Abstract
The midge Belgica antarctica is the only insect endemic to Antarctica and has the southernmost range of any insect. In its natural environment, B. antarctica frequently faces desiccating conditions, as environmental water is frozen for up to 9 months annually. The molecular mechanisms by which B. antarctica tolerates extreme dehydration are poorly understood, but recent work from our laboratory reports genome-wide expression changes in response to extreme dehydration (~40% water loss), the first genome-scale transcriptome reported for an Antarctic animal. Among transcripts differentially regulated during dehydration, there is coordinated upregulation of numerous genes involved in autophagy, including genes responsible for autophagosome synthesis and autophagy-associated transcription factors. Also, several genes and pathways that interact with and regulate autophagy, e.g., sestrins and proteasomal genes, are concurrently upregulated. This suggests that autophagy and related processes are key elements regulating stress tolerance in this extreme environment.
Keywords: antarctic midge, dehydration, environmental stress, RNA-seq, autophagy, sestrin
The Antarctic midge, Belgica antarctica, faces a number of environmental challenges, including prolonged bouts of freezing, anoxia and exposure to UV radiation. Since water is frozen and therefore unavailable for much of the year, maintaining water balance is perhaps the greatest challenge confronting this insect. Larvae of B. antarctica are extremely tolerant of dehydration, surviving a 70% loss of water under ecologically relevant conditions.
In recent years, our group described a number of mechanisms associated with dehydration tolerance in B. antarctica, including expression of heat shock proteins (Hsps) and accumulation of osmoprotective solutes (e.g., glycerol, trehalose), but we lacked a comprehensive overview of the genes and pathways required for extreme dehydration tolerance. Sequence information for this species was previously limited, with only 100 or so sequences available in GenBank. As part of an ongoing genome project for B. antarctica, we have generated ~13,500 high quality gene models, and using mRNA-seq we measured expression of these transcripts in response to dehydration (~40% water loss).
Extreme dehydration results in large-scale changes in gene expression, as roughly 25% of all transcripts are differentially expressed. To make sense of this wealth of data, we employed several enrichment techniques to identify functional groups and pathways that are coregulated in response to dehydration. What emerged is a picture suggesting that cell recycling pathways are essential for extreme dehydration tolerance, with autophagy serving as the focal point.
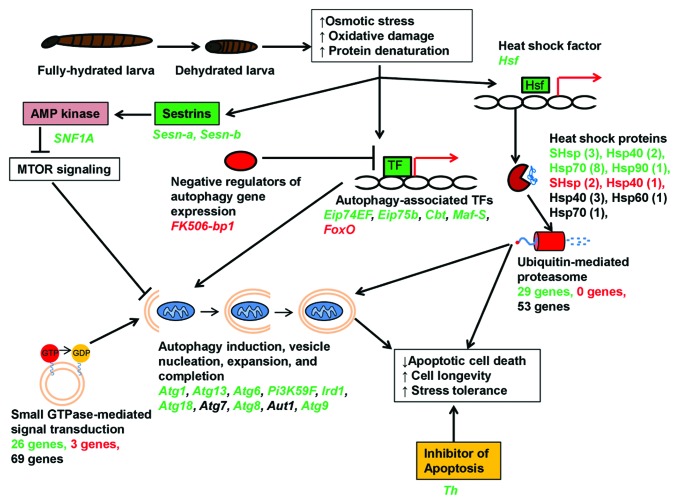
Using the technique of Gene Set Analysis, we found that the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway “Regulation of Autophagy” is positively enriched in our data set, meaning members of this pathway tend to be upregulated in response to dehydration. Indeed, out of seven autophagy-related (Atg) genes in our data set, six (Atg1, Atg6, Atg8, Atg9, Atg13 and Atg18) are significantly upregulated in response to dehydration (Fig. 1). Additionally, several other genes involved in vesicle nucleation and completion are upregulated. Upstream of autophagy induction, four transcription factors known to be positive regulators of autophagy (Eip74EF, Eip75B, Cbt and Maf-S) likewise increase in abundance (Fig. 1). Finally, the transcript FK506-bp1, encoding a negative regulator of autophagy in Drosophila, is downregulated, further supporting the activation of autophagy during dehydration.
Figure 1. Conceptual model illustrating the role of autophagy during extreme dehydration in the Antarctic midge, Belgica antarctica. Using mRNA-seq, we detected coordinated upregulation of autophagy-related genes and several other genes and functional groups known to positively regulate autophagy (e.g., genes encoding sestrins, heat shock proteins, proteasomal subunits, and those involved in GTPase signaling). In this model, dehydration stress activates cellular autophagy, which promotes cell survival and longevity during prolonged periods of osmotic stress. For each gene or functional group in the diagram, names of specific genes belonging to that group are listed below. Green indicates upregulated genes, red indicates downregulated genes, while black indicates genes that were not differentially regulated. For the heat shock proteins (Hsp), the number of individual transcripts belonging to each family is indicated in parentheses. For the proteasomal and GTPase genes, only the number of up- and downregulated genes is indicated, since there were too many to list individually. TFs, transcription factors.
Several other genes that regulate autophagy are also upregulated by dehydration (Fig. 1). Sestrins (Sesns), antioxidant proteins that promote autophagy via activation of AMP kinase, are among the most strongly induced genes in our data set. The two Sesn genes in our transcriptome are upregulated 11- and 13.5-fold, respectively. In Drosophila, Sesn has been linked to longevity and metabolic stress but has not been implicated in response to acute environmental stress. AMP kinase, the protein through which Sesn exerts its effect, is also induced at the transcript level by dehydration. Additional experiments are needed at the protein level, but upregulation of Sesn and AMP kinase transcripts is further evidence of dehydration-induced autophagy.
Aside from genes that directly regulate autophagy, several functional groups known to interact with autophagy are activated during dehydration. Genes involved in the ubiquitin-mediated proteasome, which cooperates with autophagy to remove damaged proteins, are enriched among the dehydration-upregulated genes. Transcripts of 29 proteasomal genes are upregulated, as well as 15 Hsps, chaperones that target denatured proteins to the proteasome. Furthermore, genes involved in small GTPase signal transduction are enriched in our data set. While these genes have diverse functions in the cell, several of the upregulated GTPase genes belong to the Rab family, whose members regulate vesicle formation and interact with the autophagosome.
Thus, there is clear evidence in our transcriptomic data for upregulation of autophagy and other cell recycling pathways during dehydration stress (Fig. 1). In addition to degrading damaged cellular components, we hypothesize that autophagy promotes cell survival and longevity during prolonged dehydration stress. Autophagy has two seemingly contradictory roles in relation to cell death: in some cases, autophagy serves as an alternate pathway for programmed cell death, while in others, autophagy inhibits programmed cell death and promotes cell longevity. We hypothesize that the latter is important during periods of environmental stress. The osmotic perturbations generated by dehydration cause metabolic deficits and oxidative damage, conditions known to promote apoptosis. During prolonged exposure to stress, it would be adaptive for larvae to prevent excessive cell death resulting from unregulated apoptosis, and we speculate autophagy plays a key role. In support of this hypothesis, we also see strong upregulation of Thread (Th), a negative regulator of apoptosis that prevents caspase activation (Fig. 1).
The goal of our mRNA-seq experiment was to unlock secrets of dehydration tolerance in one of the world’s most extremophilic insects. Indeed, it appears that autophagy and other cell recycling pathways are important pieces of the puzzle. While these results are preliminary and need to be validated with functional experiments, a model for the protective role of autophagy during abiotic stress is emerging (Fig. 1). There appears to be significant overlap between our data set and pathways known to be important for aging and longevity. Since aging is simply an accumulation of cellular stress, perhaps these results are unsurprising. Thus, dehydration stress in B. antarctica may elicit an effect similar to accelerated aging, with cell-recycling pathways such as autophagy coming to the rescue.
Acknowledgments
This work is supported by National Science Foundation OPP-ANT-0837613 and ANT-083559.
Glossary
Abbreviations:
- Atg
autophagy-related
- Cbt
cabut
- Eip74EF
ecdysone-induced protein 74EF
- Eip75B
ecdysone-induced protein 75B
- FK506-bp1
FK506-binding protein 1
- FoxO
forkhead box, subgroup O
- Hsf
heat shock factor
- Hsp
heat shock protein
- Ird1
immune response deficient 1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MTOR
mechanistic target of rapamycin
- Pi3K59F
phosphatidylinositol 3-kinase 59F
- Sesn
sestrin
- SHsp
small heat shock protein
- Th
thread
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23643