Abstract
Genetic variants associated with increased risk for schizophrenia (SZ) are hypothesized to be more penetrant at the level of brain structure and function than at the level of behavior. However, to date the relative sensitivity of imaging vs cognitive measures of these variants has not been quantified. We considered effect sizes associated with cognitive and imaging studies of 9 robust SZ risk genes (DAOA, DISC1, DTNBP1, NRG1, RGS4, NRGN, CACNA1C, TCF4, and ZNF804A) published between January 2005–November 2011. Summary data was used to calculate estimates of effect size for each significant finding. The mean effect size for each study was categorized as small, medium, or large and the relative frequency of each category was compared between modalities and across genes. Random effects meta-analysis was used to consider the impact of experimental methodology on effect size. Imaging studies reported mostly medium or large effects, whereas cognitive investigations commonly reported small effects. Meta-analysis confirmed that imaging studies were associated with larger effects. Effect size estimates were negatively correlated with sample size but did not differ as a function of gene nor imaging modality. These observations support the notion that SZ risk variants show larger effects, and hence greater penetrance, when characterized using indices of brain structure and function than when indexed by cognitive measures. However, it remains to be established whether this holds true for individual risk variants, imaging modalities, or cognitive functions, and how such effects may be mediated by a relationship with sample size and other aspects of experimental variability.
Keywords: endophenotype, schizophrenia, imaging genetics, neurocognitive, effect size, meta-analysis
Introduction
Schizophrenia (SZ) is a common and highly heritable psychiatric condition,1 which is associated with profound changes in brain structure and function, cognition, and perception. Despite clear evidence of SZ heritability, critical gaps in our understanding of how genetic variability increases risk for SZ psychopathology remain; perhaps as a consequence of its complex genetics.2 It has been postulated that genes do not directly encode for psychopathology in general nor for specific symptoms, but rather that genetic “risk” is mediated by effects on neural mechanisms relevant to brain structure and cognition, ie, so called “intermediate phenotypes” or “endophenotypes.”3 By moving beyond a behaviorally based diagnosis, measuring intermediate phenotypes (ie, those quantitative biological traits that occur on the pathway between genes and disease4) may: (1) help to characterize the deleterious effects of individual risk variants, (2) prove informative for preclinical studies of the genes implicated, and (3) be beneficial in therapeutic development.5
Given the relationship between cognitive function and disease-related disability in SZ,6,7 the heritability of brain volume,8 and the relationship between brain structure and cognitive function, endophenotypic investigations have considered brain and behavior. Both approaches have been useful in delineating intermediate phenotypes in SZ and in understanding the nature of apparent risk variants.9 However, because functional and/or structural variability in the brain is more proximal to underlying neurobiological mechanisms and/or pathways associated with disease risk,10 it has been suggested that phenotypic effects of risk variants for SZ may be more apparent using imaging-based indices of brain structure and function rather than overt behavioral measures of cognition. While this is an intriguing notion, whether or not this difference between measures of brain vs behavior in the penetrance of risk variants holds true has yet to be systematically evaluated.
With the aim of delineating the relative effect of known risk variants for SZ on neuroimaging and cognitive measures, we considered estimates of effect size in recently published studies considering either endophenotype class. Genes were selected that have a replicated association with increased risk of developing SZ (either in the candidate gene or genome-wide association study [GWAS] literature) and have been the subject of both cognitive and imaging studies. We included data pertaining to those candidate genes that have an established and robust relationship with SZ risk, while excluding those for which risk may not be directly related to SZ per se, but rather mediated by their impact upon aspects of the disease phenotype, such as cognitive dysfunction (eg, catechol-O-methyl transferase, brain-derived neurotrophic factor). Putative SZ candidate genes selected included common variants within disrupted in schizophrenia-1 (DISC1),11,12 dysbindin (DTNBP1),13–16 neuregulin-1 (NRG1),17,18 regulator of G-protein signaling 4 (RGS4),19,20 and D-amino acid oxidase activator (DAOA).21 For GWAS risk variants, we included all genome-wide associated variants for which a SZ-associated single-nucleotide polymorphism had been linked to variability in cognition and/or brain structure or function. GWAS variants meeting these criteria included rs1334706 on the zinc-finger protein gene ZNF804A,22 rs12807809 located upstream of the neurogranin (NRGN) gene,23 rs9960767 within intron 3 of transcription factor 4 (TCF4),23 and rs1006737 on the CACNA1C gene.24 A random effects meta-analysis was used to compare the impact of methodology on effect size for each study class. Moreover, a complementary nonparametric analysis considered the relative frequency of different effect size categories (ie, small, medium, and large) between cognitive and imaging investigations. Here, we report differences in the magnitude of observed effects between imaging and cognitive investigations of genetic risk for SZ. We also explore how experimental variation, such as sample size, might contribute to any real or apparent discrepancies and discuss potential limitations in the interpretation of these observations.
Methods
Article Selection
We searched PUBMED (http://www.ncbi.nlm.nih.gov/pubmed/) for articles published between January 2005 and November 2011 that contained the following search terms: [DISC1, DAOA, DTNBP1/dysbindin, NRG1/neuregulin, RGS4, NRGN, TCF4, CACNA1C, or ZNF804A] and [schizophrenia]. Individual abstracts were screened to determine whether the manuscript represented an original cognitive and/or imaging investigation of at least one of the genes of interest. “Imaging” studies included functional magnetic resonance imagining (fMRI), electroencephalography (EEG), or structural magnetic resonance imaging (sMRI) methods (eg, voxel-based morphometry [VBM] and other volumetric measures, diffusion tensor imaging [DTI], cortical thickness, etc.). Studies were included based upon the presentation of summary statistics that were adequate for an effect size estimate to be calculated for at least one significant effect. Using these criteria, 83 articles were selected (see online supplementary materials for details) including 23 cognitive and 51 imaging studies plus 9 providing both cognitive and imaging results (these studies are highlighted in online supplementary table S1). Within the imaging investigations, there were 5 EEG, 27 fMRI, 25 sMRI, and 3 combined f/sMRI studies. A further 8 studies (ie, 2 cognitive, 5 imaging, and 1 combined study) were initially selected but later excluded because we were unable to calculate effect sizes based upon the available summary information and efforts to contact the authors were unsuccessful. As anticipated, the studies selected for the review included a variety of patient and/or healthy control populations (see online supplementary table S1).
Effect Size Estimations
Estimated effect sizes (eg, Cohen’s d, Hedge’s g) were calculated for each reported significant outcome for each investigation. For the purposes of the nonparametric analysis, Cohen’s d was the estimate of interest since there are established criteria for categorizing d (see below). For the meta-analysis, due to software constraints, Hedge’s g was used.
Effect size calculations were performed using 2 meta-analysis software packages: (1) ClinTools25 and (2) Comprehensive Meta-Analysis v2 (CMA; www.meta-analysis.com). The effect size generator in ClinTools was used for the majority of estimations of Cohen’s d. Using this toolbox, estimates of effect size were calculated based upon either descriptive data (ie, mean, SD, and N), or inferential statistics (eg, t, F, χ2). Unless corresponding descriptive data were provided, we were unable to calculate d for F statistics pertaining to interaction effects. In those instances where the authors reported their own estimation of effect size, but using an alternative index (such as Hedge’s g, R 2, η2, Cohen’s f 2 etc.), these values were converted to Cohen’s d. The formulae that were used to perform these estimates included:
Where independent group differences were reported but only P values and N for each group were available (ie, no corresponding inferential or descriptive statistics were given), an estimate of Hedge’s g was generated based upon P, N, and whether the hypothesis was 1- or 2-tailed and subsequently converted to d.
Analysis Plan
While absolute values of d may not be directly comparable between studies of differing methodologies, it is presumed that values across studies will approximate similar effect size categories. To address this potential source of variation, we conducted an analysis of the relative frequency of different categories of effect size between endophenotype classes. This analysis served as a nonparametric complement to our meta-analysis. To account for variability in the number and range of significant effects reported in any single investigation, the mean Cohen’s d was calculated for all significant results in each study. In calculating the mean d for each study, we were interested determining the magnitude but not the direction of effects (ie, our aim was “not” to quantify the effect of a given gene or variant or variant therein). Therefore, although the direction of an effect can be indicated by reporting it as negative or positive (indeed, such information is useful in determining if and how disease risk and endophenotypic effects might be conferred by a specific gene or variant), in calculating the mean effect size for each study, all effect size estimates were considered as “positive” values. Cohen’s d may be categorized as small (d > 0.3), medium (d = 0.5), or large (d: 0.08–∞).26 Accordingly, mean d for each study was categorized as follows: small ≤ 0.4; medium=0.5–0.7; large ≥ 0.8. The relative frequency of articles meeting these criteria was evaluated between endophenotype classes, imaging modalities, and genes.
A random effects meta-analysis of relative differences in effect size between cognitive and imaging investigations was carried out in CMA. For this analysis, Hedge’s g and its associated variance were calculated for the maximal effect for each study, which was identified using preceding calculations of d. This approach was taken so as to reduce potential bias associated the variable number of significant findings reported by individual studies. For example, imaging studies have a tendency to report multiple significant results (eg, pertaining to differences in function or structure in a range of brain regions), whereas cognitive studies report comparatively fewer findings. Within the software limitations, it was not possible to impute multiple values for a single investigation and compute a mean estimate of effect size; rather we were able to select only a single effect; thus, limiting each study to one data point in the random effects model. The largest effect for each study was chosen so as to reflect the maximal sensitivity to gene effects within each investigation. As with estimates of d, calculations for g were conducted using a variety of input variables, including both descriptive and inferential statistics, and all effects were considered to be positive.
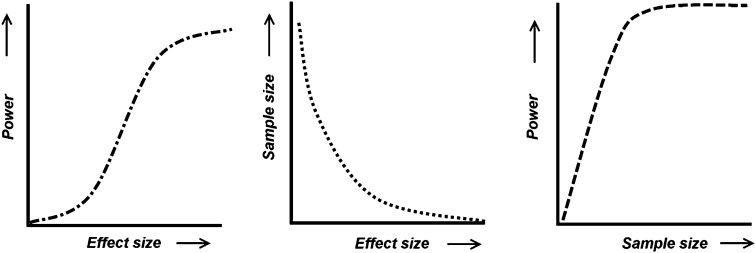
The ability to detect an effect of a given magnitude is intrinsically linked to the power of the study and proportional to the sample size. For example, with smaller sample sizes, one might anticipate lower power and the ability to detect only those effect sizes that are relatively large (figure 1). A series of linear regressions were carried out to ascertain the extent of the association between mean effect size and N in the studies included in our analyses. These regressions considered the association between N and effect size for: (1) all studies, (2) imaging studies, and (3) cognitive studies.
Fig. 1.
The relationship between “effect size,” “sample size,” and “power.”
Results
Effect Size Categorization
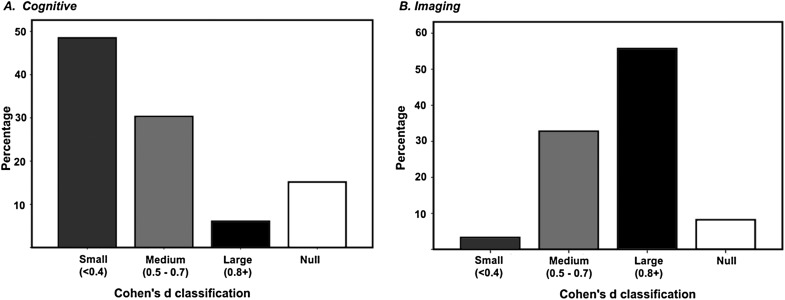
There was a significant difference in the frequency of small, medium, and large effect size estimates between cognitive and imaging investigations (χ2 = 39.47, d f = 3, P < .001). In cognitive studies, the majority of effect sizes were “small,” while outcomes in imaging studies tended to be “medium” or “large” (see online supplementary table S2 and figure 2). Conversely, the frequency of effect size categorization did not differ between imaging modalities or genes (across cognitive and imaging studies).
Fig. 2.
Percentage of results in each effect size category (ie, small, medium, or large) for (A) cognitive and (B) imaging studies.
Meta-Analysis
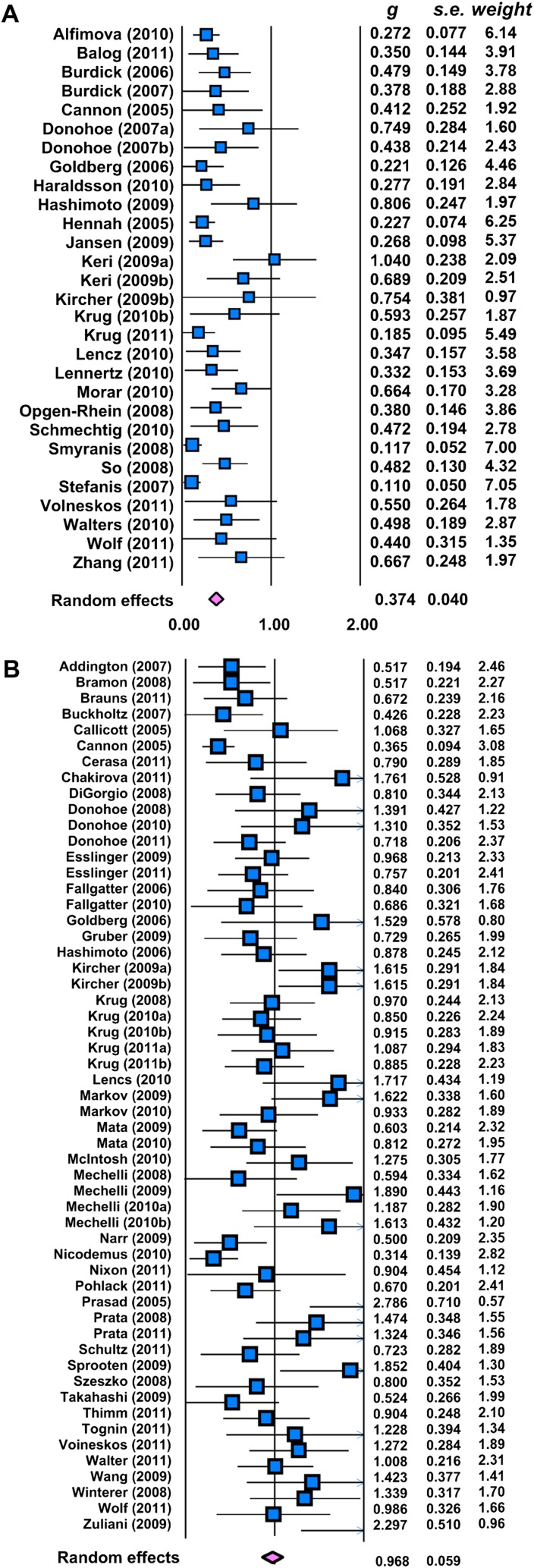
Imaging studies reported larger effects than cognitive studies (Hedge’s g (95% CI): 0.968 (0.852–1.084) vs 0.374 (0.295–0.452); Cochrane’s Q value = 69.31, df = 1, P < .001; figure 3). Again, effect size estimates did not vary as function of gene or imaging modalities.
Fig. 3.
Forest plot (random effects) showing Hedge’s g, SE and relative weight for meta-analysis in (A) cognitive and (B) imaging investigations.
Effect Size and Sample Size
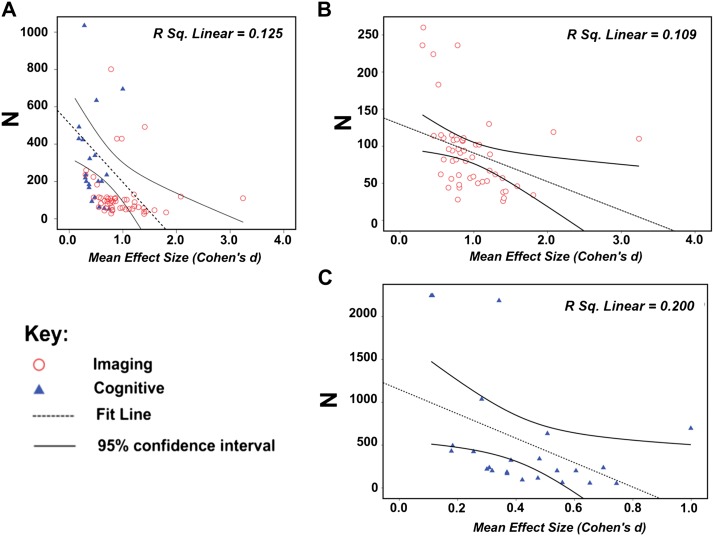
Average sample size was greater for cognitive vs imaging investigations (mean (SD): imaging = 141.82 (149.32), cognitive = 558.19 (624.32), t (90) = 4.93, P < .001). Moreover, there was a significant linear relationship between N and mean effect size ie, (1) all studies: F 1,81 = 11.64, P = .001; (2) imaging: F 1,54 = 6.48, P = .014; and (3) cognitive: F 1,26 = 6.25, P = .02 (figure 4). Across analyses, sample size was inversely proportional to the mean effect size (ie, t = −3.39, t = −2.55, and t = −2.49, respectively).
Fig. 4.
The linear regression of sample size (N) against observed mean effect size (d) in (A) imaging and cognitive studies (Note: for visualization purposes investigations where N > 1200 (ie, 3 studies) have been excluded from this graph) (B) imaging investigations, and (C) cognitive investigations.
Publication Bias
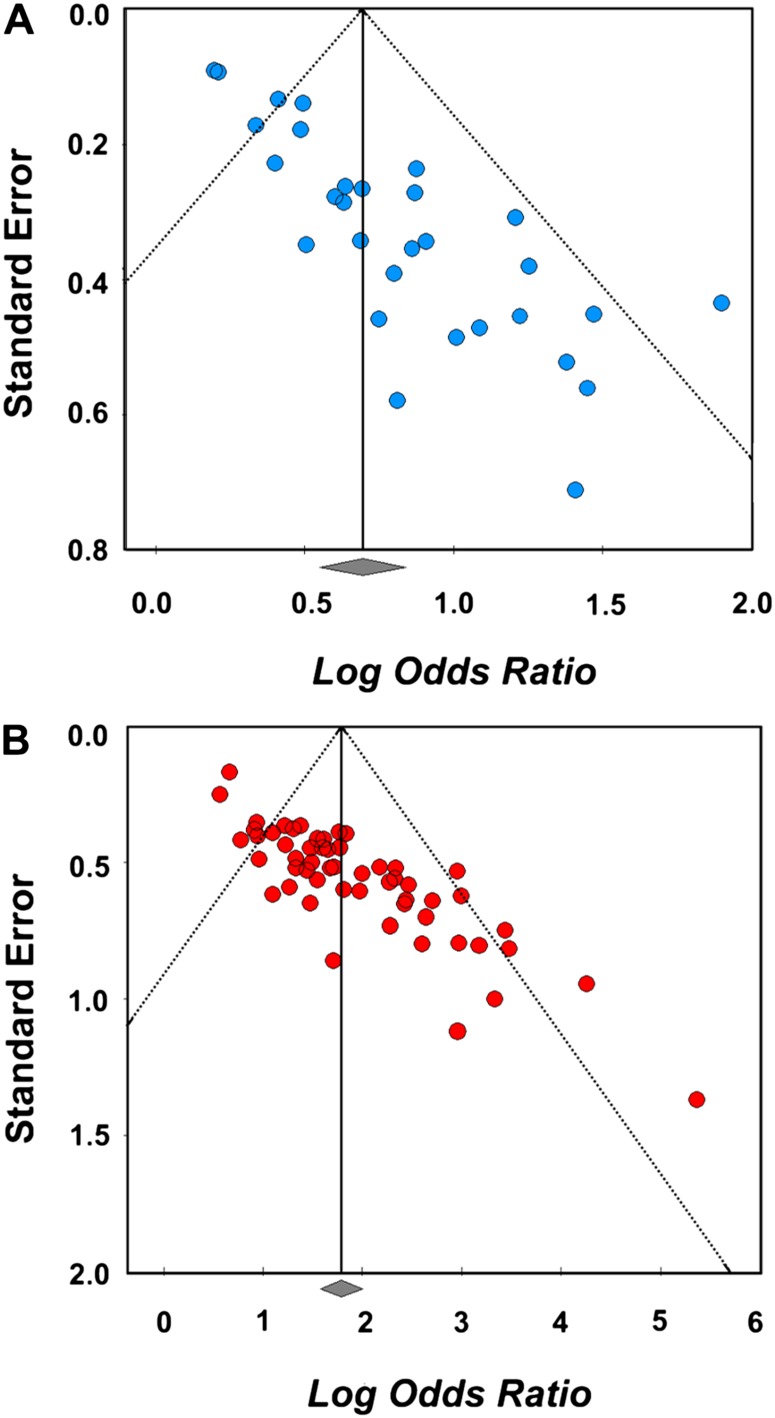
Since asymmetry in funnel plots is an indicator of biases in the data, including potential publication bias,27 we constructed funnel plots for each endophentoype class to determine whether publication bias was a contributing factor to study outcomes. In accordance with published guidelines,28 the log OR for each study was plotted again the SE of the log OR (figure 5). The extent of asymmetry was tested using a linear regression method.29 Both imaging and cognitive plots were notably asymmetrical (eg, Egger’s regression intercept: cognitive = 2.49, t (28) = 9.81, P < .001; imaging = 3.63, t (54) = 11.17, P < .001).
Fig. 5.
Funnel plots of log OR against the SE of the log OR for each study in (A) cognitive studies and (B) imaging studies. Since all effect size estimations were considered positive, these plots have been cropped at OR = 0.
Discussion and Conclusions
In this review we considered variability in effect sizes reported in recent imaging vs cognitive investigations of genes implicated in SZ risk. Outcomes in neuroimaging studies were most commonly associated with medium or large effects, while cognitive studies more frequently reported findings that constitute small effects. Furthermore, the maximal effect size reported across studies was significantly greater for imaging investigations. These observations seem to support the notion that genetic variants that confer risk for SZ are more penetrant at the level of brain structure and function than behavior. Here, we discuss the implications of our results and methodological considerations in interpreting these outcomes.
Methodological Considerations
Sample Size.
Across study types, studies with larger samples more consistently reported small effects, whereas smaller cohorts were associated with effects of larger magnitude. This relationship may be critical to our results. For example, large effects in imaging studies likely reflect the fact that these studies have comparatively smaller samples and smaller cohorts may only reliably detect effects of larger magnitude. If so, it may be inappropriate to directly compare imaging and cognitive studies, which tend to have larger samples and, thus, are able to ascertain a wider range of effects. However, despite having sufficient numbers of participants to detect large effects, cognitive studies most frequently reported small effects. This supports the notion that cognitive intermediate phenotypes are more distal from the underlying biological mechanisms and thus associated with smaller effects. Nonetheless, in the absence of larger imaging samples it is impossible to determine whether the difference between study types is reliable or whether it simply reflects experimental bias.
Publication Bias.
The extent of asymmetry in our funnel plots may be due to a range of biases (including, but not limited to, true heterogeneity, data irregularities, data artifacts, or even chance27); however, publication bias is a likely cause for this asymmetry. For example, less than 10% of the studies included failed to report positive results. It seems doubtful that this is an accurate reflection of all study outcomes. Rather, this might reflect a “file drawer” problem, where researchers either do not attempt to publish negative results or find it more difficult to have them accepted for publication. Furthermore, it is uncommon for published negative results to be accompanied by the data necessary to determine effect size. Without some measure of the observed effect, whether significant or not and no matter how small, we cannot determine whether other factors may have reduced the power to detect valid differences or to get a broader view of the impact of risk variants on intermediate phenotypes. Doing so in the future would represent an important advance in the imaging genetics field.
A bias toward publishing positive results may also have contributed to the difference in the frequency or magnitude of effects between imaging and cognitive endophenotypic studies. For example, the potential for comparisons is greater in imaging compared with cognitive studies, thus any adjustment to correct for multiple comparisons is likely to be more stringent in imaging studies. If so, imaging studies may have a greater likelihood of false negatives, which might exaggerate any potential publication bias. The rate of null results was not different between the study types (ie, cognitive ≈ 11%; imaging ≈ 9%), suggesting that the issue may not have been phenotype specific. However, these observed rates tell us nothing about the comparative rate of unpublished negative results. Without this information, it is difficult to ascertain the extent of the problem or its contribution to relative differences in the observed penetrance of cognitive and imaging endophenotypes for SZ.
False Positives.
Rather than bias in the rate of false negatives, the high rate of findings of large effect size in imaging studies might have been indicative of a failure to use appropriate procedures to control for type I error rate, ie, “false positives.” However, an empirical investigation of the rate of positive findings using commonly employed corrections in imaging genetics studies found that these methods effectively reduced the likelihood of false positives.30 Moreover, the similar rate of null findings in published imaging and cognitive endophenotype studies suggests that while the false positive rate may be a consideration in intermediate phenotypes studies of SZ, this issue is not modality specific and thus may not have been a source of significant variability in the results presented here. As with association studies focused on the broader SZ phenotype, replication of results within the same study design would represent an important step toward overcome such problems in both imaging and cognitive studies of SZ variants.
Effect Size Estimation.
Even when all the necessary information is available for effect size calculations to be accurately performed, it is reasonable to anticipate discrepancies between studies due to experimental variability. Similarly, effect size estimates are likely to be influenced by the data that are used to calculate them, such that an estimate of d based upon descriptive data will not necessarily produce exactly the same value as an estimate based upon inferential statistics for the same comparison. For example, using descriptive data from our sample size comparison (ie, cognitive vs imaging studies), we estimated d to be 0.87, whereas using t and df d was estimated as 1.06. While both values would be classified as “large,” the discrepancy between different calculation methods is noteworthy. Such discrepancies are of concern here given the considerable variability in experimental procedures and the summary data available. Despite this, we do not anticipate that such discrepancies would have contributed significantly to broad categorization and, therefore, will have not impacted upon the outcomes of the nonparametric categorical analysis.
Task Variability.
While some cognitive processes were more popular than others, there was considerable variability in the functions that were targeted and the tasks that were used across studies. Although there was some variability in imaging techniques that were used (eg, VBM, DTI, fMRI, etc.), it could be argued that the practical and statistical procedures are more standard within imaging modalities. Therefore, the increased diversification in cognitive vs imaging studies may be a potentially limiting factor in our results.
Subject Variables.
A range of subject factors, eg, study population, medication, cognitive, or physical training, have the potential to contribute to the differences seen here. Although we cannot account for all such factors in our analysis, it is somewhat reassuring that the distribution of cohort types (ie, patients vs healthy controls vs mixed) was reasonably consistent between study types (ie, cognitive: 15% vs 44% vs 41%; imaging: 14% vs 52% vs 34%). While this consistency addresses some of this potential variability, the contribution of other factors and their relative importance remains to be established.
Statistical Standards.
A final factor to note is the considerable range of analysis methods and criteria by which imaging results are considered to be significant. For instance, not all imaging results reported appear to have been corrected for multiple comparisons. Furthermore, where corrections were made a variety of correction methods were used, ranging from family-wise error or false discovery rate correction at the whole brain level or within particular a priori region-of-interest to post hoc small volume corrections centered on regions of established difference in function or structure. Similarly, acceptable levels of probability vary between studies at both uncorrected and corrected levels. Without some sort of “gold standard” for determining and reporting significant results, it is almost impossible to compare across imaging studies, let alone reliably compare imaging results to cognitive or other measures of risk. Such a standard would almost certainly remove at least one potential source of experimental bias, ie, rate of false positives.
Differences in Cognitive/Imaging Findings Between and Within Risk Genes
Comparative Effect Size.
In determining the implications of our results, it is important to consider the relative impact of genes/variants that were associated with large or small effect sizes within modalities. Intriguingly, for cognitive studies, the smallest (d = 0.11) and largest measured effects (d = 1.00) were reported for the same gene, DTNBP1. The smallest effect was associated with the impact of a DTNBP1 risk haplotype on IQ and attention,31 while the largest effect size was noted for the impact of the P1763 variant on memory.32 Both studies included large samples (N = 2243 and 695) but differed with regards to the study populations (ie, healthy individuals vs mixed sample). Comparison of these studies suggests that even within a gene, all variants are not created equal. Indeed, different risk variants within a gene may differ in their molecular mechanisms and hence the nature or extent of their functional impact. Moreover, even if different risk variants have similar functional profiles, study population may be a critical factor in determining intermediate phenotypes such that examination of the same gene in different populations may give rise to diverse results.
For imaging studies, the smallest effect (d = 0.30) was seen for DISC1, whereas the largest effects (d = 2.08 and 3.24) were related to DAOA. Interestingly, studies at both ends of the spectrum employed sMRI, included patients, and had relatively large samples (DISC1, N = 263; DAOA, N = 119 and 110). These observations qualitatively support our observation that imaging modality was not related to effect size variability. Alternatively, they may simply reflect the preference of sMRI methods over other imaging techniques. Nonetheless, it is reassuring that largest and smallest effects were associated with imaging studies of relatively large sample size, which indicates that imaging investigations with adequate numbers might indeed have the capacity to detect effects across a range of magnitudes.
Combined Studies.
Considering different classes of intermediate phenotype in the same individuals has the benefit of reducing sources of variability that might contribute to analyses between cognitive and imaging studies. Across those studies that considered both endophenotype classes, effect size differences between methods mimicked the overall differences observed in the meta-analysis, ie, cognitive measures were more typically associated with small effects, while imaging measures were associated with large effect sizes. Reflecting this, no association was observed between effect sizes for cognitive and imaging measures within studies (P > .05). This may be indicative of a functional distinction in the pathways that give rise to different endophenotypes. Alternatively, it may suggest interactions with other variables (eg, environmental factors) that are more likely to impact those indices that are more distal from the underlying molecular biology. Identifying those genes and/or variants that have similar effects on brain and behavior and delineating other factors that might impact upon the relationship between genetic risk and individual phenotypes is a crucial next step in determining the relative impact of SZ risk genes.
Optimizing Endophenotypes.
Determining optimal endophenotypes is critical, particularly with regards to the utility of the endophentoype approach in clinical practice. However, identifying those characteristics that are most likely to mediate the effects of risk variants on disease phenotype is complex. Our data represent the first empirical validation of the notion that imaging measures are more proximal to underlying genetic risk and thus may be more appropriate indices to consider. As such, these outcomes represent an important first step. This relative difference between cognitive and imaging measures is likely to reflect proximity to the biological pathways that mediate genetic risk and give rise to disease symptoms.33,34 However, not all of the genes/variants considered here have clearly delineated biological functions or pathways. Determining the underlying molecular functions and consequences of individual variants that are implicated in SZ risk will be integral to the development of this field.
While differences in effect magnitude between imaging and cognitive intermediate phenotypes may reflect proximity to biological mechanisms, imaging intermediate phenotypes may also simply be more reproducible or less amenable to environmental influence. Despite the need to more fully delineate these factors, alternative explanations do not alter the fact that imaging phenotypes appear to permit a more sensitive endophenotypic exploration of SZ risk.
Summary and Future Directions
In sum, our results support the notion that genetic risk for SZ is more closely indexed by measures of brain structure and function than cognition. However, a number of methodological concerns must be addressed before concluding that one approach holds inherent value over the other. Increased sample sizes, standardized statistical procedures, and determining the rate of null results in imaging studies will aid in delineating those measures, cognitive or imaging, which are truly more sensitive to genetic variation and should be considered as optimal intermediate phenotypes for SZ.
Of all these issues, sample size may be the most critical. Smaller samples in imaging studies are likely to contribute to overestimating the true population mean. Tackling this issue may be logistically or financially impractical on a site-by-site basis, however, steps are being taken toward addressing it in the form of large multisite consortia, eg, ENIGMA (http://enigma.loni.ucla.edu/). These projects face considerable logistical issues and the implications they will have on investigations conducted by individual groups with limited resources remains to be established. Nonetheless, they have the potential to include imaging and genetics information from thousands of participants and are primed to pave the way in imaging genetics methods and for delineating the limitations of genetic effects that can be detected at the level of brain structure and function. Interestingly, this trajectory toward large multicenter studies closely reflects the development of the genetic association literature, with both gene discovery and gene characterization requiring large sample sizes to understand the real but subtle effects of common variants on SZ risk. Perhaps most importantly, these studies will be well situated to determine which endophentoypes truly mediate aspects of the disease phenotype.
Funding
Science Foundation Ireland (SFI08/IN.1/B1916-Corvin).
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Acknowledgments
We would like to thank the following individuals for kindly agreeing to provide unpublished supplementary data: Drs A. McIntosh and E. Sprooten (University of Edinburgh, UK); Drs A. Mechelli and D. Prata (Kings College London, UK); Drs T. Kircher, A. Jansen, and A. Krug (Philipps-University Marburg, Germany); Dr B. Crespo-Facorro (University of Cantabria, Spain); and Dr W. Hennah (University of Helsinki, Finland). The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1.Owen MJ, O'Donovan MC, Gottesman II. Schizophrenia. In: McGuffin P, Owen MJ, Gottesman II, editors. Psychiatric Genetics and Genomics. Oxford, UK: Oxford University Press; 2002. pp. 247–266. [Google Scholar]
- 2.Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 3.Tan HY, Callicott JH, Weinberger DR. Intermediate phenotypes in schizophrenia genetics redux: is it a no brainer? Mol Psychiatry. 2008;13:233–238. doi: 10.1038/sj.mp.4002145. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 5.Berrettini WH. Genetic bases for endophenotypes in psychiatric disorders. Dialogues Clin Neurosci. 2005;7:95–101. doi: 10.31887/DCNS.2005.7.2/wberrettini. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green MF, Kern RS, Heaton RK. Longitudinal studies of cognition and functional outcome in schizophrenia: implications for MATRICS. Schizophr Res. 2004;72:41–51. doi: 10.1016/j.schres.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Toulopoulou T, Picchioni M, Rijsdijk F, et al. Substantial genetic overlap between neurocognition and schizophrenia—genetic modeling in twin samples. Arch Gen Psychiatry. 2007;64:1348–1355. doi: 10.1001/archpsyc.64.12.1348. [DOI] [PubMed] [Google Scholar]
- 8.Bartley AJ, Jones DW, Weinberger DR. Genetic variability of human brain size and cortical gyral patterns. Brain. 1997;120:257–269. doi: 10.1093/brain/120.2.257. [DOI] [PubMed] [Google Scholar]
- 9.O'Donovan MC, Craddock N, Owen MJ. Schizophrenia: complex genetics, not fairy tales. Psychol Med. 2008;38:1697–1699. doi: 10.1017/S0033291708003802. ; discussion 1818–1820. [DOI] [PubMed] [Google Scholar]
- 10.Rasetti R, Weinberger DR. Intermediate phenotypes in psychiatric disorders. Curr Opin Genet Dev. 2011;21:340–348. doi: 10.1016/j.gde.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porteous DJ, Thomson P, Brandon NJ, Millar JK. The genetics and biology of DISC1-An emerging role in psychosis and cognition. Biol Psychiatry. 2006;60:123–131. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Blackwood DHR, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ. Schizophrenia and affective disorders—cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funke B, Finn CT, Plocik AM, et al. Association of the DTNBP1 locus with schizophrenia in a U.S. population. Am J Hum Genet. 2004;75:891–898. doi: 10.1086/425279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley B, Kendler KS. Molecular genetic studies of schizophrenia. Eur J Hum Genet. 2006;14:669–680. doi: 10.1038/sj.ejhg.5201571. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- 16.Tosato S, Ruggeri M, Bonetto C, et al. Association study of dysbindin gene with clinical and outcome measures in a representative cohort of Italian schizophrenic patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:647–659. doi: 10.1002/ajmg.b.30484. [DOI] [PubMed] [Google Scholar]
- 17.Hall J, Whalley HC, Job DE, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. [DOI] [PubMed] [Google Scholar]
- 18.Stefansson H, Sigurdsson E, Steinthorsdottir V, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:110. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 20.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry. 2006;60:106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 22.O'Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 23.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green EK, Grozeva D, Jones I, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ClinTools Software for Windows: Version 4 [computer program] Psytek Ltd; 2005. http://www.clintools.com. Accessed August 29, 2011. [Google Scholar]
- 26.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 27.Sterne JA, Harbord RM. Funnel plots in meta-analysis. Stata J. 2004;4:127–141. [Google Scholar]
- 28.Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54:1046–1055. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 29.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer-Lindenberg A, Nicodemus KK, Egan MF, Callicott JH, Mattay V, Weinberger DR. False positives in imaging genetics. Neuroimage. 2008;40:655–661. doi: 10.1016/j.neuroimage.2007.11.058. [DOI] [PubMed] [Google Scholar]
- 31.Stefanis NC, Trikalinos TA, Avramopoulos D, et al. Impact of schizophrenia candidate genes on schizotypy and cognitive endophenotypes at the population level. Biol Psychiatry. 2007;62:784–792. doi: 10.1016/j.biopsych.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Alfimova MV, Monakhov MV, Abramova LI, Golubev SA, Golimbet VE. Polymorphism of serotonin receptor genes (5-HTR2A) and Dysbindin (DTNBP1) and individual components of short-term verbal memory processes in Schizophrenia. Neurosci Behav Physiol. 2010;40:934–940. doi: 10.1007/s11055-010-9348-7. [DOI] [PubMed] [Google Scholar]
- 33.Walters JT, Owen MJ. Endophenotypes in psychiatric genetics. Mol Psychiatry. 2007;12:886–890. doi: 10.1038/sj.mp.4002068. [DOI] [PubMed] [Google Scholar]
- 34.Kendler KS, Neale MC. Endophenotype: a conceptual analysis. Mol Psychiatry. 2010;15:789–797. doi: 10.1038/mp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.