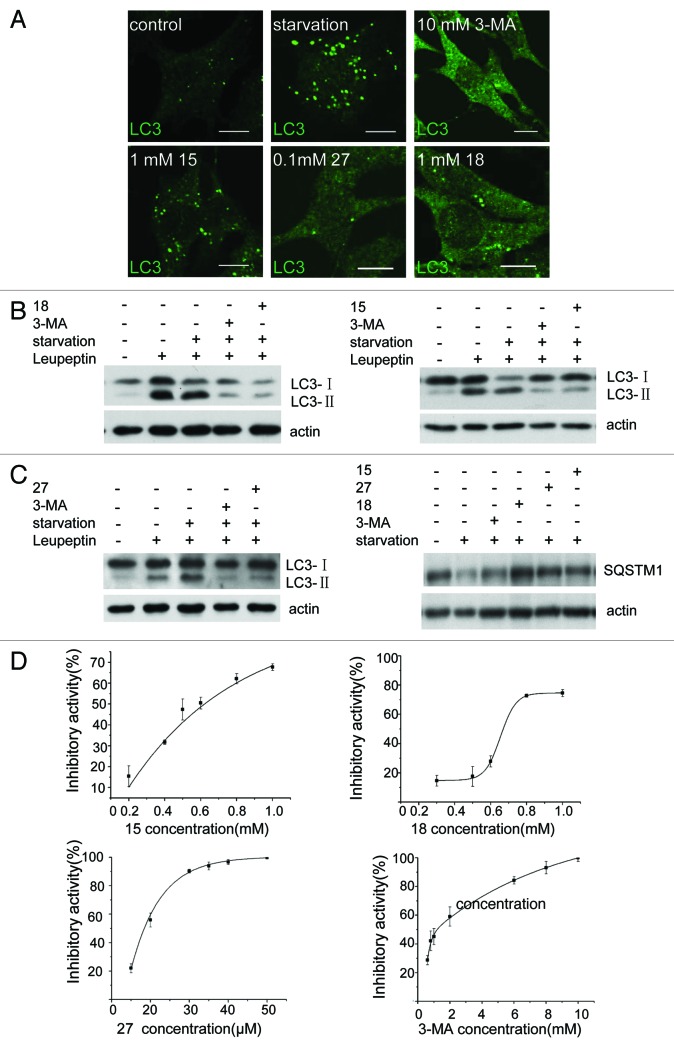
To confirm the effectiveness of 15, 18 and 27 in inhibition of autophagy, we tested endogenous LC3 puncta formation (Fig. 3A), LC3 processing and SQSTM1/p62 degradation (Fig. 3B and C).14-16 All three assays indicated that these compounds were able to block autophagy induced by nutrient starvation. Next, we measured the effective concentrations of these compounds. We found that 27 can block > 90% of starvation-induced autophagy at 30 μM, whereas 15 and 18 block 70% to 80% of starvation-induced autophagy at 0.8 mM and 1 mM respectively (Fig. 3D). In contrast, the concentration of 3-MA required for > 80% inhibition of autophagy is 6 mM. The IC50 values are 1.21 mM for 3-MA, 0.62 mM for 15, 0.67 mM for 18 and 18.5 μM for 27. Our data shows that these compounds are more effective inhibitors of autophagy than 3-MA.
Figure 3. Derivatives 15, 18 and 27 inhibit autophagy in NRK cells. (A) NRK cells were starved for 4 h with or without 10 mM 3-MA, 0.1 mM 27, 1 mM 18 or 1 mM 15. Cells were stained with an anti-LC3 antibody and observed by confocal microscopy. Scale bar: 10 μm. (B) Leupeptin (10 μg/ml)-treated NRK cells were starved for 4 h with or without 10 mM 3-MA, 1 mM 18, 1 mM 15 or 0.1 mM 27, and processing of LC3 was measured by western blot using anti-LC3 antibody. (C) NRK cells were starved for 4 h with or without 10 mM 3-MA, 1 mM 18, 1 mM 15 or 0.1 mM 27 and the level of SQSTM1 protein was measured by anti-SQSTM1 antibody. (D) Dose-response of the three compounds compared with 3-MA. GFP-LC3-NRK cells were starved for 4 h with or without the indicated compounds. The level of LC3-positive puncta per cell was quantified in at least 30 cells. Autophagy inhibition index = % (1 − (a − b) / (c − b)), where “a” means the number of GFP-LC3-positive puncta per cell in cells treated with compound + DPBS; “b” means the number of GFP-LC3-positive puncta per cell in DMSO-treated cells; “c” means the number of GFP-LC3-positive puncta per cell in DPBS-treated cells. Results were analyzed with OriginPro8 software.
Derivatives 15, 18 and 27 inhibit omegasome formation
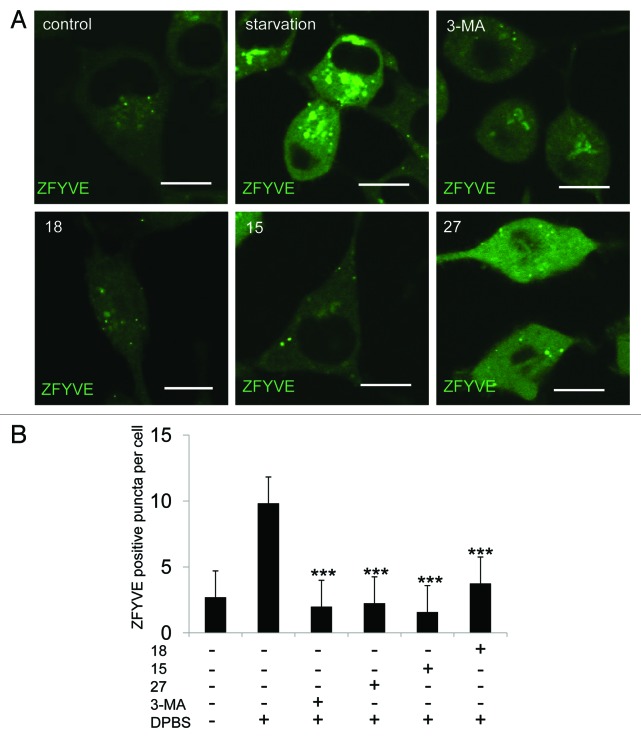
It is well established that 3-MA inhibits autophagy by inhibiting the class III PtdIns3K.9 During autophagy, phosphatidylinositol 3-phosphate (PtdIns3P)-enriched structures named omegasomes are formed on the endoplasmic reticulum.17 The formation of omegasomes is completely dependent on PIK3C3/VPS34 and can be inhibited by treating cells with 3-MA.17 To test whether these three new compounds can also inhibit omegasome formation, we added them to an NRK cell line, which stably expresses GFP-ZFYVE/DFCP1 (green fluorescent protein-zinc finger, FYVE domain containing 1), a specific marker for omegasomes. We found that all three compounds blocked starvation-induced omegasome formation (Fig. 4A and B). Thus, we concluded that it is likely these compounds block autophagy by inhibiting PIK3C3.
Figure 4. Derivatives 15, 18 and 27 inhibit omegasome formation. (A) GFP-ZFYVE NRK cells were unstarved or starved for 2 h with or without 10 mM 3-MA, 1 mM 15, 1 mM 18 or 0.1 mM 27, and imaged by confocal microscopy. Scale bars: 10 μm. (B) Cells from (A) were quantified for the number of omegasomes. At least 30 cells were counted. Error bars, s.d.; ***p = 0.0001 compared with starved cells by Student’s t-test.
Derivatives 15, 18 and 27 do not inhibit class I PI3K
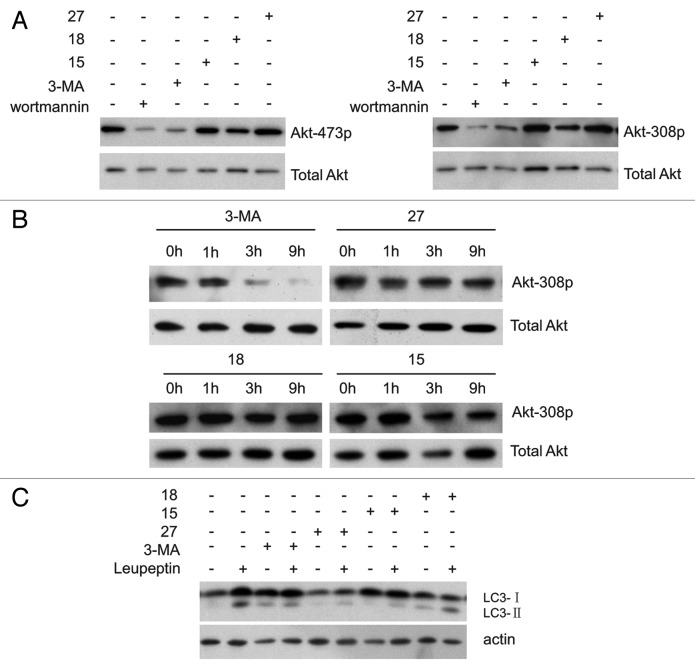
It has been reported that 3-MA inhibits the activity of the class I PI3K in addition to the class III PtdIns3K.10 AKT, a downstream target of the class I PI3K, is activated by insulin and various growth and survival factors.18,19 AKT can be activated by phosphorylation at Thr308 by phosphoinositide-dependent kinase 1 (PDPK1) and Ser473 by the MTOR complex, and these phosphorylation events are blocked by the PI3K and PIK3C3 inhibitor wortmannin.20 It has also been reported that 3-MA inhibits the phosphorylation of AKT at Thr308 and Ser473.21 To determine whether these compounds inhibit the activity of class I PI3K, we tested the phosphorylation of AKT at Thr308 and Ser473 by immunoblotting. AKT phosphorylation was decreased in NRK cells treated for 4 h with wortmannin and 3-MA, but was unchanged in cells treated with 15, 18 or 27 (Fig. 5A), phosphorylation of AKT is further decreased after prolonged treatment with 3-MA, but not with 15, 18 or 27 (Fig. 5B). It has been confirmed that 3-MA causes increased levels of LC3-II by blocking the activity of the class I PI3K.10 As shown in Figure 5C, the protein level of LC3-II in the 3-MA-treated cells is higher than in cells treated with 15, 18 and 27. Taken together, these results indicated that 15, 18 and 27 do not inhibit class I PI3K.
Figure 5. Derivatives 15, 18 and 27 do not inhibit AKT phosphorylation. (A) NRK cells were incubated with 2 μM wortmannin, 10 mM 3-MA, 0.1 mM 27, 1 mM 15 or 1 mM 18 for 4 h and total cell extracts were immunoblotted with antibodies against AKT and phospho-AKT. (B) NRK cells were incubated with 10 mM 3-MA, 0.1 mM 27, 1 mM 15 or 1 mM 18 for the indicated time and total cell extracts were immunoblotted with antibodies against AKT and phospho-308 AKT. (C) Leupeptin (10 μg/ml)-treated NRK cells were incubated with or without 10 mM 3-MA, 1 mM 18, 1 mM 15 or 0.1 mM 27 for 4 h, and the processing of LC3 was measured by western blot using an anti-LC3 antibody.
The effect of derivatives 15, 18 and 27 on viability, cellular ATP level, endocytosis and proteasome activity
To determine the effect of these compounds on various important cellular functions, we compared viability, cellular ATP, protein level, proteasomal degradation and endocytosis. We found that compared with 3-MA, cells treated with 27 and 18 have reduced viability while cells treated with 15 have greater viability (Fig. S1A). Cells treated with 27 and 18 had reduced ATP levels, whereas cells treated with 15 have increased ATP levels (Fig. S1B). Cells treated with compound 15 and 18 had similar or increased protein levels compared with 3-MA, whereas cells treated with 27 had decreased protein levels (Fig. S1C).
The effect of 3-MA on endocytosis is controversial.6,22 To investigate whether endocytosis is affected by 15, 18 and 27 NRK cells were treated with these compounds for 4 h and incubated with Alexa 488-conjugated transferrin (TF), which is internalized by endocytosis. After washing away the surface TF, cells were imaged by confocal microscopy and the fluorescence intensity was measured by flow cytometry. We found that endocytosis is not inhibited in cells treated with these compounds for 0.5 h. However, in cells treated with 3-MA for 4 h, the internalization of Alexa 488-conjugated TF was partially blocked. The imaging data also show that 3-MA, and to a lesser degree 27, but not 15 or 18, blocked the internalization of TF and caused accumulation of TF on the plasma membrane (Fig. S2A and S2B).
Ub-G76V-GFP is a short-lived green fluorescent protein for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Previous literature has reported that prolonged treatment with 3-MA inhibits proteasome degradation.23 Similarly, we found that treatment with 3-MA, 15, 18 and 27 for 4 h all caused increased Ub-G76V-GFP levels. Fifteen and 18 had a similar effect to 3-MA, while 27 caused slightly more accumulation of Ub-G76V-GFP than 3-MA (Fig. S3A and S3B).
Discussion
In this work, we described an approach to generate autophagy inhibitors by optimizing an existing autophagy inhibitor, 3-MA. By modifying the C6 and N3 side chains, we generated a small library of 3-MA derivatives, and performed imaging-based screening to test the inhibitory properties of each compound. Promising candidates were further validated by additional autophagy assays. Through this approach, we generated three new autophagy inhibitors, which have better solubility and effectiveness than 3-MA. More importantly, these 3-MA derivatives do not inhibit class I PI3K. We also tested the effect of these compounds on various cellular functions including viability, cellular ATP level, protein level, endocytosis and proteasome activity. We found that for most of these cellular functions, the new compounds have a similar or milder effect than 3-MA. However, some effects of the new compounds on key cellular functions are slightly more severe; for example, 27 caused a greater loss of viability than 3-MA. Further optimization may increase the effectiveness and reduce the adverse consequences of these compounds.
Materials and Methods
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde then washed with PBS twice. After blocking with 10% FBS in PBS for 20 min, cells were incubated with anti-LC3 primary antibody and secondary antibody (Alexa Fluor546 goat anti-rabbit, Invitrogen, A11035), then washed with PBS three times. Imaging was performed with an Olympus FV1000 confocal microscope.
Western blotting
NRK cells were lysed in 2% SDS and the extracts were separated by SDS-PAGE, then transferred to polyvinylidene difluoride membrane. After blocking with 5% milk in Tris-buffered saline containing 0.1% Tween for 1 h, the membrane was incubated with primary antibodies and secondary antibodies (Goat anti-mouse IgG1, Southern Biotech, 1070-05; goat anti-rabbit, Southern Biotech, 4050-05). ECL western blotting detection reagents (Pierce, 32106) were used for the detection step.
Establishing NRK cell lines stably expressing GFP-LC3 (GFP-LC3-NRK cells)
GFP-LC3 was amplified from pEGFP-LC3 and cloned into an entry vector using a pENTR Directional Top Clone Kit (Invitrogen, K2400-20). The promoter of EEF1D (kindly provided by Professor Kehkooi Kee, Tsinghua University) was cloned into pENTR5′-TOPOTA with the pENTR5′-TOPOTA Cloning Kit (Invitrogen, K591-10). These two entry vector inserts were recombined into the lentiviral vector p2K7 using LR Clonase II Enzyme Mix (Invitrogen, 17791-020). Lentiviral stocks were obtained by cotransfecting the plasmids delta 8.9, p2K7-GFP-LC3 and VSVG into 293FT cells. After infecting NRK for 48 h, cell lines stably expressing GFP-LC3 were obtained.
Establishing an NRK cell line stably expressing GFP-ZFYVE1
ZFYVE1 was amplified from HeLa cDNA and inserted into pEGFP-C2. pEGFP-ZFYVE1 was transfected into NRK cells using Amaxa nucleofection solution T. One mg/ml G418 was then added to generate the cell line stably expressing GFP- ZFYVE1.
Live cell imaging
GFP-LC3 or GFP-ZFYVE1 stable cell lines were seeded at 37°C with 5% CO2 in a PeCon open chamber (PeCon). Images were acquired by confocal microscopy (Olympus FV1000).
Reagents and antibodies
3-MA (M9281) and leupeptin (103476-89-7) were purchased from Sigma. Anti-LC3 polyclonal antibody (PM036) for immunofluorescence staining was obtained from MBL. Anti-LC3 polyclonal antibody (PM046) and anti-SQSTM1 antibody (PM045) for western blotting were obtained from MBL. Anti-total protein AKT antibody (9272), anti-phospho-AKT (Ser473) antibody (4070) and anti-phospho-AKT (Ser308) antibody (9275) were from Cell Signaling. Anti-GFP monoclonal antibody was from Roche (11814460001). DPBS (Dulbecco’s phosphate-buffered saline) (D4031) was from Sigma. The BCA protein assay kit was from Pierce (23227) and Ub-G76V-GFP was from Addgene (11941). Anti-actin antibody (A2066) was from Sigma.
Cell culture
NRK cells were grown in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10% FBS, 2 mM glutamine and antibiotics.
Measuring intracellular TF
NRK cells were rinsed to remove any residual TF and were then exposed to 5 μg/ml TF conjugated with Alexa Fluor 488 (Invitrogen,T11342) at 37°C for 15 min. Internalization was stopped by chilling the cells on ice. External TF was removed by washing with ice-cold serum-free DMEM and PBS, whereas bound TF was removed by washing in PBS at pH 5.0 followed by a wash with PBS at pH 7.0. The fluorescence intensity of internalized TF was measured for 10,000 cells by flow cytometry using a FACSCalibur (BD Biosciences) instrument.24 For imaging, the cells were treated as previously reported.25 Briefly, NRK Cells were rinsed to remove any residual TF and were then exposed to 5 μg/ml TF conjugated with Alexa Fluor 488 at 37°C for the 15 min. Internalization was stopped by chilling the cells on ice and washing six times with 2 ml prechilled neutral pH 7.4 buffer (150 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 20 mM HEPES acid). Cells were then incubated with 1 ml prechilled pH 2.0 buffer (500 mM NaCl, 0.2 N glacial acetic acid) for 5 min at 4°C. After removing and discarding the pH 2.0 buffer, the cells were washed twice with prechilled neutral buffer, fixed with 4% paraformaldehyde and washed three times with PBS.
Solubility testing
Half a milligram of 3-MA, 15, 18 and 27 were added to 1 ml water and the absorption at 254 nm was determined using a UV-spectrophotometer. 100 mg 15, 18, 27 and 3-MA were added to 100 µl DMSO or 15 mg were added to 100 µl water and magnetically stirred at 37°C for 10 min. The solutions were filtered to obtain saturated solutions of 15, 18, 27 and 3-MA. For 15, 18 and 27, at least six different concentrations were tested by HPLC and a linear relation between peak area and concentration was obtained. For each saturated solution, the peak area was determined by HPLC. The solubility of the three compounds was calculated according to the linear relation between peak area and concentration. For 3-MA, at least six different concentrations were tested by UV-spectrophotometry and a linear correlation was observed between the absorption value at 254 nm and the concentration. The absorption value at 254 nm of the saturated solution was determined by UV-spectrophotometry and the solubility of 3-MA was calculated according to the linear relation between peak area and concentration.
Supplementary Material
Acknowledgments
The p2K7 and VSVG plasmids and the 293FT cell line were kindly provided by Professor Kehkooi Kee. This work was supported by a grant from Tsinghua-Bayer Collaboration.
Glossary
Abbreviation:
- ATG7
autophagy-related 7
- BECN1
Beclin 1
- ATG16L1
autophagy-related 16-like 1
- EGFP
enhanced green fluorescent protein
- LC3
microtubule-associated protein 1 light chain 3
- MTOR
mechanistic target of rapamycin
- SQSTM1
sequestosome 1
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- class I PI3K
phosphoinositide 3-kinase
- 3-MA
3-methyladenine
- 15
(4-(3-methyl-3H-purin-6-yl)thiomorpholine)
- 18
3-methyl-6-(3-methylpiperidin-1-yl)-3H-purine
- 27
(6-(4-(3,4-dichlorophenyl)piperazin-1-yl)-3-methyl-3H-purine)
- DMSO
dimethyl sulfoxide
- PtdIns3P
phosphatidylinositol 3-phosphate
- ZFYVE/DFCP1
zinc finger FYVE domain containing 1
- PDPK1
phosphoinositide-dependent kinase
- PIK3C3/VPS34
phosphatidylinositol 3-kinase, catalytic subunit type3
- PtdIns3K
class III phosphatidylinositol 3-kinase
- NRK
normal rat kidney
- TF
transferrin
- DMEM
Dulbecco's modified Eagle’s medium
- DPBS
Dulbecco's phosphate-buffered saline
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/23641
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23641
References
- 1.Grinde B. Autophagy and lysosomal proteolysis in the liver. Experientia. 1985;41:1089–95. doi: 10.1007/BF01951685. [DOI] [PubMed] [Google Scholar]
- 2.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurent A, Nicco C, Chéreau C, Goulvestre C, Alexandre J, Alves A, et al. Controlling tumor growth by modulating endogenous production of reactive oxygen species. Cancer Res. 2005;65:948–56. [PubMed] [Google Scholar]
- 4.He C, Klionsky DJ. Autophagy and neurodegeneration. ACS Chem Biol. 2006;1:211–3. doi: 10.1021/cb600182h. [DOI] [PubMed] [Google Scholar]
- 5.Massey DC, Parkes M. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn’s disease. Autophagy. 2007;3:649–51. doi: 10.4161/auto.5075. [DOI] [PubMed] [Google Scholar]
- 6.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–92. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–8. [PubMed] [Google Scholar]
- 8.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–6. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 10.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285:10850–61. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergmann F, Levin G, Kalmus A, Kwietny-Govrin H. Synthesis and Properties of 3-Methylpurines1. J Org Chem. 1961;26:1504–8. doi: 10.1021/jo01064a047. [DOI] [Google Scholar]
- 12.Wright AE, Roth GP, Hoffman JK, Divlianska DB, Pechter D, Sennett SH, et al. Isolation, synthesis, and biological activity of aphrocallistin, an adenine-substituted bromotyramine metabolite from the Hexactinellida sponge Aphrocallistes beatrix. J Nat Prod. 2009;72:1178–83. doi: 10.1021/np900183v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo PW, Kostlan CR, Sircar JC, Dong MK, Gilbertsen RB. Inhibitors of human purine nucleoside phosphorylase. Synthesis and biological activities of 8-amino-3-benzylhypoxanthine and related analogues. J Med Chem. 1992;35:1451–7. doi: 10.1021/jm00086a014. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 17.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 19.Franke TF, Yang SI, Chan TO, Datta K, Kazlauskas A, Morrison DK, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–36. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 20.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–51. [PMC free article] [PubMed] [Google Scholar]
- 21.Farkas T, Daugaard M, Jäättelä M. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem. 2011;286:38904–12. doi: 10.1074/jbc.M111.269134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendil KB, Lauridsen AM, Seglen PO. Both endocytic and endogenous protein degradation in fibroblasts is stimulated by serum/amino acid deprivation and inhibited by 3-methyladenine. Biochem J. 1990;272:577–81. doi: 10.1042/bj2720577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–27. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padrón D, Tall RD, Roth MG. Phospholipase D2 is required for efficient endocytic recycling of transferrin receptors. Mol Biol Cell. 2006;17:598–606. doi: 10.1091/mbc.E05-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGraw TE, Subtil A. Endocytosis: Biochemical Analyses. Current Protocols in Cell Biology: John Wiley & Sons, Inc., 2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.