Abstract
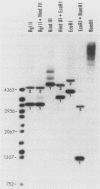
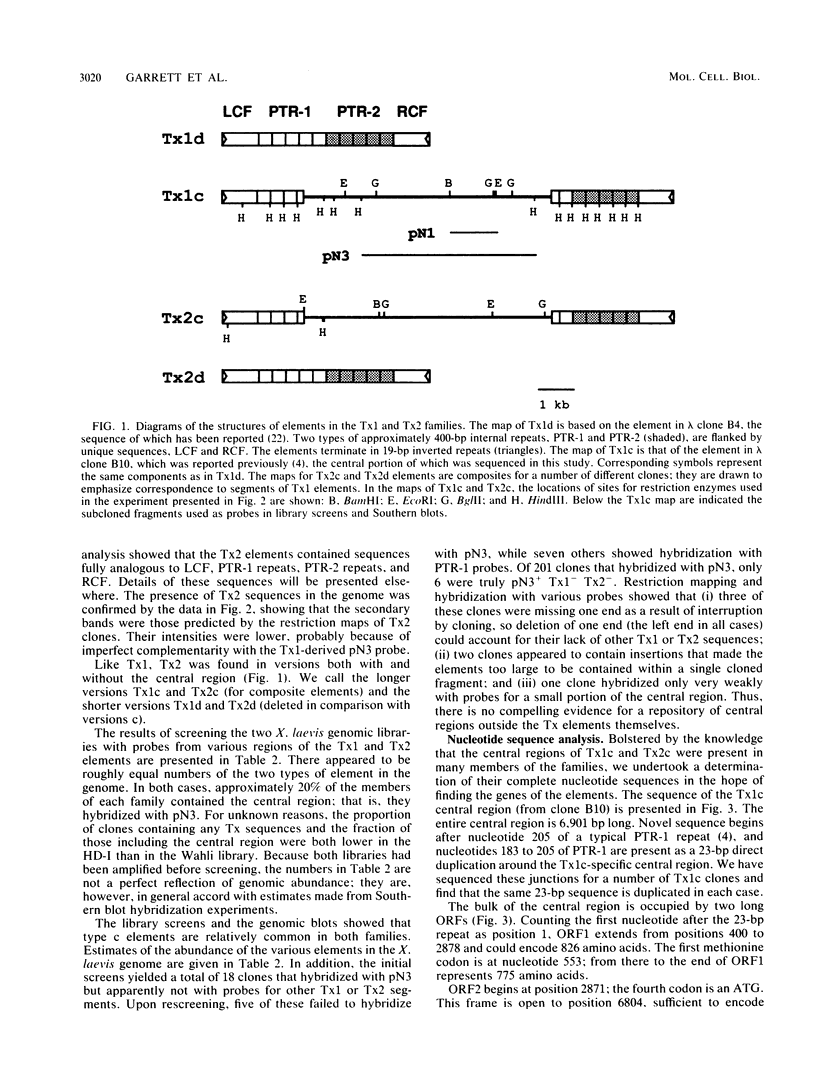
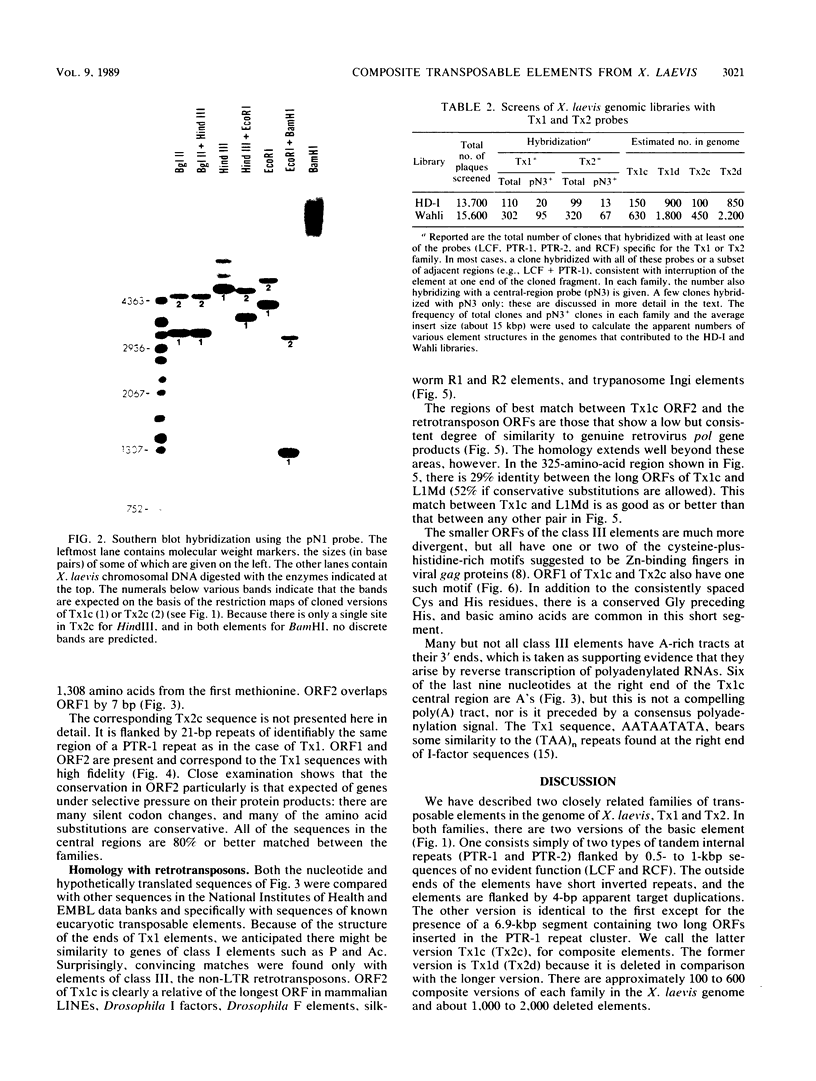
Members of two related families of transposable elements, Tx1 and Tx2, were isolated from the genome of Xenopus laevis and characterized. In both families, two versions of the elements were found. The smaller version in each family (Tx1d and Tx2d) consisted largely of two types of 400-base-pair tandem internal repeats. These elements had discrete ends and short inverted terminal repeats characteristic of mobile DNAs that are presumed to move via DNA intermediates, e.g., Drosophila P and maize Ac elements. The longer versions (Tx1c and Tx2c) differed from Tx1d and Tx2d by the presence of a 6.9-kilobase-pair internal segment that included two long open reading frames (ORFs). ORF1 had one cysteine-plus-histidine-rich sequence of the type found in retroviral gag proteins. ORF2 showed more substantial homology to retroviral pol genes and particularly to the analogs of pol found in a subclass of mobile DNAs that are supposed retrotransposons, such as mammalian long interspersed repetitive sequences, Drosophila I factors, silkworm R1 elements, and trypanosome Ingi elements. Thus, the Tx1 elements present a paradox by exhibiting features of two classes of mobile DNAs that are thought to have very different modes of transposition. Two possible resolutions are considered: (i) the composite versions are actually made up of two independent elements, one of the retrotransposon class, which has a high degree of specificity for insertion into a target within the other, P-like element; and (ii) the composite elements are intact, autonomous mobile DNAs, in which the pol-like gene product collaborates with the terminal inverted repeats to cause transposition of the entire unit.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker R. F., Thompson D. V., Talbot D. R., Swanson J., Bennetzen J. L. Nucleotide sequence of the maize transposable element Mul. Nucleic Acids Res. 1984 Aug 10;12(15):5955–5967. doi: 10.1093/nar/12.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Burke W. D., Calalang C. C., Eickbush T. H. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol Cell Biol. 1987 Jun;7(6):2221–2230. doi: 10.1128/mcb.7.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Garrett J. E., Lam B. S. Isolated clusters of paired tandemly repeated sequences in the Xenopus laevis genome. Mol Cell Biol. 1984 Feb;4(2):254–259. doi: 10.1128/mcb.4.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Wright S. H., Wolff R. K., Grzesiuk E., Maryon E. B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986 Jun;6(6):2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S. N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986 Jan 24;14(2):623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio E., Waitzkin S. D., Witney F. R., Salemme A., Furano A. V. Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol Cell Biol. 1986 Feb;6(2):411–424. doi: 10.1128/mcb.6.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Casari G. Related polypeptides are encoded by Drosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5843–5847. doi: 10.1073/pnas.84.16.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Barbara McClintock's controlling elements: now at the DNA level. Cell. 1984 Dec;39(2 Pt 1):253–259. doi: 10.1016/0092-8674(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Molecular genetics of transposable elements in plants. Annu Rev Genet. 1986;20:175–200. doi: 10.1146/annurev.ge.20.120186.001135. [DOI] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L. High-frequency excision of transposable element Tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984 Mar;36(3):599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V. About maize transposable elements and development. Cell. 1989 Jan 27;56(2):181–191. doi: 10.1016/0092-8674(89)90891-x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J. Transposable elements in eukaryotes. Int Rev Cytol. 1985;93:281–326. doi: 10.1016/s0074-7696(08)61376-5. [DOI] [PubMed] [Google Scholar]
- Furano A. V., Robb S. M., Robb F. T. The structure of the regulatory region of the rat L1 (L1Rn, long interspersed repeated) DNA family of transposable elements. Nucleic Acids Res. 1988 Oct 11;16(19):9215–9231. doi: 10.1093/nar/16.19.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Boeke J. D., Fink G. R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985 Sep;42(2):507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- Garrett J. E., Carroll D. Tx1: a transposable element from Xenopus laevis with some unusual properties. Mol Cell Biol. 1986 Mar;6(3):933–941. doi: 10.1128/mcb.6.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984 Dec 3;145(2):203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Greenblatt I. M. Movement of modulator in maize: a test of an hypothesis. Genetics. 1974 Aug;77(4):671–678. doi: 10.1093/genetics/77.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J. B., Wickens M. P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986 Jun 5;321(6070):625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kimmel B. E., ole-MoiYoi O. K., Young J. R. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol Cell Biol. 1987 Apr;7(4):1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper M. T., Lambowitz A. M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988 Nov 18;55(4):693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. II. Dispersed clusters of a 388 base-pair repeating unit. J Mol Biol. 1983 Apr 25;165(4):587–597. doi: 10.1016/s0022-2836(83)80268-x. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Penkala J. E., Peterson C. A., Wright D. A. Repair of UV-induced lesions in Xenopus laevis oocytes. Mol Cell Biol. 1987 Dec;7(12):4317–4323. doi: 10.1128/mcb.7.12.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984 Nov 16;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Mellor J., Malim M. H., Gull K., Tuite M. F., McCready S., Dibbayawan T., Kingsman S. M., Kingsman A. J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature. 1985 Dec 12;318(6046):583–586. doi: 10.1038/318583a0. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988 Aug 26;54(5):685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur I., Pascale E., Furano A. V. The left end of rat L1 (L1Rn, long interspersed repeated) DNA which is a CpG island can function as a promoter. Nucleic Acids Res. 1988 Oct 11;16(19):9233–9251. doi: 10.1093/nar/16.19.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Padgett R. W., Hutchison C. A., 3rd, Edgell M. H. The F-type 5' motif of mouse L1 elements: a major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucleic Acids Res. 1988 Jan 25;16(2):739–749. doi: 10.1093/nar/16.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Leclercq L., Göbel E., Saedler H. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 1987 Dec 20;6(13):3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Saigo K. Retrovirus-like particles containing RNA homologous to the transposable element copia in Drosophila melanogaster. Nature. 1983 Mar 10;302(5904):119–124. doi: 10.1038/302119a0. [DOI] [PubMed] [Google Scholar]
- Truett M. A., Jones R. S., Potter S. S. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981 Jun;24(3):753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- Voliva C. F., Jahn C. L., Comer M. B., Hutchison C. A., 3rd, Edgell M. H. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res. 1983 Dec 20;11(24):8847–8859. doi: 10.1093/nar/11.24.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahli W., Dawid I. B. Isolation of two closely related vitellogenin genes, including their flanking regions, from a Xenopus laevis gene library. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1437–1441. doi: 10.1073/pnas.77.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988 Oct 21;55(2):235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988 Nov;5(6):675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988 Jan;8(1):114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]