Abstract
The lack of consistency of genetic associations in highly heritable mental illnesses, such as schizophrenia, remains a challenge in molecular psychiatry. Because clinical phenotypes for psychiatric disorders are often ill defined, considerable effort has been made to relate genetic polymorphisms to underlying physiological aspects of schizophrenia (so called intermediate phenotypes), that may be more reliable. Given the polygenic etiology of schizophrenia, the aim of this work was to form a measure of cumulative genetic risk and study its effect on neural activity during working memory (WM) using functional magnetic resonance imaging. Neural activity during the Sternberg Item Recognition Paradigm was measured in 79 schizophrenia patients and 99 healthy controls. Participants were genotyped, and a genetic risk score (GRS), which combined the additive effects of 41 single-nucleotide polymorphisms (SNPs) from 34 risk genes for schizophrenia, was calculated. These risk SNPs were chosen according to the continuously updated meta-analysis of genetic studies on schizophrenia available at www.schizophreniaresearchforum.org. We found a positive relationship between GRS and left dorsolateral prefrontal cortex inefficiency during WM processing. GRS was not correlated with age, performance, intelligence, or medication effects and did not differ between acquisition sites, gender, or diagnostic groups. Our study suggests that cumulative genetic risk, combining the impact of many genes with small effects, is associated with a known brain-based intermediate phenotype for schizophrenia. The GRS approach could provide an advantage over studying single genes in studies focusing on the genetic basis of polygenic conditions such as neuropsychiatric disorders.
Keywords: schizophrenia, DLPFC, working memory, intermediate phenotype, fMRI, genetic risk score
Introduction
Family, twin, and adoption studies support a genetic basis for schizophrenia and are broadly consistent with an estimated heritability of 80%–85%.1 Genetic association studies have nonetheless yielded only weak effects for specific points of genomic variation to be related to schizophrenia. Evidence for an association is often inconsistent, ie, genetic polymorphisms identified as risk variants by some studies are often not associated with schizophrenia in other studies. For example, the catechol-O-methyltransferase (COMT) Val108/158Met polymorphism, the most-studied common variant in schizophrenia with profound effects on COMT enzyme activity and the dopaminergic tone, failed to be associated with the disorder on meta-analytic review.2 A more general review of 27 schizophrenia linkage studies reported that no genomic region was implicated in more than 4 of 27 studies.1 Even gene variants, which have been repeatedly linked to schizophrenia, such as zinc finger binding protein 804A (ZNF804A), confer only a small increment in risk (OR∼1.09).3 The reasons for weak and inconsistent specific genetic effects may be 2-fold. First, schizophrenia is a polygenic disease, in which not a single gene, but a combination of several genes with small effects contributes to an overall risk. For polygenic traits such as height single genes could only explain 0.3%–0.5% of the phenotypic variance,4 whereas combining the effects of more than 180 markers predicted more than 10 %.5 Second, schizophrenia is an illness with a wide range of symptoms and neuropsychological impairments, which vary within and across individuals. The practice of grouping a diverse patient population into a single diagnostic category based on clinical observation and patient reports, which may not necessarily relate to better-defined biological constructs, may reduce the statistical power to detect associated risk variants. Furthermore, the complex clinical phenotype and associated comorbidities frequently lead to diagnostic uncertainties. This may be one of the reasons why some genetic factors identified to date confer comparable risk for both schizophrenia and bipolar disorder.6 On the other hand, different etiological pathways could lead to similar phenotypic expressions creating even more difficulties in mapping specific genetic effects in schizophrenia. In the current study, we aimed to combine 2 new strategies that may help address the aforementioned problems associated with polygenic diseases and phenotypic complexity: an intermediate phenotype approach and the implementation of a genetic risk score (GRS).
“Intermediate phenotypes” (also known as “endophenotypes”) are heritable, disease-associated neurophysiological, cognitive, or neurobiological traits.7 They are thought to be more closely related to the pathophysiology of a disease than clinically observable constructs such as signs, symptoms, or diagnosis and therefore more proximal to the genetic substrate. Intermediate phenotypes may limit phenotypic complexity and reduce genetic heterogeneity thereby facilitating identification of susceptibility genes that underlie pathophysiological aspects of mental disorders.7
Dorsolateral prefrontal cortex (DLPFC) dysfunction during working memory (WM) processing is a widely acknowledged intermediate phenotype for schizophrenia.8 Compared with matched healthy controls, patients display abnormal DLPFC functioning across task difficulties.9 More specifically, DLPFC activity is assumed to follow an inverted U-shaped curve with increasing task difficulty. However, the activity pattern in schizophrenia appears to be shifted slightly so that compared with healthy controls, persons with schizophrenia show more activation for easy tasks but less activation for difficult tasks.10 Thus, patients may exhibit an “inefficiency” of the prefrontal cortex and need to recruit more neural resources than controls for the same task and may show decreased neural activity (hypofrontality) when task difficulty becomes too great and performance declines.10–12 Numerous studies have found associations between genetic risk and DLPFC inefficiency, providing evidence for the heritability of DLPFC function. Two meta-analyses on unaffected relatives of schizophrenia patients and healthy controls reported abnormalities in DLPFC functioning in the majority of reported experiments, substantiating that DLPFC inefficiency is heritable and related to elevated risk for schizophrenia.13,14 Karlsgodt et al12 observed that unaffected cotwins were intermediate in performance and prefrontal activity compared with comparison subjects and patients, who showed clear signs of DLPFC inefficiency. Investigating the impact of the COMT Val108/158Met polymorphism on DLPFC activity in schizophrenia patients, unaffected siblings, and healthy controls, Egan et al15 detected an allele dosage–dependent association with DLPFC inefficiency. Prefrontal activity was greater and performance worse with increasing number of Val alleles with siblings being intermediate in performance when compared with patients and controls. Similar results were found in a study by Bertolino et al,16 who showed a COMT genotype–dependent improvement of DLPFC dysfunction after antipsychotic treatment with Val-carriers benefiting the least from the intervention, thus demonstrating a clinical relevance of assessing genetic risk.
To address the polygenic nature of the disorder, we derived a cumulative summary score, which we call a GRS. This score measures risk that is based on the additive effects of several known genetic susceptibility loci for schizophrenia. The implementation of a GRS is a new and promising approach to identify people at risk and has so far been applied in other diseases such as rheumatoid arthritis,17 multiple sclerosis,18 and coronary heart disease.19 A similar approach was used in a genetic association study comparing schizophrenia patients with healthy controls.6 However, at a conventional threshold, the cumulative risk measure explained only 1% of the variance of disorder occurrence. A GRS used to predict an intermediate phenotype of schizophrenia may explain more variance than the prediction of case-control status. Given the polygenic basis and complexity of symptoms in schizophrenia, the combination of an intermediate phenotype and a GRS may improve the statistical power for detecting associations between schizophrenia-related genetic variation and pathophysiological aspects of schizophrenia. The goals of this study were (1) to derive a GRS that conveys the combined risk of several previously identified risk variants for schizophrenia and (2) to investigate the association between the risk score and whole-brain activity during a WM task. We expected an association between the risk score and DLPFC inefficiency.
Methods
Participants
Imaging genetic and behavioral data from participants of the Mind Clinical Imaging Consortium (MCIC) study of schizophrenia from 4 participating sites (the University of New Mexico [UNM], the University of Minnesota [UMN], Massachusetts General Hospital [MGH], and the University of Iowa [UI]) were used to determine genetic polymorphisms in cryo-conserved blood samples and to analyze whole-brain neural activity during a WM task. Out of a total of 285 participants, who passed imaging quality control procedures (see below), blood samples of 180 participants were available for genetic analysis, resulting in a final dataset of 79 schizophrenia patients and 99 healthy controls after genetic quality control steps (see below). Patients had a Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) diagnosis of schizophrenia or schizophreniform disorder, established using a Structured Clinical Interview for DSM disorders and a review of case files by trained clinicians. In the initial cohort, controls were matched to the patient group for age, gender, and parental education and were excluded if they had a history of a medical or axis I psychiatric diagnosis. The majority of participants were of Caucasian descent (94 healthy controls and 60 patients). For additional details about the participants and clinical measures, see online supplementary methods (SM) 1.1, SM 1.2, and references.20–22
Behavioral Task
The Sternberg Item Recognition Paradigm (SIRP) is a WM task, previously shown to consistently activate the DLPFC in healthy controls and schizophrenia patients.11 The SIRP was administered during six 46-s blocks per run for three 360-s runs. In each block, a memory set, composed of 1 (load 1), 2 (load 3), or 5 (load 5) digits, was presented (2 blocks per load condition). The Encode phase was followed by a presentation of 14 digits, one at a time (the probe phase) and participants responded to each probe to indicate whether or not the probe digit was in the memory set. Participants were instructed to respond as quickly and accurately as possible and were given a bonus of 5 cents for each correct response. This bonus was provided after completion of the scan. For additional details about the paradigm, see reference.20,21 The stimuli and responses were presented and collected using E-prime software (EPrime v1.1, Psychology Software Tools, Inc., Pittsburg, PA). Participants were excluded from further analysis, if they completed a block with less than a 78% accuracy rate and/or with more than 6 probes not answered within a block.
Image Acquisition and Preprocessing
Structural magnetic resonance imaging (MRI) data were acquired with either a 1.5T Siemens Sonata (UNM, MGH, and UI) or a 3T Siemens Trio (UMN). Functional MRI (fMRI) data were acquired with either a 1.5T Siemens Sonata (UNM) or a 3T Siemens Trio (UMN, MGH, and UI). For additional details about data acquisition, see online supplementary methods, SM 1.3. Cortical reconstruction and volumetric segmentation based on high resolution structural MRI scans were performed with the FreeSurfer surface reconstruction software http://surfer.nmr.mgh.harvard.edu. Functional data were analyzed using fMRIB Software Library (FSL) (http://www.fmrib.ox.ac.uk/fsl). We fit a general linear model to the fMRI time course at each voxel in a whole-brain model to estimate the average activation during the 3 loads of the probe condition in all trials. Equal weight was given to all loads. Quality assurance steps included checks for whole-brain coverage of brain masks, motion and global mean intensity outlier timepoints, alignment of structural and functional scans, and registration problems (Echo Planar Imaging to T1 and T1 to template).
Genotyping
Blood samples were obtained from each participant and sent to the Harvard Partners Center for Genetics and Genomics for DNA extraction. All DNA extraction and genotyping were done blind to group assignment. Genotyping was performed at the Mind Research Network Neurogenetics Core Lab using the Illumina HumanOmni-Quad BeadChip. Further genotyping steps are described in online supplementary methods, SM 1.5. Quality control steps included common standard procedures.23 Single-nucleotide polymorphisms (SNPs) were checked for a genotyping rate of less than 90% and deviation from Hardy-Weinberg Equilibrium with a threshold of 10−6. No SNPs failed these tests, and the total genotyping rate was 99.7%. We removed participants with a genotyping rate of less than 90%. In 2 other cases, where the genotyping rate was above 90% but less than 100%, missing values (one SNP per participant) were replaced with the group mean.18 Analyses were carried out with PLINK, 1.06 (http://pngu.mgh.harvard.edu/purcell/plink).
GRS Calculation
SNPs were selected based on the continuously updated meta-analysis of genetic studies on schizophrenia, available at www.schizophreniaresearchforum.orgupdated on February 24, 2010. We selected all significant SNPs listed under the “Top Results.” For significance value calculation, see http://www.szgene.org/methods.asp. Given a large proportion of Caucasians in our sample, we excluded SNPs, which were significant for non-Caucasian groups only (n = 5). Furthermore, we excluded common variants other than SNPs (in this case, the Variable Number Tandem Repeat in the haptoglobin gene). The apolipoprotein E. (APOE) risk variants for schizophrenia represent a combination of 2 SNPs, rs429358 and rs7412. We therefore calculated APOE haplotypes in PLINK.
Based on these criteria, 55 SNPs were selected, out of which only 37 were present on the Illumina HumanOmni1-Quad BeadChip. Four more target SNPs, which were not on the Illumina chip, were replaced using tagging SNPs with a linkage disequilibrium (LD) r 2 > .8 within a 200 kbp window (based on phase 3 HapMap data). The reference allele of the tagSNP corresponding to the risk allele of the target SNP was also identified using HapMap. This resulted in a total of 41 SNPs in 34 genes, which were used for the GRS calculation (see online supplementary table 1 in SM 1.6). Furthermore, we analyzed the LD structure between these 41 SNPs in our own dataset using Haploview 4.2. Based on a threshold of r 2 > .8, only 2 SNPs (TPH1 gene on chromosome 11) were in LD suggesting a high degree of independence between all other SNPs in the GRS. An additive GRS for each participant was calculated based on the following formula:
where w is the log-transformed OR for each SNP with w = ln(ORSNP) and X is the number of risk alleles (0, 1, or 2).18
Allelic ORs for each SNP were taken from the SZGene database. They describe the association between schizophrenia and an allele by comparing the odds of disease in an individual carrying the wild type allele to the odds of disease in an individual carrying the mutation. If ORs were significant for several populations, we used either the overall OR (all) or, if not available, the OR for Caucasians. If an OR for the protective allele was reported, the reciprocal of this reported OR was used. The GRS was weighted by multiplying the number of risk alleles with the logarithmized OR of each SNP to take different effect sizes of SNPs into account. Supplementary table 1 in section SM 1.6 shows the log-transformed OR for each SNP.
Statistical Models
We performed whole-brain analyses investigating the relationship between GRS-and WM-induced brain activity for patients and controls using mixed effects models in FSL. All models were cluster-corrected according to FSL default settings with a z value of 2.3 and a P value of .05 and controlled for acquisition site. To account for nonrandom sampling of schizophrenia patients, we explicitly modeled the effects of diagnosis in our main model (model 1). In a second model, we pooled controls and patients (model 2).
To control for population stratification, we used linkage agglomerative clustering, based on pairwise identity by state (IBS) distance. In detail, we first merged the MCIC dataset with the Hapmap phase II dataset, containing 502 participants of the 4 Hapmap populations: Utah residents with ancestry from Northern and Western Europe (CEU), Han Chinese in Beijing, China (CHB), Japanese in Tokyo, Japan (JPT), and Yoruban in Ibadan, Nigeria west Africa (YRI). A symmetric 680 × 680 matrix of the IBS distances for all pairs of individuals was created and used for multidimensional scaling analysis. Six dimensions were extracted and included in the first 2 FSL models as covariates to correct for residual population substructure. We also ran a third model (model 3), where we included only participants of Caucasian descent, controlling for diagnosis and acquisition site. Caucasians were defined as ±0.05 units of deviation from the Hapmap CEU cluster in a plot with the first 2 principal components on the x- and y-axis. Sample characteristic analyses were carried out with SPSS 17.0.
Results
Sample Characteristics
Patients and controls did not differ in age, handedness, parental socioeconomic status (SES), or in their GRS (table 1). Patients, had a significantly lower IQ (Welch test F = 28.86; P < .001), were less likely to be female (Pearson Chi-square test χ2 = 5.665; P = .017) and included fewer Caucasians (Pearson Chi-square test χ2 = 13.60; P < .001). Patients and controls were distributed evenly across acquisition sites (table 1). Patients’ and controls’ accuracy on the SIRP were above 78% at all 3 WM loads but on average schizophrenia patients were less accurate (95%) than healthy controls (98%; Welch test due to unequal variances between groups with F = 29.54; P < .001).
Table 1.
Demographic and Clinical Results
| Site | Sample | Gender (Female) | Race (Caucasians) | Age (y) | Cognitive Function (WRAT-III) | Parental SES | Handedness (0–12) | GRS | ||||||||
| n | n | % | n | % | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| UI | HC | 49a | 23a | 46.9 | 48 | 98.0 | 30.27 | 10.47 | 49.98 | 4.11 | 2.86b | 0.46 | 0.73 | 2.64 | 6.83 | 0.76 |
| SCZ | 22a | 3a | 13.6 | 20 | 90.9 | 30.82 | 9.14 | 48.14 | 5.07 | 2.38b | 0.74 | 0.82 | 2.81 | 6.94 | 0.72 | |
| MGH | HC | 19 | 8 | 42.1 | 16 | 84.2 | 40.11 | 10.53 | 52.58b | 3.36 | 2.84 | 0.90 | 1.26 | 3.19 | 6.87 | 0.89 |
| SCZ | 26 | 7 | 26.9 | 16 | 61.5 | 38.58 | 9.94 | 45.04b | 8.45 | 3.38 | 1.10 | 0.83 | 2.18 | 6.94 | 0.65 | |
| UMN | HC | 15 | 7 | 46.7 | 15a | 100.0 | 33.00 | 11.40 | 51.08b | 3.94 | 2.50 | 0.91 | 0.67 | 0.89 | 6.93 | 0.51 |
| SCZ | 18 | 6 | 33.3 | 11a | 61.1 | 31.28 | 10.62 | 45.56b | 5.44 | 2.67 | 0.69 | 2.50 | 4.22 | 6.91 | 0.80 | |
| UNM | HC | 16 | 4 | 25.0 | 15 | 93.8 | 30.42 | 12.77 | 51.37b | 3.93 | 2.05b | 0.52 | 1.21 | 2.57 | 7.11 | 1.18 |
| SCZ | 13 | 4 | 30.8 | 13 | 100.0 | 34.08 | 13.79 | 46.17b | 6.83 | 2.92b | 1.24 | 1.92 | 3.57 | 7.16 | 0.60 | |
| Total | HC | 99 | 42a | 42.4 | 94a | 94.9 | 32.52 | 11.54 | 50.88b | 3.99 | 2.66 | 0.70 | 0.92 | 2.58 | 6.90 | 0.85 |
| SCZ | 79 | 20a | 25.3 | 60a | 75.9 | 34.01 | 10.93 | 46.21b | 6.67 | 2.87 | 1.02 | 1.40 | 3.18 | 6.97 | 0.69 | |
Note: Means and SDs are given, unless otherwise indicated. UI/MGH/UMN/UNM represent the 4 different study sites. Race and GRS were defined as described under “Methods” section. Parental SES was classified according to Hollingshead and handedness, determined using the Annett Scale of Hand Preference. WRAT-III, Wide Range Achievement Test; SCZ, patient with schizophrenia; HC, healthy controls; UMN, University of Minnesota; UNM, the University of New Mexico; MGH, Massachusetts General Hospital; UI, the University of Iowa; GRS, genetic risk score; SES, socioeconomic status.
Significantly different between SCZ patients and healthy normal controls on the basis of a Chi-square (P < .05).
Significantly different between SCZ patients and healthy normal controls on the basis of a one-way ANOVA (P < .05).
GRS Characteristics
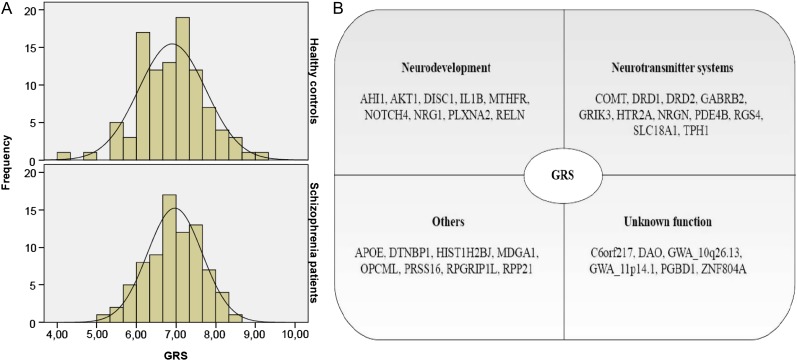
GRS did not correlate with age, SIRP performance, parental SES, or IQ. Furthermore, there were no significant correlations between GRS and cumulative and current antipsychotic drug dose (for details, see online supplementary methods, SM 1.2), as well as negative and positive symptoms in the patient group (see online supplementary table 2). GRS did not differ between acquisition sites, gender, diagnostic group, or race (table 1) and followed a normal distribution both in patients and in controls (figure 1A). Forty-one SNPs from 34 different genes were included in the GRS. These genes have many different functions but can be categorized into genes involved in neurodevelopment (26%), neurotransmitter systems (32%), and other functions such as cell adhesion, cell cycle, immune response, and transcription (24%; figure 1B).
Fig. 1.
(A) The genetic risk score (GRS) follows a normal distribution both in controls and in patients. (B) GRS comprises genes, which play a role in neurodevelopment, neurotransmitter systems, and other functions, such as cell adhesion, cell cycle, immune response, and transcription. It also includes genes of yet unknown function.
WM-Related Neural Activity
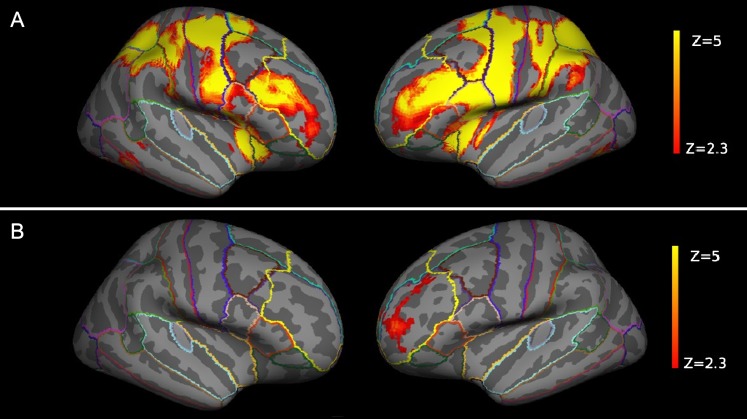
The SIRP task reliably activated WM-associated brain regions such as the DLPFC and parietal regions (figure 2A). As predicted, we could show a positive association between GRS and neural activity in the left DLPFC (model 1, z-max = 3.84, P = .0252, cluster-corrected; figure 2B), which was the only surviving cluster in a whole-brain analysis for a GRS main effect in a model covarying for the effects of acquisition site, diagnosis, and population stratification. This finding remained significant in a pooled analysis without controlling for diagnosis (model 2, z-max = 3.93, P = .007, cluster-corrected). GRS, which combined the impact of many genes with small effects (see online supplementary figure 1), accounted for 3.6% of the total variance (adjusted R2, model 1) at the most activated DLPFC locale (x, y, z: −26, 50, 8), corresponding to a significant R 2 change of .04 (P = .008). There were no significant diagnosis by GRS interaction effects.
Fig. 2.
(A) The Sternberg paradigm successfully activated the dorsolateral prefrontal cortex (DLPFC) in both hemispheres (cluster-corrected with z = 2.3 and P < .05). (B) Genetic risk correlated with activity in the left DLPFC (cluster-corrected with z = 2.3 and P < .05, displayed are the results of model 1). The DLPFC labels are displayed for illustrative purposes only—please refer to online supplementary material, SM 1.4, for further details. The z values are represented according to the color code.
As an alternative way to control for possible bias due to population stratification, we ran a subsequent whole-brain model including only Caucasians (n = 154) and covarying for acquisition site and diagnosis (model 3). The effect of GRS on left DLPFC activity remained significant (z-max = 3.84, P = .018). We also found a second significant cluster in the left pars triangularis with the effect of GRS pointing in the same direction as in the DLPFC cluster (z-max = 4.58, P = .005; see online supplementary figure 2). As in the first model, there were no significant diagnosis by GRS interaction effects.
Discussion
Summary of Findings
In this large multicenter study, we found an overall positive correlation between additive GRS for schizophrenia and increased WM-related DLPFC activity. This finding was highly specific for left DLPFC activity during mental scanning of WM items—a widely acknowledged intermediate phenotype for schizophrenia. The association between the GRS and DLPFC activation was not attributable to symptom severity, population stratification, or group differences in WM performance.
Schizophrenia and Prefrontal Inefficiency
As expected, the SIRP task induced reliable bilateral activations in the DLPFC and parietal lobe, regions typically found in WM tasks.24 The association between cumulative genetic risk for schizophrenia and WM-induced prefrontal inefficiency is in line with a large body of literature from previous neurophysiology and neuroimaging studies demonstrating DLPFC dysfunction during executive functioning in schizophrenia patients. Frontal abnormalities in schizophrenia patients during rest as well as executive functioning are well established. More than 10 years ago, Manoach et al11 confirmed that schizophrenia patients showed a significantly greater magnitude of activation than controls in the DLPFC during WM processing using fMRI. These findings were interpreted as a deficit in automation, ie, patients may fail to automate WM tasks, which in turn leads to decreased efficiency.11 The inefficiency hypothesis was supported by Callicott et al,10 who found that high-performing patients used greater resources in parts of the left DLPFC if compared with healthy controls. Similarly, Potkin et al25 observed that patients had an increased DLPFC response already during moderate memory loads when compared with healthy controls. Effects were stronger on the left side. A study on medication-naive patients and controls compared DLPFC activity before and after practice of a WM task.26 As expected, activity decreased with practice in all groups, but this decrease was smaller in patients in the left DLPFC, even after controlling for behavioral differences. This illustrates nicely the inefficiency hypothesis—even with training patients continue to recruit more neural resources than controls to perform the same tasks. Accordingly, we observed greater prefrontal neural activity with increasing risk for schizophrenia, although task performance was high in healthy controls and schizophrenia patients.
In an additional exploratory model, where we excluded all non-Caucasian participants, we also found a second significant association between genetic risk and WM-induced activation in the left pars triangularis in the same direction as for the DLPFC cluster. This region is part of the midventrolateral prefrontal cortex, which mediates interference resolution during WM tasks.27 Activation in the same region has also been found to correlate with increasing task demand during a WM task and was interpreted as a compensatory response.28 Compensatory activation in the ventral prefrontal cortex was further closely associated with DLPFC inefficiency in schizophrenia patients.29
Genetic Risk and Prefrontal Inefficiency
The fact that GRS was linked to brain function but not to task performance or symptom severity suggests that the GRS reflects unique genetic aspects of aberrant neural responses related to schizophrenia that are perhaps not well represented in the clinical or cognitive presentations of patients or high-risk individuals. Such a finding bodes well for the validity of intermediate neural phenotypes suggesting that genetic effects are more clearly discerned through biological constructs than descriptions of complex behavior. Effect sizes for genetic associations with an intermediate phenotype have been found to be consistently higher than with psychiatric diagnoses indicating that gene effects may have a higher penetrance at the level of brain physiology than at the level of higher order constructs such as behavior.30 Associations between WM-induced abnormalities in prefrontal function and genetic risk variants for schizophrenia have been repeatedly documented in studies. The early studies from the Weinberger group established relationships between genes with functions in the dopaminergic system and prefrontal inefficiency during executive functioning: COMT Val108/158Met risk allele carriers were found to have increased WM-induced prefrontal activation compared with nonrisk homozygotes.31 Furthermore, risk variants of the dopamine D1 and D2 receptor and the gamma amino butyric acid (GABA) receptor B2 were all associated with prefrontal inefficiency.32–34 In addition, risk variants in genes with functions in brain development such as AKT1, NRG1, DISC1, MTHFR, and DTNBP1 and genes related to other complex functions such as RGS4, APOE, and ZNF804A were also associated with prefrontal dysfunction during executive tasks.20,35–40
Combining the effects of several risk variants for schizophrenia predicted WM-related DLPFC activity and explained a total amount of variance that exceeded that of traditional case-control studies.6,41 Thus, by studying the effects of many markers for schizophrenia on a brain-based intermediate phenotype, we were able to detect the additive effects of different biological pathways and systems that may impinge on prefrontal cortical function but perhaps are below the level of detection in behavior and clinical symptomatology. We did not find significant differences in GRS, when comparing cases and controls. Considering that the sample size is comparatively small and that the GRS was derived from only a few previously studied SNPs, this could be due to a lack of power. Given that the GRS was derived from risk genes for schizophrenia found in previous genetic association studies and that DLPFC dysfunction is a well-validated intermediate phenotype for schizophrenia, our imaging genetics results indicate an important association with the disorder that warrants further investigation.
Our GRS comprised genes from several major gene families, such as those involved in neurodevelopment (AHI1, AKT1, DISC1, IL1B, MTHFR, NOTCH4, NRG1, PLXNA2, and RELN) and in the regulation of neurotransmitter systems like dopamine (COMT, SLC18A1, DRD1, and DRD2), serotonin (HTR2A and TPH1), and GABA (GABRB2). Other genes are involved in processes such as cell adhesion (MDGA1), immune responses (OPCML and PRSS16), cell cycle (RPGRIP1L), or transcription (RPP21) (http://www.ncbi.nlm.nih.gov/gene). Our SNP selection was based on a meta-analysis of previously published schizophrenia case-control association studies, and conclusions about underlying pathological pathways should be considered with caution. The fact that these SNPs have been previously linked to schizophrenia and, in combination, predict a widely acknowledged imaging phenotype for schizophrenia lends support to the hypothesis that abnormal neurodevelopment processes and dysfunctional neurotransmitter systems may have causal effects in the pathogenesis of this burdensome disorder. However, it should be noted that we did not explicitly select for WM-related SNPs and that the level of evidence for each schizophrenia risk gene varies depending on the currently available case-control studies. For some of the SNPs included into our GRS, the preexisting evidence might therefore be only moderate and warrants replication in independent cohorts. Also, lacking a complete understanding of the molecular pathways for these traits and the relevant gene-gene interaction effects, our genetic model of risk was additive assuming a linear increase in disease susceptibility per risk allele. With the advancements in computational modeling, future studies should go beyond the additive model underlying our risk score and try to incorporate existing biological knowledge and gene-gene interactions.
Conclusions
We derived a GRS, which combined the effects of 41 genetic risk variants for schizophrenia, and demonstrated that this score predicted DLPFC inefficiency during a WM task, a common intermediate phenotype in schizophrenia. The finding is in line with a growing number of reports demonstrating associations between single genetic risk variants and schizophrenia-related DLPFC dysfunction and supports an additive genetic risk model for a polygenic phenotype. A neural characterization of genetic risk could help to define system neuroscience models of schizophrenia. Ongoing work will show, if a GRS approach provides an advantage over studying single genes in predicting intermediate phenotypes for neuropsychiatric disorders and if it can be used to estimate the risk for schizophrenia in susceptible populations.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
National Institutes of Health/National Center for Research Resources (NIH/NCRR P41RR14075); Department of Energy (DE-FG02-99ER62764); Mind Research Network, Morphometry BIRN (1U24, RR021382A); Function Biomedical Informatics Research Network (U24RR021992-01, NIH/NCRR MO1 RR025758-01, National Institute of Mental Health 1RC1MH089257 to V.D.C.); the Deutsche Forschungsgemeinschaft (Research Fellowship to S.E.); the Deutscher Akademischer Auslandsdienst; and the Friedrich-Ebert Stiftung (stipends to E.W.).
Supplementary Material
Acknowledgments
All authors declare no biomedical financial interest or other potential conflict of interest.
References
- 1.Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munafo MR, Bowes L, Clark TG, Flint J. Lack of association of the COMT (Val158//108 Met) gene and schizophrenia: a meta-analysis of case-control studies. Mol Psychiatry. 2005;10:765–770. doi: 10.1038/sj.mp.4001664. [DOI] [PubMed] [Google Scholar]
- 3.O’Donovan MC, Craddock NJ, Owen MJ. Genetics of psychosis; insights from views across the genome. Hum Genet. 2009;126:3–12. doi: 10.1007/s00439-009-0703-0. [DOI] [PubMed] [Google Scholar]
- 4.Weedon MN, Lettre G, Freathy RM, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 8.Hall MH, Smoller JW. A new role for endophenotypes in the GWAS era: functional characterization of risk variants. Harv Rev Psychiatry. 2010;18:67–74. doi: 10.3109/10673220903523532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberger DR, Egan MF, Bertolino A, et al. Prefrontal neurons and the genetics of schizophrenia. Biol Psychiatry. 2001;50:825–844. doi: 10.1016/s0006-3223(01)01252-5. [DOI] [PubMed] [Google Scholar]
- 10.Callicott JH, Mattay VS, Verchinski BA, Marenco S, Egan MF, Weinberger DR. Complexity of prefrontal cortical dysfunction in schizophrenia: more than up or down. Am J Psychiatry. 2003;160:2209–2215. doi: 10.1176/appi.ajp.160.12.2209. [DOI] [PubMed] [Google Scholar]
- 11.Manoach DS, Press DZ, Thangaraj V, et al. Schizophrenic subjects activate dorsolateral prefrontal cortex during a working memory task, as measured by fMRI. Biol Psychiatry. 1999;45:1128–1137. doi: 10.1016/s0006-3223(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 12.Karlsgodt KH, Glahn DC, van Erp TGM, et al. The relationship between performance and fMRI signal during working memory in patients with schizophrenia, unaffected co-twins, and control subjects. Schizophr Res. 2007;89:191–197. doi: 10.1016/j.schres.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald AW, III, Thermenos HW, Barch DM, Seidman LJ. Imaging genetic liability to schizophrenia: systematic review of FMRI studies of patients’ nonpsychotic relatives. Schizophr Bull. 2009;35:1142–1162. doi: 10.1093/schbul/sbn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goghari VM. Executive functioning-related brain abnormalities associated with the genetic liability for schizophrenia: an activation likelihood estimation meta-analysis. Psychol Med. 2011;41:1239–1252. doi: 10.1017/S0033291710001972. [DOI] [PubMed] [Google Scholar]
- 15.Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertolino A, Caforio G, Blasi G, et al. Interaction of COMT Val108/158 Met genotype and olanzapine treatment on prefrontal cortical function in patients with schizophrenia. Am J Psychiatry. 2004;161:1798–1805. doi: 10.1176/ajp.161.10.1798. [DOI] [PubMed] [Google Scholar]
- 17.Karlson EW, Chibnik LB, Kraft P, et al. Cumulative association of 22 genetic variants with seropositive rheumatoid arthritis risk. Ann Rheum Dis. 2010;69:1077–1085. doi: 10.1136/ard.2009.120170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jager PL, Chibnik LB, Cui J, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8:1077–1079. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison AC, Bare LA, Chambless LE, et al. Prediction of coronary heart disease risk using a genetic risk score: the Atherosclerosis Risk in Communities Study. Am J Epidemiol. 2007;166:28–35. doi: 10.1093/aje/kwm060. [DOI] [PubMed] [Google Scholar]
- 20.Roffman JL, Gollub RL, Calhoun VD, et al. MTHFR 677C → T genotype disrupts prefrontal function in schizophrenia through an interaction with COMT 158Val → Met. Proc Natl Acad Sci U S A. 2008;105:17573–17578. doi: 10.1073/pnas.0803727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehrlich S, Brauns S, Yendiki A, et al. Associations of cortical thickness and cognition in patients with schizophrenia and healthy ontrols [published online ahead of print March 24, 2011] Schizophr Bull. 2011 doi: 10.1093/schbul/sbr018. doi:10.1093/schbul/sbr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrlich S, Morrow EM, Roffman JL, et al. The COMT Val108/158Met polymorphism and medial temporal lobe volumetry in patients with schizophrenia and healthy adults. Neuroimage. 2010;53:992–1000. doi: 10.1016/j.neuroimage.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teo YY. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Curr Opin Lipidol. 2008;19:133–143. doi: 10.1097/MOL.0b013e3282f5dd77. [DOI] [PubMed] [Google Scholar]
- 24.Manoach DS, Schlaug G, Siewert B, et al. Prefrontal cortex fMRI signal changes are correlated with working memory load. Neuroreport. 1997;8:545–549. doi: 10.1097/00001756-199701200-00033. [DOI] [PubMed] [Google Scholar]
- 25.Potkin SG, Turner JA, Brown GG, et al. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009;35:19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Veelen NMJ, Vink M, Ramsey NF, Kahn RS. Left dorsolateral prefrontal cortex dysfunction in medication-naive schizophrenia. Schizophr Res. 2010;123:22–29. doi: 10.1016/j.schres.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Badre D, Wagner AD. Left ventrolateral prefrontal cortex and the cognitive control of memory. Neuropsychologia. 2007;45:2883–2901. doi: 10.1016/j.neuropsychologia.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 28.Wolf RC, Vasic N, Walter H. Differential activation of ventrolateral prefrontal cortex during working memory retrieval. Neuropsychologia. 2006;44:2558–2563. doi: 10.1016/j.neuropsychologia.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Tan H-Y, Sust S, Buckholtz JW, et al. Dysfunctional prefrontal regional specialization and compensation in schizophrenia. Am J Psychiatry. 2006;163:1969–1977. doi: 10.1176/ajp.2006.163.11.1969. [DOI] [PubMed] [Google Scholar]
- 30.Meyer-Lindenberg A. From maps to mechanisms through neuroimaging of schizophrenia. Nature. 2010;468:194–202. doi: 10.1038/nature09569. [DOI] [PubMed] [Google Scholar]
- 31.Meyer-Lindenberg A, Nichols T, Callicott JH, et al. Impact of complex genetic variation in COMT on human brain function. Mol Psychiatry. 2006;11:867–877. doi: 10.1038/sj.mp.4001860. [DOI] [PubMed] [Google Scholar]
- 32.Tura E, Turner JA, Fallon JH, Kennedy JL, Potkin SG. Multivariate analyses suggest genetic impacts on neurocircuitry in schizophrenia. Neuroreport. 2008;19:603–607. doi: 10.1097/WNR.0b013e3282fa6d8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertolino A, Fazio L, Caforio G, et al. Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain. 2009;132:417–425. doi: 10.1093/brain/awn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winterer G, Smolka M, Samochowiec J, et al. Association analysis of GABAAbeta2 and gamma2 gene polymorphisms with event-related prefrontal activity in man. Hum Genet. 2000;107:513–518. doi: 10.1007/s004390000401. [DOI] [PubMed] [Google Scholar]
- 35.Markov V, Krug A, Krach S, et al. Impact of schizophrenia-risk gene dysbindin 1 on brain activation in bilateral middle frontal gyrus during a working memory task in healthy individuals. Hum Brain Mapp. 2010;31:266–275. doi: 10.1002/hbm.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicodemus KK, Law AJ, Radulescu E, et al. Biological validation of increased schizophrenia risk with NRG1, ERBB4, and AKT1 epistasis via functional neuroimaging in healthy controls. Arch Gen Psychiatry. 2010;67:991–1001. doi: 10.1001/archgenpsychiatry.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prata DP, Mechelli A, Fu CHY, et al. Effect of disrupted-in-schizophrenia-1 on pre-frontal cortical function. Mol Psychiatry. 2008;13:915–917. doi: 10.1038/mp.2008.76. , 909. [DOI] [PubMed] [Google Scholar]
- 38.Filippini N, MacIntosh BJ, Hough MG, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esslinger C, Walter H, Kirsch P, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- 40.Buckholtz JW, Sust S, Tan HY, et al. fMRI evidence for functional epistasis between COMT and RGS4. Mol Psychiatry. 2007;12:893–895. doi: 10.1038/sj.mp.4002008. , 885. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda M, Aleksic B, Kinoshita Y, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.