Abstract
Background:
Rare copy number variations (CNVs) were involved in the etiology of neuropsychiatric disorders, and some of them appeared to be shared risk factors for several different diseases. One of those promising loci is the CNV at 15q11.2, including 4 genes, TUBGCP5, CYFIP1, NIPA2, and NIPA1. Several studies showed that microdeletions at this locus were significant associated with schizophrenia. In the current study, we investigated the role of both rare CNVs and common single nucleotide polymorphisms (SNPs) at 15q11.2 in schizophrenia in the Chinese Han population.
Methods:
We screened deletions at 15q11.2 in 2058 schizophrenia patients and 3275 normal controls in Chinese Han population by Affymetrix 500K/6.0 SNP arrays and SYBR green real-time polymerase chain reaction and then validated deletions by multiplex ligation-dependent probe amplification and Taqman real-time assays. We successfully genotyped 27 tag SNPs in total and tested associations in 1144 schizophrenia cases and 1144 normal controls.
Results:
We found a triple increase of deletions in cases over controls, with OR = 4.45 (95% CI = 1.36–14.60) and P = .014. In the analysis of common SNPs, we found that the most significant SNP in schizophrenia was rs4778334 (OR = .72, 95% CI = 0.60–0.87, allelic P = .0056 after permutation, genotypic P = .015 after permutation). We also found SNP rs1009153 in CYFIP1 was associated with schizophrenia (OR = 0.82, 95% CI = 0.73–0.93, allelic P = .044 after permutation).
Conclusion:
We found that both rare deletions and common variants at 15q11.2 were associated with schizophrenia in the Chinese Han population.
Keywords: 15q11.2, copy number variation (CNV), schizophrenia, tag SNP
Introduction
Schizophrenia is a severe mental disorder marked by hallucinations, delusions, cognitive deficits, and apathy, with a heritability estimated at 73%–90%.1 Recently, copy number variations (CNVs) have been found to present throughout the human genome.2–7 CNVs, especially rare ones, have emerged as important risk factors for a number of major psychiatric disorders including schizophrenia, autism, and mental retardation.8,9 CNVs on chromosomes 15q11.2, 15q13.3, 16p11.2, 22q11, and NRXN1 have been reported to be associated with a broad range of psychiatric phenotypes.10–19
The 15q11–13 region is one of the most complicated, unstable, and variable regions of the human genome. It harbors multiple low copy repeats which probably mediate the diverse range of deletions, duplications, and inversions in the region. Two well recognized syndromes, Prader-Willi syndrome (PWS) and Angelman syndrome (AS), appear to share the same distal breakpoint (BP3) at 15q13.3 among all cases, whereas the proximal breakpoints differ (BP1 and BP2 at 15q11.2).20–22 Another study22 concluded that AS children with the deletion including 15q11.2 were more likely to meet criteria for autism, had lower cognitive scores and lower expressive language scores. The deletions, which affected genes CYFIP1, NIPA1, NIPA2, and TUBGCP5, were reported in one AS case with mental retardation and severe speech impairment.23 Moreover, PWS with deletions of this region were associated with increased risks of preservative/obsessive-compulsive behavior, deficits in adaptive skills and lower intellectual ability.24 Thus, the autistic features in AS and the preservative behavior of PWS may arise from deletion of the genes in the proximal portion at 15q11.2.
Recently, Stefansson et al19 found microdeletions at 15q11.2 (chr15: 20306549-20777695) to be associated with schizophrenia in Caucasian population. Moreover, deletions at this locus were 2-fold excess in cases more than in controls in the analysis of the International Schizophrenia Consortium (ISC) data14 although this region was filtered out because deletions and duplication affected >1% of the sample. Data from Kirov et al25 also showed a trend for support. However, there was no support for this locus in a Japanese study, in which the authors found 3 deletions in 543 controls and only 1 deletion in 519 cases.26 CNV data in the Molecular Genetics of Schizophrenia Study27 also revealed no increase in frequency of deletions at 15q11.2 in cases. There are still some uncertainties for the association.
In our study, we tried to investigate the susceptibility of both CNVs and tag single nucleotide polymorphisms (SNPs) at 15q11.2 (chr15:20.3–20.8 Mb; March 2006 (hg18) assembly, University of California, Santa Cruz Genome Browser; the region covered by the 4 genes) in schizophrenia in the Han Chinese population.
Methods
Subjects
CNVs were screened in 2058 schizophrenia cases (1152 males and 906 females with mean age 35.5 ± 7.5 y) and 3275 controls (1695 males and 1580 females with mean age 30.9 ± 11.1 y) in our analysis panel.
Tag SNPs were genotyped in 1144 schizophrenia cases (636 men and 508 women with mean age 35.4 ± 7.2 y and mean onset age 20.4 ± 8.7 y) and 1144 controls (378 men and 766 women, mean age 58.7 ± 9.9 y).
A standard informed consent, which was reviewed and approved by the local Ethical Committee of Human Genetics Resources, was signed by all the participants after the nature of study had been fully explained.
All cases were outpatients or stable inpatients. The inclusion criteria for cases were (1) all cases were interviewed by 2 independent psychiatrists and were diagnosed strictly according to the Diagnostic and Statistical Manual for Mental Disorders, Fourth Edition criteria; (2) the age of all cases was from 18 to 65 y; and (3) all cases had at least 2-year psychiatry history. The subjects were excluded if (1) they had other diseases, such as diabetes and hypertension; or (2) they were biologically related to another study participant; or (3) if they had IQ < 70.
Normal controls were chosen from random population. Volunteers who replied to a written invitation completed the evaluation of medical history, supplemented by questions about psychosis and other major complex diseases. The questionnaires were screened for potentially suitable volunteers to exclude subjects with major mental illness in themselves or their first-degree relatives and those taking neuroleptic medication. Moreover, subjects were excluded if they were clearly diagnosed for other diseases, such as cancer, diabetes, and hypertension etc. Controls with IQ < 70 were excluded.
DNA was extracted from peripheral blood samples of the subjects using commercial kits (QuickGene-610L, Fujifilm). Collection, storage, and extraction of samples were identical between cases and controls.
CNV Screening Strategies
Affymetrix 500K and SNP6.0 Chip.
Part of our schizophrenia cases and controls were genotyped by GeneChip Mapping 500K Assay (Affymetrix) with more than 500 000 SNPs and Genome-Wide Human SNP Array 6.0 (Affymetrix) with more than 906 600 SNP probes and more than 946 000 copy number probes. All of chip data were generated in-house, and experiments were executed strictly in accordance with manufacturer’s protocol. Additionally, we discarded those samples with SNP call rate less than 95%. We also tested for possible contamination and cryptic relatedness by PLINK and removed susceptible samples. Finally, 155 cases and 187 controls genotyped by Affymetrix 500K chips with an average success rate 98.64% of genotyping were included in the following CNV calling by software Partek and double checked by Software Genotyping Console (GTC2.1, Affymetrix).28 Five hundred and forty-two cases and 2961 controls were genotyped by Affymetrix SNP6.0 chips, and CNVs were called by PennCNV. Five hundred and one cases and 2830 controls remained in the following analysis after excluding samples that had LRR (the log R Ratio) SD > 0.35, BAF (the B Allele Frequency) median > 0.6 or < 0.4, and BAF drift > 0.01. We excluded any CNV calls overlapping with chr15: 13.6–18.9 Mb by more than 50% of its length. We also ignored CNV calls less than 100 kb in length and with less than 10 probes.
Real-Time PCR (SYBR Green-Based RT-PCR).
A primer list can be found in supplementary table S1. HBB gene was chosen as endogenous control in this study. A standard curve for each primer was determined by a set of diluted standard DNA in order to make sure the efficiency of primers (Supplementary figure S1). polymerase chain reaction (PCR) reactions using SYBR Green Dye were run according to standard protocol. For each primer, 4 independent replication experiments of 1 sample were carried out to ensure the results. One thousand four hundred and sixty-two cases and 262 controls were screened, and 1402 cases and 258 controls were successfully genotyped.
CNVs from RT-PCR ΔΔCt data were detected as follow: the relative quantity is denoted by ΔCt of a certain pair of primers subtracting that of the HBB gene of the same sample DNA. Empirically, ΔCt data from a certain 384-well plate obey the normal distribution whose mean and SD can be calculated. If the cumulative probability of a ΔΔCt was higher than 0.99, we concluded a deletion at the probe. If 3 probes consistently meet these criteria, we concluded a deletion in 15q11.2 was present.
All deletions at 15q11.2 were double checked by the following multiplex ligation-dependent probe amplification (MLPA) step and Taqman assay-based RT-PCR.
Validation Methods
Multiplex Ligation-Dependent Probe Amplification.
The deletions observed by RT-PCR and SNP chips were validated by MLPA methods.29 Probes and figures were listed in the supplementary file.
We designed 12 primers, 10 of which were specific for those 4 genes and 2 were endogenous control probes. DNA preparation, ligation, PCR amplification, and capillary electrophoresis were carried out according to the methods of Schouten et al.29 The sequence of the labeled primer is 5′-GGGTTCCCTAAG GGTTGGA-3′ and that of the unlabeled primer is 5′-GTGCCAGCAAGATCCA ATCTAGA-3′.
Analysis of the MLPA results was performed according to the criteria of Nygren et al.30 For a certain sample, the relative signal of a target probe was defined by dividing its peak area by that of control probes. Relative copy number ratio was obtained by comparing a relative signal with the mean of corresponding relative signal of 7 normal control samples. A ratio lower than 0.7 was considered as the signal of deletion (supplementary figure S2).
Real-Time PCR (Taqman Assay).
Two sets of primers and probes targeting the 15q11.2 region were designed using Primerexpress software. Probes (in supplementary file) were labeled with 5′FAM, 3′ Eclipse (Shanghai Sangon Biological Engineering Technology and Servie Co., Ltd.), which allowed each assay to be multiplexed with the control assay, RNase P labeled with 5′VIC, 3′ TAMRA (Applied Biosystems). An absolute quantification RT-PCR was performed for each test assay, multiplexed with the control assay, on all samples with an expected CNV event at that site and minimum of 40 controls. All samples were plated in quadruplicate using 2 ng of DNA with a final reaction volume of 5 μl. RT-PCR results were analyzed by Copycaller software (Applied Biosystems).
Selection of Tag SNPs and Genotyping
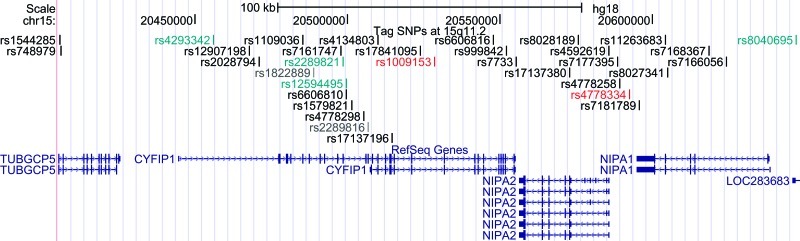
The pattern of linkage disequilibrium was evaluated using genotype data from HapMap (phase 2, release 21) unrelated CHB + JPT subjects. Tagging SNPs in the 4 genes were chosen using the software Haploview 4.231 with a pairwise tagging strategy based on r 2 ≥ .5 and minimum minor allele frequency ≥ 0.05. Primers and probes of 33 tag SNPs (as shown in figure 1, these SNPs are rs1544285, rs748979, rs4293342, rs12907198, rs2028794, rs1109036, rs1822889, rs7161747, rs2289821, rs12594495, rs6606810, rs1579821, rs4778298, rs2289816, rs4134803, rs17137196, rs17841095, rs1009153, rs6606816, rs999842, rs7733, rs17137380, rs8028189, rs4592619, rs7177395, rs4778258, rs4778334, rs7181789, rs11263683, rs8027341, rs7168367, rs7166056, and rs8040695) were then synthesized by Applied Biosystems as TaqMan predesigned assays. Among the 33 SNPs, 2 were in TUBGCP5, 19 were in CYFIP1, 3 were in NIPA2, 5 were in NIPA1, and 4 in intergenic regions.
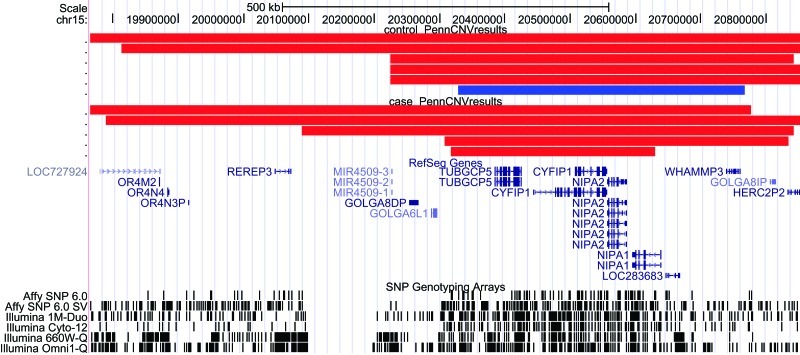
Fig. 2.
Deletions at 15q11.2 identified by Affymetrix 500K (in blue) and Affymetrix SNP6.0 (in red) in the University of California, Santa Cruz Genome Browser (Human March 2006 assembly, hg18).
The standard of 5 μl PCR reaction of each SNP using TaqMan Universal PCR Master Mix was carried out and then genotyped on the ABI 7900 DNA detection system (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol.
Statistical Analysis
For the deletions, 2 types of association test were performed in SAS to evaluate the risk of CNVs: 2-tailed Fisher’s exact tests and the stratified Cochran-Mantel-Haenszel (CMH) exact tests. For the CMH test, we stratified our samples by different platforms to the overall association. The Breslow-Day test was also performed to assess the homogeneity of the ORs across the strata.
For each SNP, deviation of genotype frequencies from Hardy-Weinberg equilibrium (HWE) in controls was assessed by a χ2 test with one degree of freedom (1 df). Risks were evaluated using ORs with 95% CIs. Associations of case/control status were evaluated by χ2 tests with 1 df for alleles and 2 df respectively for genotypes.
Allele and genotype frequencies, Hardy-Weinberg equilibrium, ORs with 95% CIs, allelic and genotypic associations, were calculated on SHEsis (http://analysis2.bio-x.cn/myAnalysis.php),32 an online user-friendly platform with integrated analysis tools particularly suited to association studies. SNPs with P ≤ .05 were subjected to 10 000 times permutation on PLINK 33.
Results
Association Between 15q11.2 Deletions and Schizophrenia
After validation, we confirmed 1 deletion in 187 controls (figure 2) in Affy 500K chip experiments; we confirmed 5 deletions in 501 schizophrenia cases and 5 deletions in 2830 controls (figure 2) in Affy SNP6.0 chip experiments; and we confirmed 7 deletions in 1402 schizophrenia patients in RT-PCR experiments (table 1).
Fig. 1.
Locations of all 33 tag SNPs in the human genome browser. Top: the coordinate in the human genome (March 2006 [NCBI36/hg18] assembly); middle: tag SNPs chosen in our study; and bottom: reference genes (TUBGCP5, CYFIP1, NIPA2, and NIPA1). Two SNPs in dark gray failed in the test experiments; 4 SNPs in green deviated from Hardy-Weinberg equilibrium in controls; and 2 SNPs in red remained positive in schizophrenia after permutation.
Table 1.
Frequencies of Deletions at 15q11.2 in Schizophrenia Cases and Controls Stratified by Platforms
| CMH | ||||||||
| Total | Deletion | Deletion (%) | OR (95% CI) | P (2 Sided) | OR (95% CI) | P | Breslow-Day P | |
| Schizophrenia | ||||||||
| RT-PCR | 1402 | 7 | 0.50 | |||||
| Chip (500K) | 155 | 0 | 0 | |||||
| Chip (6.0) | 501 | 5 | 1.00 | |||||
| Control | 3.2 (1.2–8.5) | .026 | 4.45 (1.36–14. 60) | .014 | .13 | |||
| RT-PCR | 258 | 0 | 0 | |||||
| Chip (500K) | 187 | 1 | 0.53 | |||||
| Chip (6.0) | 2830 | 5 | 0.18 | |||||
Note: CMH, Cochran-Mantel-Haenszel; RT-PCR, real-time PCR.
A 3-fold increase of deletions were observed in cases over controls with OR = 3.2 (95% CI = 1.2–8.5) and P = .026. We also stratified our data by different platforms and calculated CMH P = .014 and OR = 4.45 (95% CI = 1.36–14.6). The Breslow-Day test (P = .13) showed that there was no OR heterogeneity within the strata.
Up to now, there have been several reports on the association between 15q11.2 deletions and schizophrenia, which were summarized in table 2 for comparison. Although frequencies varied across studies, populations, and platforms, the average frequency of deletions in schizophrenia cases across different studies is ∼0.6% and in controls is ∼0.2%.
Table 2.
Deletion Frequency Distribution Between Schizophrenia Cases and Controls Reported in Published Literature
| References | Ethnicity | Platform | Deletions in Cases | Deletion in Controls | OR (95% CI) | P |
| Current study | Chinese | Diverse | 12 of 2058 | 6 of 3275 | 4.45 (1.36–14.60) | .014 |
| Ikeda et al26 | Japanese | Affy SNP5.0 | 1 of 519 | 3 of 543 | 0.3 (0.04–3.35) | .625 |
| ISC14 | Caucation | Affy SNP5.0/6.0 | 26 of 3391 | 11 of 3181 | 2.2 (1.1–4.5) | .031 |
| Steffanson et al19 | Diverse | Diverse | 26 of 4718 | 79 of 41 194 | 2.73 (1.5–4.9) | .0006 |
| Kirov et al25 | Bulgarian | Affy 500K (Agilent 444K arrays) | 4 of 471 | 14 of 2792 | 1.7 (0.6–5.2) | .316 |
| Levinson et al27 | European/African-American | Affy SNP6.0 | 19 of 3945 | 17 of 3611 | 1.02 (0.5–2.1) | 1 |
| In totala | 81 of 14 440 | 129 of 53 926 |
Associations Between Tag SNPs and Schizophrenia
Two SNPs (rs1822889 and rs2289816) failed in the experiments and 4 SNPs (rs4293342, rs2289821, rs12594495, and rs8040695) were found to be in deviation from HWE in the control samples and were therefore discarded. Two SNPs in TUBGCP5, 14 SNPs in CYFIP1, 3 SNPs in NIPA2, 5 in NIPA1, and 3 in intergenic regions remained. P values less than .05 were adjusted by 10 000 times permutation. Data of all genotyped SNPs were summarized in the supplementary table S2.
The most significant SNP (as shown in table 3) in schizophrenia was rs4778334, which located between NIPA2 and NIPA1 (OR = 0.72, 95% CI = 0.60–0.87, allelic P = .0056 after permutation, genotypic P = .015 after permutation). We also found rs1009153 (table 3) in CYFIP1 remained significance after permutation (OR = 0.82, 95% CI = 0.73–0.93, allelic P = .044 after permutation).
Table 3.
Allelic and Genotypic Distribution of All Genotyped 31 Tag SNPs at 15q11.2 in Cases and Controls. (For More Details, Please Refer to Online Supplementary Table S2)
| SNP ID | Allele Frequency | OR | P Value* | Genotype Frequency | H-W P | P Value* | |||
| rs1009153 | A | G | A/A | A/G | G/G | ||||
| SZ cases | 1101 (52.8%) | 983 (47.2%) | 0.82 (0.73–0.93) | .0013 | 296 (28.4%) | 509 (48.8%) | 237 (22.7%) | .52 | .0036 |
| Control | 1289 (57.7%) | 945 (42.3%) | (.044) | 366 (32.8%) | 557 (49.9%) | 194 (17.4%) | .47 | (.090) | |
| HapMap | 58.90% | 41.10% | 35.60% | 46.70% | 17.80% | ||||
| rs4778334 | C | T | C/C | C/T | T/T | ||||
| SZ cases | 1764 (86.0%) | 288 (14.0%) | 0.72 (0.60–0.87) | .00065 | 763(74.4%) | 238 (23.2%) | 25 (2.4%) | .22 | .0013 |
| Control | 1880 (89.4%) | 222 (10.6%) | (.0056) | 838 (79.7%) | 204 (19.4%) | 9 (0.9%) | .37 | (.015) | |
| HapMap | 84.40% | 15.60% | 73.30% | 22.20% | 4.40% | ||||
*SNPs with P ≤ .05 were subjected to 10 000 times permutation. P values were in parentheses.
Discussion
The interval of the deletions at 15q11.2 forms part of a larger region of chromosomal instability, which has known associations with mental retardation, severe speech impairment, AS and PWS. At present, the difficulty of determining whether a given CNV is pathological arises from our rudimentary knowledge of mutation rates and population-distribution statistics of CNVs. By screening a large number of schizophrenia cases and population controls, our data suggest deletions at 15q11.2 (chr15:20.3–20.8 Mb; hg18) were associated with Schizophrenia in the Chinese Han population.
A limitation of our study is that we screened deletions by 3 platforms. However, in order to minimize the false positive calls, we validated each CNV by another 2 independent experimental methods, as described in the “Methods” section.
There is a consensus that several CNVs can be shared by clinically different psychiatric disease. The pleiotropic effects and phenotypic diversity of the CNV may be considerable (data are summarized in table 4). Doornbos et al reported 9 cases with a microdeletion at 15q11.2 between BP1–BP2.34 They carefully evaluated the clinical features of the 9 patients and found several shared features, including delayed motor and speech development, dysmorphisms such as long fingers and behavioral problems (ADHD, autism, obsessive–compulsive behavior). De Kovel et al35 reported that microdeletions at 15q11.2 were observed 12 times in 1234 idiopathic generalized epilepsy case and 6 times in 3022 controls. Screening of target CNV loci in children with unexplained intellectual disability by Mefford et al36 also showed that a 6.5-fold increase of the 15q11.2 deletion in cases. The role of 15q11.2 CNVs in other psychiatric disorders like ADHD, major depressive disorder, and bipolar disorder needed further studies.
Table 4.
Deletion Frequency Distribution Between Other Phenotype Cases and Controls Reported in Published Literature
| References | Ethnicity/Phenotype | Platform | Deletion in Cases | Deletion in Controls | OR (95%) | P (2 Sided) |
| Doornbos et al34 | Dutch patients with mental retardation and/or multiple congenital abnormalities | MLPA, array CGH, SNP array (validated by FISH) | 9 of 1576 | 0 of 350 | — | |
| De kovel et al35 | Case: North-Western European | Affy SNP6.0 (validated by qPCR) | 12 of 1234 | 6 of 3022 | 4.9 (1.8–13.2) | 4.2 × 10−4 |
| Control: German idiopathic generalized epilepsy | ||||||
| Mefford et al36 | Children with unexplained intellectual disability | Illumina BeadXpress SNP assay | 8 of 1010 | 3 of 2493 | 6.6 (1.8–25.0) | .003 |
Note: MLPA, multiplex ligation-dependent probe amplification.
We also tested the associations of common variants at 15q11.2 by genotyping tag SNPs in 1144 schizophrenia cases and 1144 normal controls. Twenty-seven SNPs were successfully genotyped and analyzed in the final association tests.
It is very interesting that rs4778334, which located between NIPA2 and NIPA1, was the most positive SNPs in schizophrenia. Up to now, there has been limited literature on the function of NIPA2 and NIPA1 in psychiatric disease. Chai et al37 mapped and characterized these 2 genes at 15q11.2 and suggested that these encode for a receptor or transporter. Follow-up studies38,39 proved that both NIPA2 and NIPA1encode for Mg2+ transporters.
We also found that a common SNP in CYFIP1 was associated with schizophrenia. The most significant SNP in CYFIP1 for schizophrenia was rs1009153 (OR = 0.82, 95% CI = 0.73–0.93, allelic P = .044 after permutation). Since CNVs at 15q11.2 have been reported to be associated with psychiatric phenotypes, CYFIP1 has been highlighted8,19,25,40 because of functional relevance. By tagging common SNPs in the gene, our study has provided further evidence of the involvement of CYFIP1 in schizophrenia. CYFIP1 interacts with both Fragile X mental retardation protein (FMRP) and Rho GTPase Rac1, which is involved in regulating axonal and dendritic outgrowth and in the development and maintenance of neuronal structures. Napoli et al41 reported that CYFIP1 was a binding partner of FMRP and directly binds the translation initiation factor eIF4E through a domain that is structurally related to those present in 4E-BP translational inhibitors, to repress translation of several mRNAs in the brain, including MAP1B, APP, and aCaMKII. They also pointed out that the complex responded to synaptic stimuli such as brain-derived neurotrophic factor that stimulates translation via mTOR and ERK-MAPK pathway at synapses and dihydroxyphenylglycine. Interestingly, a recent study42 identified the AKT-mTOR signaling pathway as a critical target of DISC1, which had long been recognized as a predisposing gene in schizophrenia in regulating neuronal development. These findings provided a framework for understanding how multiple susceptibility genes identified in association studies functionally converge into a common pathway and contributed to the etiology of certain psychiatric disorders.42
In conclusion, we confirmed the association between deletions at 15q11.2 and schizophrenia in the Chinese Han population and provided first evidence that common variants in this region were also associated with schizophrenia.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
National 863 program (2009AA022701); 973 program (2010CB529600); National Natural Science Fundation of China (81130022, 81121001, 31000553); the Foundation for the Author of National Excellent Doctoral Dissertation of China (201026); the Program for New Century Excellent Talents in University (NCET-09-0550); Shanghai Municipal Health Bureau program (2008095).
Supplementary Material
Acknowledgments
We sincerely thank all the participants for their participation in this study and all the medical staff involved in sample collection and diagnosis. We report no biomedical financial interests or potential conflicts of interest.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 3.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 4.Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- 5.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 6.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu Rev Genomics Hum Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 7.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St Clair D. Copy number variation and schizophrenia. Schizophr Bull. 2009;35:9–12. doi: 10.1093/schbul/sbn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 10.Kirov G, Gumus D, Chen W, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 11.Ousley O, Rockers K, Dell ML, Coleman K, Cubells JF. A review of neurocognitive and behavioral profiles associated with 22q11 deletion syndrome: implications for clinical evaluation and treatment. Curr Psychiatry Rep. 2007;9:148–158. doi: 10.1007/s11920-007-0085-8. [DOI] [PubMed] [Google Scholar]
- 12.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone JL, O'Donovan MC, Gurling H, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 17.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 18.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 19.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christian SL, Robinson WP, Huang B, et al. Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet. 1995;57:40–48. [PMC free article] [PubMed] [Google Scholar]
- 21.Amos-Landgraf JM, Ji YG, Gottlieb W, et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahoo T, Peters SU, Madduri NS, et al. Microarray based comparative genomic hybridization testing in deletion bearing patients with Angelman syndrome: genotype-phenotype correlations. J Med Genet. 2006;43:512–516. doi: 10.1136/jmg.2005.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murthy SK, Nygren AO, El Shakankiry HM, et al. Detection of a novel familial deletion of four genes between BP1 and BP2 of the Prader-Willi/Angelman syndrome critical region by oligo-array CGH in a child with neurological disorder and speech impairment. Cytogenet Genome Res. 2007;116:135–140. doi: 10.1159/000097433. [DOI] [PubMed] [Google Scholar]
- 24.Bittel DC, Kibiryeva N, Butler MG. Expression of 4 genes between chromosome 15 breakpoints 1 and 2 and behavioral outcomes in Prader-Willi syndrome. Pediatrics. 2006;118:e1276–e1283. doi: 10.1542/peds.2006-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirov G, Grozeva D, Norton N, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda M, Aleksic B, Kirov G, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 27.Levinson DF, Duan JB, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi YY, He G, Zhang Z, et al. A study of rare structural variants in schizophrenia patients and normal controls from Chinese Han population. Mol Psychiatry. 2008;13:911–913. doi: 10.1038/mp.2008.69. [DOI] [PubMed] [Google Scholar]
- 29.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nygren AO, Ameziane N, Duarte HM, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CAL, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur J Med Genetics. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 35.de Kovel CGF, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mefford HC, Cooper GM, Zerr T, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai JH, Locke DP, Greally JM, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet. 2003;73:898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goytain A, Hines RM, Quamme GA. Functional characterization of NIPA2, a selective Mg2+ transporter. Am J Physiol Cell Physiol. 2008;295:C944–C953. doi: 10.1152/ajpcell.00091.2008. [DOI] [PubMed] [Google Scholar]
- 39.Goytain A, Hines RM, El-Husseini A, Quamme GA. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J Biol Chem. 2007;282:8060–8068. doi: 10.1074/jbc.M610314200. [DOI] [PubMed] [Google Scholar]
- 40.Tam GW, Redon R, Carter NP, Grant SG. The role of DNA copy number variation in schizophrenia. Biol Psychiatry. 2009;66:1005–1012. doi: 10.1016/j.biopsych.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 41.Napoli I, Mercaldo V, Boyl PP, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Kim JY, Duan X, Liu CY, et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.