Abstract
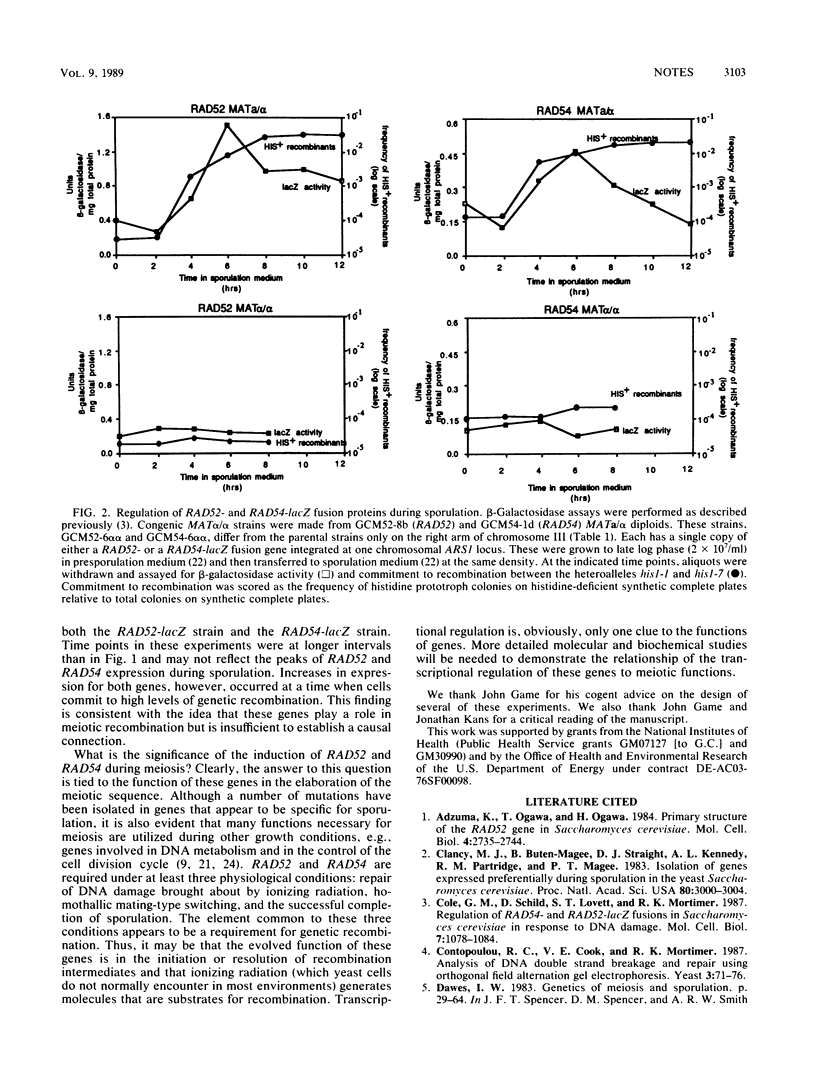
The DNA repair and recombination genes of Saccharomyces cerevisiae, RAD52 and RAD54, were transcriptionally induced approximately 10- to 15-fold in sporulating MATa/alpha cells. Congenic MATa/a cells, which did not sporulate, did not show similar increases. Assays of beta-galactosidase activity in strains harboring either a RAD52- or RAD54-lacZ gene fusion indicated that this induction occurred at a time concomitant with a commitment to meiotic recombination, as measured by prototroph formation from his1 heteroalleles.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adzuma K., Ogawa T., Ogawa H. Primary structure of the RAD52 gene in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2735–2744. doi: 10.1128/mcb.4.12.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy M. J., Buten-Magee B., Straight D. J., Kennedy A. L., Partridge R. M., Magee P. T. Isolation of genes expressed preferentially during sporulation in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 May;80(10):3000–3004. doi: 10.1073/pnas.80.10.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1987 Mar;7(3):1078–1084. doi: 10.1128/mcb.7.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contopoulou C. R., Cook V. E., Mortimer R. K. Analysis of DNA double strand breakage and repair using orthogonal field alternation gel electrophoresis. Yeast. 1987 Jun;3(2):71–76. doi: 10.1002/yea.320030203. [DOI] [PubMed] [Google Scholar]
- Esposito R. E., Esposito M. S. Genetic recombination and commitment to meiosis in Saccharomyces. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Game J. C., Zamb T. J., Braun R. J., Resnick M., Roth R. M. The Role of Radiation (rad) Genes in Meiotic Recombination in Yeast. Genetics. 1980 Jan;94(1):51–68. doi: 10.1093/genetics/94.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlin-Ninfa E., Kaback D. B. Isolation and functional analysis of sporulation-induced transcribed sequences from Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2185–2197. doi: 10.1128/mcb.6.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Feldberg L. R. Saccharomyces cerevisiae exhibits a sporulation-specific temporal pattern of transcript accumulation. Mol Cell Biol. 1985 Apr;5(4):751–761. doi: 10.1128/mcb.5.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S. M., Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974 Apr;118(1):8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraig E., Haber J. E. Messenger ribonucleic acid and protein metabolism during sporulation of Saccharomyces cerevisiae. J Bacteriol. 1980 Dec;144(3):1098–1112. doi: 10.1128/jb.144.3.1098-1112.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Lindquist S. Changing patterns of gene expression during sporulation in yeast. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7323–7327. doi: 10.1073/pnas.81.23.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A., Segall J. Isolation of DNA sequences preferentially expressed during sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):142–150. doi: 10.1128/mcb.4.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L., Burke W., Montelone B. A. Effects of the RAD52 Gene on Recombination in SACCHAROMYCES CEREVISIAE. Genetics. 1980 Jan;94(1):31–50. doi: 10.1093/genetics/94.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W., Murray A. W. Cloning regulated yeast genes from a pool of lacZ fusions. Methods Enzymol. 1983;101:253–269. doi: 10.1016/0076-6879(83)01019-8. [DOI] [PubMed] [Google Scholar]
- SHERMAN F., ROMAN H. Evidence for two types of allelic recombination in yeast. Genetics. 1963 Feb;48:255–261. doi: 10.1093/genetics/48.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Byers B. Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma. 1978 Dec 21;70(1):109–130. doi: 10.1007/BF00292220. [DOI] [PubMed] [Google Scholar]
- Simchen G. Are mitotic functions required in meiosis? Genetics. 1974 Apr;76(4):745–753. doi: 10.1093/genetics/76.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. T., Frackman S., Kowalisyn J., Esposito R. E., Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987 Apr;7(4):1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]