Abstract
Mitochondrial dysfunction is a contributor to diabetic cardiomyopathy. Previously, we observed proteomic decrements within the inner mitochondrial membrane (IMM) and matrix of diabetic cardiac interfibrillar mitochondria (IFM) correlating with dysfunctional mitochondrial protein import. The goal of this study was to determine whether overexpression of mitochondria phospholipid hydroperoxide glutathione peroxidase 4 (mPHGPx), an antioxidant enzyme capable of scavenging membrane-associated lipid peroxides in the IMM, could reverse proteomic alterations, dysfunctional protein import, and ultimately, mitochondrial dysfunction associated with the diabetic heart. MPHGPx transgenic mice and controls were made diabetic by multiple low-dose streptozotocin injections and examined after 5 wk of hyperglycemia. Five weeks after hyperglycemia onset, in vivo analysis of cardiac contractile function revealed decreased ejection fraction and fractional shortening in diabetic hearts that was reversed with mPHGPx overexpression. MPHGPx overexpression increased electron transport chain function while attenuating hydrogen peroxide production and lipid peroxidation in diabetic mPHGPx IFM. MPHGPx overexpression lessened proteomic loss observed in diabetic IFM. Posttranslational modifications, including oxidations and deamidations, were attenuated in diabetic IFM with mPHGPx overexpression. Mitochondrial protein import dysfunction in diabetic IFM was reversed with mPHGPx overexpression correlating with protein import constituent preservation. Ingenuity Pathway Analyses indicated that oxidative phosphorylation, tricarboxylic acid cycle, and fatty acid oxidation processes most influenced in diabetic IFM were preserved by mPHGPx overexpression. Specific mitochondrial networks preserved included complex I and II, mitochondrial ultrastructure, and mitochondrial protein import. These results indicate that mPHGPx overexpression can preserve the mitochondrial proteome and provide cardioprotective benefits to the diabetic heart.
Keywords: mitochondria, proteomics, diabetes mellitus, oxidative stress, heart
cardiac failure and heart disease are the leading cause of morbidity and mortality among diabetic patients (18). Multiple studies have specifically implicated the mitochondrion as a participant in the mechanisms contributing to cardiac dysfunction in the Type 1 diabetic heart (14, 18, 56). Mitochondrial analyses are complicated by the fact that two spatially distinct mitochondrial subpopulations exist in cardiac tissue, subsarcolemmal (SSM), and interfibrillar mitochondria (IFM). Literature suggests that mitochondrial subpopulations are differentially affected by pathological insults (2, 14, 29, 35, 44). We have previously shown that cardiac IFM exhibit greater dysfunction following Type 1 diabetic insult compared with SSM, which included enhanced oxidative stress, decreased oxidative phosphorylation (OXPHOS), and diminished nuclear-encoded mitochondrial protein import (2, 14).
Broad-scale proteomic analyses are becoming increasingly prevalent and are paramount for understanding mitochondrial dysfunction associated with the diabetic heart (2, 6, 22, 47, 51). In general, diabetes mellitus leads to significant alterations to proteins directly involved in mitochondrial energy production, including processes such as fatty acid oxidation (FAO), OXPHOS, and the tricarboxylic acid cycle (TCA). Furthermore, proteins involved in mitochondrial antioxidant defense, nuclear-encoded mitochondrial protein import, and protein folding are also highlighted as key loci affected by diabetes mellitus (2, 30, 51). We previously reported proteomic differences in spatially distinct mitochondrial subpopulations of which cardiac IFM displayed decrements to proteins involved in FAO, OXPHOS, the TCA cycle, mitochondrial structure (mitofilin), and nuclear-encoded mitochondrial protein import. Posttranslational modifications (PTM), such as oxidations and deamidations, were also elevated within the diabetic IFM (2).
Proteins and lipids within the inner mitochondrial membrane (IMM) are suspected to act as targets for oxidative damage based on their composition and close proximity to reactive oxygen species-generating sources. Phospholipid hydroperoxide glutathione peroxidase (PHGPx; GPx4) is an antioxidant enzyme that exists as a monomeric structure and has the ability to reduce peroxidized acyl groups in phospholipids, fatty acid hydroperoxides, and cholesterol peroxides within subcellular membranes (38). The mitochondrial-specific isoform mPHGPx is located primarily within the inner membrane space (IMS) (37). MPHGPx overexpression has been shown to be protective against simulated (26) and global ischemia-reperfusion injury (15). Essential mitochondrial phospholipids, such as cardiolipin, are known to surround electron transport chain (ETC) complexes as well as protein import machinery (19, 28, 41) and are suggested to be required for proper function of these processes. The goal of the current study was to determine whether overexpression of mPHGPx provides cardioprotective benefit to the Type 1 diabetic heart and whether the protective profile is associated with specific benefit to IMM functional processes including mitochondrial proteomic integrity and nuclear-encoded mitochondrial protein import. We hypothesized that mPHGPx overexpression would preserve the essential mitochondrial protein import process, leading to restoration of nuclear-encoded mitochondrial protein import and proteomic alterations associated with the diabetic heart.
MATERIALS AND METHODS
Experimental Animals
The animal experiments conformed to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the West Virginia University Animal Care and Use Committee. MPHGPx transgenic mouse lines were generated, screened, and characterized as previously described (15). Briefly, the pCAGGS vector places the mPHGPx cDNA under the control of the human cytomegalovirus (24, 27). All littermate control and transgenic mice were generated using an FVB background, and experimental procedures were performed on animals of ∼8 wk of age. Briefly, mouse litters were delivered after 19–20 days of gestation. To verify if the chimeric transgene was present in the genome, DNA from 4-wk-old mice was isolated from tail clips using a Qiagen DNeasy tissue kit (Qiagen, Valencia, CA). Transgene screening was performed using a quantitative PCR (qPCR) approach, in which we probed for the gpx4 cDNA using a fluorometric probe (Applied Biosystems, Foster City, CA). The probe spanned the exon 1 and exon 2 boundary such that only the nongenomic cDNA, void of an intron, was detected. Because the inherent genomic DNA encoding for mPHGPx contains an intron between exons 1 and 2, the ability for the fluorometric probe to bind across the exon 1 and 2 junction is prohibited. This approach ensured that only the transgenic cDNA encoding for gpx4 was detected and did not enable genomic gpx4 DNA to be detected. Isolated tail DNA, probe, primers, and universal master mix were brought up to 25 μl, and qPCR was performed in a 96-well plate using an Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems). All samples were run in duplicate. A reaction time versus cycle number amplification plot was generated, and transgene-positive and -negative animals were determined.
Diabetes Induction
Mice were housed in the West Virginia University Health Sciences Center animal facility and given unlimited access to a rodent diet and water. Four separate groups were utilized for the studies: 1) littermate control; 2) mPHGPx control; 3) diabetes mellitus; and 4) mPHGPx diabetes mellitus. Type 1 diabetes mellitus was induced in mixed sex control and transgenic mPHGPx mice following the protocol of the Animal Models of Diabetic Complications Consortium using multiple low-dose streptozotocin (STZ; Sigma, St. Louis, MO) injections. Briefly, injections of 50 mg/kg body wt STZ dissolved in sodium citrate buffer (pH 4.5) were performed daily for 5 consecutive days after 6 h of fasting. Mice that served as vehicle controls were given the same volume per body weight of sodium citrate buffer. Three days after the last injection, hyperglycemia was confirmed in all animals by measuring blood glucose levels (Contour Blood Glucose Test Strips, Bayer Healthcare, Mishawaka, IN), where the level of >250 mg/dl was considered diabetic. Animals were then maintained for 5 wk in a hyperglycemic state. Five weeks posthyperglycemia onset, animals were euthanized for further experimentation.
Echocardiographic Assessment
Transthoracic echocardiography was performed following the protocols provided by the Animal Models of Diabetic Complications Consortium. Briefly, mice were anesthetized in a knockdown box with inhalant isofluorane at 3% with 100% oxygen. When the mouse was anesthetized, it was transferred to dorsal recumbency and placed onto a heated imaging platform where electrocardiogram and rectal temperature were monitored. Before imaging was started, the isofluorane was reduced to 1% to minimize the anesthesia effects. Microultrasound images were acquired using a Vevo 2100 Imaging System (Visual Sonics, Toronto, Canada) and a 32- to 55-MHz linear array transducer. M-mode images were acquired via the parasternal short axis at midpapillary level with all images acquired at the highest possible frame rate (233–401 frames/s). Measurements obtained from left ventricular M-mode images included end-diastolic and end-systolic diameters and volumes, stroke volume, ejection fraction, fractional shortening, and cardiac output. All echocardiographic measurements were performed under the supervision of the West Virginia University Animal Models of Imaging Core Facility.
Preparation of Individual Mitochondrial Subpopulations
At 5 wk posthyperglycemia onset, control, diabetic, mPHGPx, and mPHGPx diabetic mice were euthanized and their hearts excised. Hearts were rinsed in PBS (pH 7.4) and then blotted dry. IFM were isolated as previously described following the methods of Palmer et al. (39) with minor modifications (2, 12, 14). Protein concentrations were determined using the Bradford method with bovine serum albumin as a standard (5).
Electron Transport Chain Respiration
State 3 and state 4 respiration rates were assessed as previously described (9, 10, 25) with slight modifications (15). Isolated mitochondria were resuspended in respiration buffer containing (in mmol/l) 80 KCl, 50 MOPS, 1 EGTA, 5 KH2PO4, and 1 mg/ml BSA, and equal volumes were loaded into a Gilson chamber (Gilson, Middleton, WI) attached to a Yellow Springs Instruments 5300 biological oxygen monitor (YSI, Yellow Springs, OH). The substrates glutamate (5 mM) plus malate (5 mM) were used to initiate respiration, and measurements of state 3 (1 mM ADP) and state 4 (ADP limited) respiration were made as previously described (13–15).
Electron Transport Chain Complex Activities
Electron Transport Chain (ETC) complexes I, III, and IV activities were measured spectrophotometrically as previously described (13–15, 50). Briefly, complex I activity was determined by measuring the oxidation of NADH at 340 nm, complex III activity was determined by following the reduction of cytochrome c at 550 nm in the presence of reduced decylubiquinone (50 μM), and complex IV activity was determined by measuring the oxidation of cytochrome c at 550 nm. Protein content was determined as described above (5), and values were expressed as activities in nanomoles substrate consumed per minute per milligram of protein. ATP synthase activity was measured as oligomycin-sensitive ATPase activity using an assay coupled with pyruvate kinase, which converts ADP to ATP and produces pyruvate from phosphoenolpyruvate as previously described (13, 16, 42, 45). Protein content was assessed as above (5) with final values expressed as nanomoles consumed per minute per milligram of protein, which was equal to the nanomoles of NADH oxidized per minute per milligram of protein.
Hydrogen Peroxide Production
H2O2 production was determined using the oxidation of the fluorogenic indicator amplex red in the presence of horseradish peroxidase (HRP) (57). The concentrations of HRP and amplex red in the incubation were 0.1 U/ml and 50 μm, and detection of fluorescence was assessed on a Molecular Devices Flex Station 3 fluorescence plate reader (Molecular Devices, Sunnyvale, CA) with 530-nm excitation and 590-nm emission wavelengths. Standard curves were obtained by adding known amounts of H2O2 to the assay medium in the presence of the substrates amplex red and HRP. H2O2 production was initiated in mitochondria using glutamate-malate as substrates. The addition of catalase diminished fluorescence confirming dissipation of H2O2 presence in the sample. Final values were calculated as pmoles per milligram of protein.
Lipid Peroxidation Products
Lipid peroxidation by-products malondialdehyde (MDA) and 4-hydroxyalkenal (4-HAE), stable end products formed from the oxidation of polyunsaturated fatty acids and esters, were assessed as previously described (13–15). Absorbance was measured on a Molecular Devices Flex Station 3 spectrophotometer (Molecular Devices, Sunnyvale, CA), and protein content was assessed as described above (5) with final values expressed per milligram of protein.
iTRAQ Labeling
Proteomic analyses were performed as previously described (2). Pooled IFM (n = 4) from control, diabetic, mPHGPx, and mPHGPx diabetic mice were lysed and precipitated overnight in acetone at −20°C, and pellets were resuspended in 20 μl of 0.5 M triethylammonium bicarbonate (TEAB; pH 8.5). Protein contents were determined using a 2-D Quant Kit (Amersham, Piscataway, NJ), and 100 μg of each pooled sample were then denatured with 0.1% SDS and reduced with 5 mM tris-(2-carboxyethyl) phosphine. After incubation for 1 h at 60°C, cysteines were blocked with 10 mM methyl methane thiosulfonate in isopropanol, and the samples were incubated at room temperature for 10 min. Sequencing grade trypsin (10 μl, Applied Biosystems) was added in a trypsin-to-protein ratio of 1:20, and the samples were incubated at 37°C overnight. Digested samples were labeled with the iTRAQ reagents following the protocol provided by the vendor (Applied Biosystems).
After digestion and iTRAQ labeling, the ultracomplex protein digests were combined to create a 400-μg pooled protein digest sample that contained equal fractions of each of the four labeled samples for subsequent Multidimensional Protein Identification Technology (MudPIT) analysis (36). After lyophilization, the digest mixture was reconstituted in strong cation exchange (SCX) loading buffer (5 mM ammonium formate in 20% acetonitrile; pH 3.0) to be fractionated with SCX SpinTips (Protea Biosciences, Morgantown, WV) per the manufacturer's protocol. Briefly, the sample solution was loaded centrifugally onto the SCX SpinTip. The nonadsorbing fraction that passed through the SCX SpinTip was collected. Eight different elution solutions were used to fractionate the peptides (20, 60, 100, 150, 200, 250, 400, and 500 mM ammonium formate in 10% acetonitrile) in a step-wise manner for a total of nine sample fractions. The collected fractions were lyophilized and reconstituted in a 0.1 M acetic acid solution and then lyophilized to dryness. The fractions were then submitted for LC-MALDI TOF/TOF mass spectral analysis for protein identification, characterization, and differential expression analysis.
Mass Spectrometry Analyses With iTRAQ Labeling
The LC-MALDI mass spectrometry system utilized was an ABI Tempo LC MALDI spotter with Tempo LC MALDI version 2.00.09 data acquisition and processing software. Lyophilized SCX sample fractions were reconstituted in LC aqueous run buffer (0.1% trifluoroacetic acid, 2% acetonitrile) and were injected onto a Zorbax C18 chromatographic column (150 × 0.3 mm; Agilent Technologies, Wilmington, DE). The peptides were eluted from the column using an acetonitrile-trifluoroacetic acid gradient (2–72% acetonitrile in 35 min) and spotted directly onto a MALDI plate in 6-s spot fractions. The MALDI spots were analyzed using an ABI 4800 MALDI TOF/TOF analyzer operated with 4000 Series Explorer software. The mass spectrometry (MS) acquisition was in positive ion reflector mode with 400 laser shots per spectrum performed. The 15 strongest precursors per spot were chosen for MS/MS and the MALDI spot was interrogated until at least 4 peaks in the MS/MS spectra achieved a signal/noise ≥ 70.
The resulting MS/MS spectra were analyzed using ABI Protein ProteinPilot software 2.0 (Applied Biosystems). The spectral data were searched against the mouse protein database (NCBI nr. fasta database customized to select for all mouse proteins) for identification of the peptides and corresponding proteins. In ProteinPilot, the sample type was selected as iTRAQ 4Plex for retrieval of the isotopic tag information from the mass spectra. After database correlation analysis, the proteins were grouped, scored, and normalized against one to four isotope correction factors. The Pro Group algorithm of ProteinPilot generated a ProtScore that is a cumulative score from each of the peptides used by the algorithm in the protein identification. Protein scores (ProtScore) above 2.0, 1.0, and 0.47 expressed the percent confidence levels of >99, >90, and >66%, respectively. Each peptide match showed the iTRAQ isotopic labels, carbamidomethylated cysteines, and other posttranslational modifications present as mass spectral shifts identified during the database correlation analysis. Each protein identified also showed the differential protein expression compared against the other iTRAQ-labeled samples for relative quantitation.
Canonical Pathways and Molecular Network Analysis
After protein identification/quantification through iTRAQ proteomic analyses, the accession numbers and fold changes of the differentially expressed proteins were tabulated and imported into Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) for canonical pathway analyses. Results were visualized using bar graphs, and the software was utilized for construction of interacting protein networks identified within diabetic IFM and mPHGPx diabetic IFM groups. IPA contains a database that uses the most current knowledge available on genes, proteins, disease processes, signaling, and metabolic pathways needed for protein network construction.
Protein Import
Plasmid construction.
The fusion protein pAcGFP1-Mito (Clontech Laboratories, Mountain View, CA) containing the precursor subunit VIII of human cytochrome c oxidase and the green fluorescent protein (GFP) from Aequorea coerulescens (AcGFP1) were cloned into pIVEX2.3d (Roche Applied Science, Indianapolis, IN) as previously described (2). pMitoGFP1 was grown to a concentration of 500 ng/μl and isolated using miniprep plasmid DNA isolations (Qiagen, Valencia, CA).
In vitro synthesis of mitochondrial protein.
In vitro transcription/translation of mitoGFP1 was performed using the S30 T7 protein expression system (Promega, Madison, WI) per manufacturer's protocol as previously described (2). MitoGFP1 lysate was subsequently used as substrates for the in vitro protein import process.
Mitochondrial protein import.
The mitochondrial protein import procedure was performed following the protocol from Stojanovski et al. (49) with modifications as previously described (2). Briefly, 40 μg of mitochondria were resuspended in 100 μl of import buffer with addition of ATP and Na-succinate. Lysate containing MitoGFP1 protein was added, and protein import was performed at increasing time intervals of 1 and 2 min at 25°C. Valinomycin was added to stop the import reaction, and samples were centrifuged at 4°C. The supernatant was discarded, and the pellet resuspended in SEM (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, at pH 7.2) buffer containing trypsin, on ice. Trypsin was subsequently inhibited by protease inhibitor cocktail (Biovision, Mountain View, CA), and samples were centrifuged again at 4°C. Pellets were then resuspended in lysis buffer (Biovision), subjected to SDS-PAGE, and subsequent Western blot analyses. Quantification of blots was performed using Pierce ECL Western Blotting Substrate (Pierce Biotech, Rockford, IL). The primary antibody was an anti-GFP monoclonal antibody raised in a mouse host (Clontech Laboratories). The secondary antibody was an anti-mouse IgG horseradish peroxidase conjugate (Sigma). Quantification of chemiluminescent signals were detected using a G:BOX Bioimaging System (Syngene, Frederick, MD), and data were expressed as arbitrary optical density units.
Western Blot Analyses
SDS polyacrylamide gel electrophoresis (SDS-PAGE) was run on 4–12% gradient gels as previously described (34, 54) with equal amounts of protein loaded for each study treatment. Relative amounts of subpopulation-specific mitochondrial heat shock protein 70 (mtHsp70), translocase of the outer membrane 20 (Tom20), and translocases of the inner membrane 23 (Tim23) and 50 (Tim50) were analyzed using the following primary antibodies: anti-mtHsp70 mouse antibody (product no. SPA-825; Stressgen, Ann Arbor, MI), anti-Tom20 mouse antibody (product no. 612278; BD Biosciences, San Jose, CA), anti-Tim23 mouse antibody (product no. 611222; BD Biosciences), and anti-Tim50 goat antibody (product no. ab23938; Abcam, Cambridge, MA). The secondary antibodies used in the analyses were donkey anti-goat IgG HRP conjugate (product no. sc-2020; Santa Cruz Biotech, Santa Cruz, CA) for Tim50; and goat-anti mouse conjugate (product no. 31430; Pierce Biotech, Rockford, IL) for Tom20, Tim23, and mtHsp70. Anti-Cox IV rabbit antibody (product no. ab16056; Abcam) was used as a loading control with secondary antibody anti-rabbit IgG HRP conjugate (product no. 31463; Pierce Biotech). Detection of signal was performed according to the manufacturer's instructions using Pierce ECL Western Blotting Substrate (Pierce Biotech). Autoradiographic signals were assessed using a G:Box Bioimaging System (Syngene), and the data were captured and analyzed using GeneSnap/GeneTools software (Syngene).
Blue Native Gel
To assess mtHsp70 abundance within the presequence translocase-associated motor (PAM) complex, Native polyacrylamide gel electrophoresis (NativePAGE) was performed in control, diabetic, mPHGPx, and diabetic mPHGPx IFM according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). Briefly, 20 μg of isolated mitochondria were solubilized with 1% digitonin for 15 min on ice. After addition of 1.25 μl of Coomassie G-250, samples were run on 4–16% NativePAGE gels at 120 V for 90 min at room temperature. Gels were transferred to PDVF membranes and subsequently probed with mtHSP70 antibody as above.
Statistics
Means ± SE were calculated for all data sets. Data were analyzed using a two-way ANOVA method to evaluate the main treatment effect, diabetes induction, and mPHGPx transgene presence (GraphPad Software, La Jolla, CA). Fisher's Least Significant Difference (LSD) post hoc tests were performed to determine the significant differences among means. When appropriate a Student's t-test was employed. P < 0.05 was considered significant.
RESULTS
MPHGPx Transgenic Mice
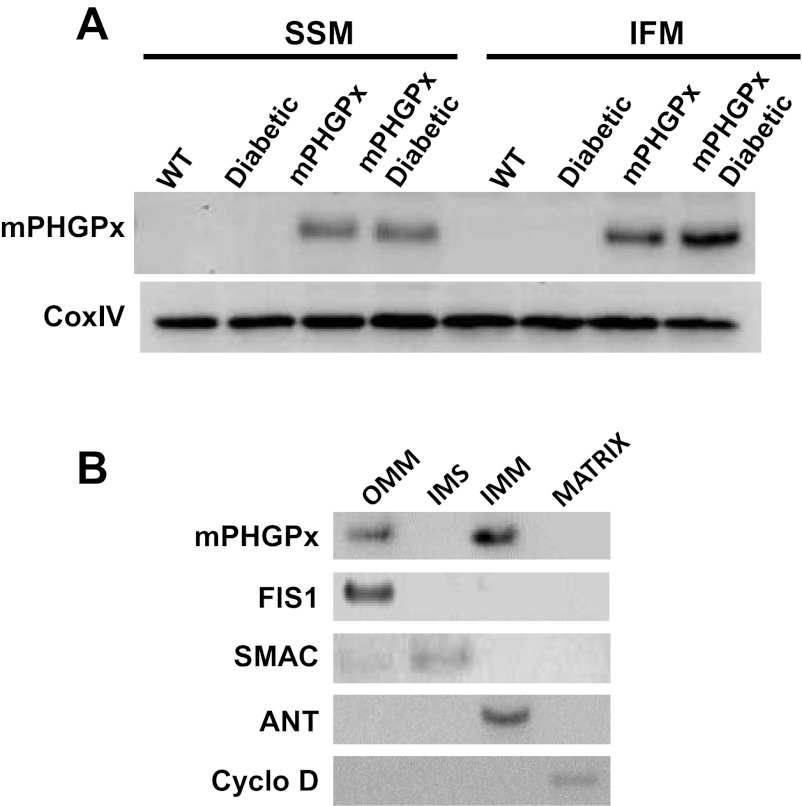
Mitochondrial gpx4 protein levels were significantly higher in both subpopulations of transgenic-positive mice relative to wild-type controls and despite the diabetic state (Fig. 1A). Evaluation of protein localization revealed gpx4 presence in the OMM and IMM mitochondrial subfractions (Fig. 1B) consistent with reports suggesting that mPHGPx exists primarily at contact sites between the OMM and IMM (21, 23). Previous data from our laboratory has indicated increased phospholipid hydroperoxide and hydrogen peroxide scavenging in mPHGPx transgenic mice (15).
Fig. 1.
Mitochondria phospholipid hydroperoxide glutathione peroxidase 4 (mPHGPx) transgenic mouse screening. A: representative Western blot analysis of mPHGPx protein expression in individual mitochondrial subpopulations of mPHGPx transgenic (mPHGPx) and wild-type control (WT) mouse hearts. Pon, Ponceau staining loading control. B: representative Western blot analysis of mPHGPx protein expression in mitochondrial subcompartments. Mitochondrial subcompartments were fractionated and probed for mPHGPx presence. Individual fractions were probed with subcompartment-specific proteins to determine purity of the subfractionation process, outer mitochondrial membrane (OMM) (mitochondrial fission 1 protein, FIS1), inner membrane space (IMS) (second mitochondria-derived activator of caspases, SMAC), inner mitochondrial membrane (IMM) (adenine nucleotide translocase, ANT), and matrix (cyclophilin D).
Cardiac Contractile Function
Cardiac contractile function was significantly altered in diabetic hearts compared with control hearts. Ejection fraction, fractional shortening, and cardiac output were significantly decreased in the diabetic heart compared with control (Table 1, P < 0.05). With overexpression of mPHGPx in the diabetic heart, ejection fraction, fractional shortening, and cardiac output were restored back to control values (Table 1, P < 0.05). In addition, stroke volume was significantly greater in mPHGPx diabetic animals compared with control diabetic animals (Table 1, P < 0.05).
Table 1.
Cardiac contractile function
| Contractile Parameter | Control | Control Diabetic | mPHGPx | mPHGPx Diabetic |
|---|---|---|---|---|
| BW, g | 29.7 ± 2.1 | 26.2 ± 1.0 | 28.7 ± 2.0 | 25.1 ± 1.0 |
| HW, mg | 102.8 ± 2.7 | 94.4 ± 4.5 | 101.9 ± 5.4 | 93.4 ± 3.6 |
| HW/BW, mg/g | 3.5 ± 0.1 | 3.6 ± 0.1 | 3.6 ± 0.1 | 3.7 ± 0.2 |
| Diameter; s, mm | 2.0 ± 0.1 | 2.0 ± 0.2 | 1.9 ± 0.1 | 2.1 ± 0.2 |
| Diameter; d, mm | 3.2 ± 0.3 | 3.1 ± 0.2 | 3.3 ± 0.2 | 3.5 ± 0.2 |
| Volume; s, μl | 11.2 ± 1.3 | 13.1 ± 1.2 | 11.5 ± 0.9 | 13.9 ± 1.2 |
| Volume; d, μl | 42.4 ± 2.6 | 37.5 ± 1.6 | 45.3 ± 1.5 | 50.2 ± 1.7 |
| Stroke volume, μl | 27.9 ± 1.8 | 24.4 ± 1.2 | 33.8 ± 1.2 | 36.3 ± 1.4† |
| Ejection fraction, % | 74.1 ± 0.5 | 65.5 ± 0.6* | 74.8 ± 0.2 | 72.5 ± 0.5 |
| Fractional shortening, % | 41.9 ± 0.6 | 34.9 ± 0.6* | 42.5 ± 0.2 | 40.8 ± 0.6 |
| Cardiac output, ml/min | 15.1 ± 1.5 | 9.7 ± 0.8* | 16.9 ± 0.8 | 14.7 ± 1.2 |
Cardiac functional measurements were assessed using echocardiography. Values are expressed as means ± SE.
BW, body weight; HW, heart weight; mPHGPx, mitochondria phospholipid hydroperoxide glutathione peroxidase 4; s, systole; d, diastole.
P < 0.05 for diabetic vs. all groups.
P < 0.05 for mPHGPx diabetic vs. control diabetic.
Mitochondrial Subpopulation Functional Assessment
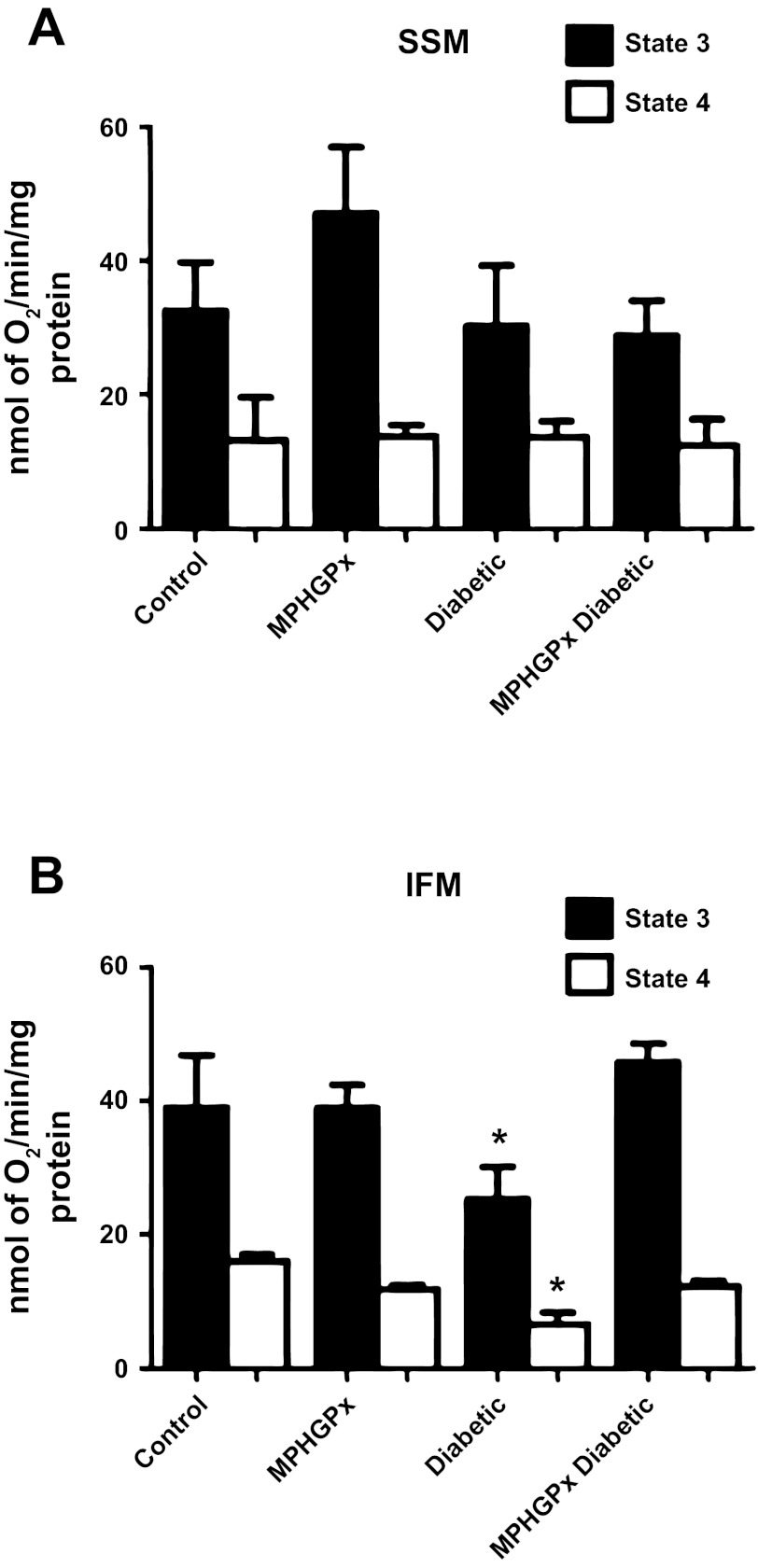
Assessment of state 3 and state 4 respiration rates with complex I substrates glutamate/malate revealed a significant decrease in diabetic IFM compared with control IFM (Fig. 2B; P < 0.05), with no significant changes between control and diabetic SSM (Fig. 2A). Overexpression of mPHGPx in the diabetic heart restored state 3 and state 4 respiration rates in the IFM (Fig. 2B) with no significant effects observed in the SSM (Fig. 2A). ETC complexes I, III, and IV activities were also significantly decreased in diabetic IFM compared with control IFM (Table 2; P < 0.05), with no significant differences between control SSM and diabetic SSM (Table 2). MPHGPx overexpression in the diabetic heart preserved ETC complex I, III, and IV activities in the IFM (Table 2; P < 0.05). Alterations to ETC function can contribute to changes in oxidative phosphorylation. ATP synthase activity (complex V) was significantly decreased in diabetic IFM compared with control IFM (Table 2; P < 0.05), with no significant differences observed between diabetic SSM and control SSM (Table 2). Overexpression of mPHGPx enhanced ATP synthase activity in diabetic IFM (Table 2, P < 0.05), with no significant effects on any of the SSM groups (see also Fig. 4A).
Fig. 2.
Mitochondrial respiration. State 3 and state 4 respiration rates with glutamate-malate as substrates in subsarcolemmal membrane (SSM) (A) and interfibrillar mitochondria (IFM) (B). State 3 and state 4 respiration rates were determined in the presence of the substrates glutamate-malate, and state 3 respiration was examined on addition of ADP. Values are ± SE. *P < 0.05 for diabetic vs. all groups.
Table 2.
Electron transport chain complex activities
| Group | Complex I | Complex III | Complex IV | Complex V |
|---|---|---|---|---|
| SSM Control | 66 ± 5 | 335 ± 51 | 1,215 ± 39 | 76 ± 17 |
| SSM mPHGPx | 58 ± 7 | 351 ± 48 | 1,053 ± 129 | 68 ± 22 |
| SSM Diabetic | 68 ± 6 | 302 ± 29 | 1,126 ± 94 | 89 ± 10 |
| SSM mPHGPx Diabetic | 56 ± 5 | 290 ± 28 | 1,158 ± 55 | 95 ± 8 |
| IFM Control | 90 ± 7 | 392 ± 29 | 2,179 ± 264 | 267 ± 43 |
| IFM mPHGPx | 96 ± 16 | 434 ± 59 | 2,560 ± 334 | 199 ± 8 |
| IFM Diabetic | 70 ± 12* | 301 ± 22* | 1,386 ± 45* | 115 ± 25* |
| IFM mPHGPx Diabetic | 102 ± 10 | 496 ± 30 | 1,937 ± 283 | 230 ± 13 |
Enzymatic activities are expressed as activity·min−1·mg−1 of protein ± SE. Electron transport chain (ETC) function was examined in mitochondrial subpopulations from control, mPHGPx, diabetic, mPHGPx diabetic hearts. ETC complex I, III, IV, and V activities were assessed spectrophotometrically by measuring the oxidation of NADH (complex I), reduction of cytochrome c (complex III), oxidation of cytochrome c (complex IV), and an assay coupled with pyruvate kinase, which converts ADP to ATP and produces pyruvate from phosphoenolpyruvate (complex V).
SSM, subsarcolemmal membrane: IFM, interfibrillar mitochondria.
P < 0.05 for diabetic vs. all groups.
Fig. 4.
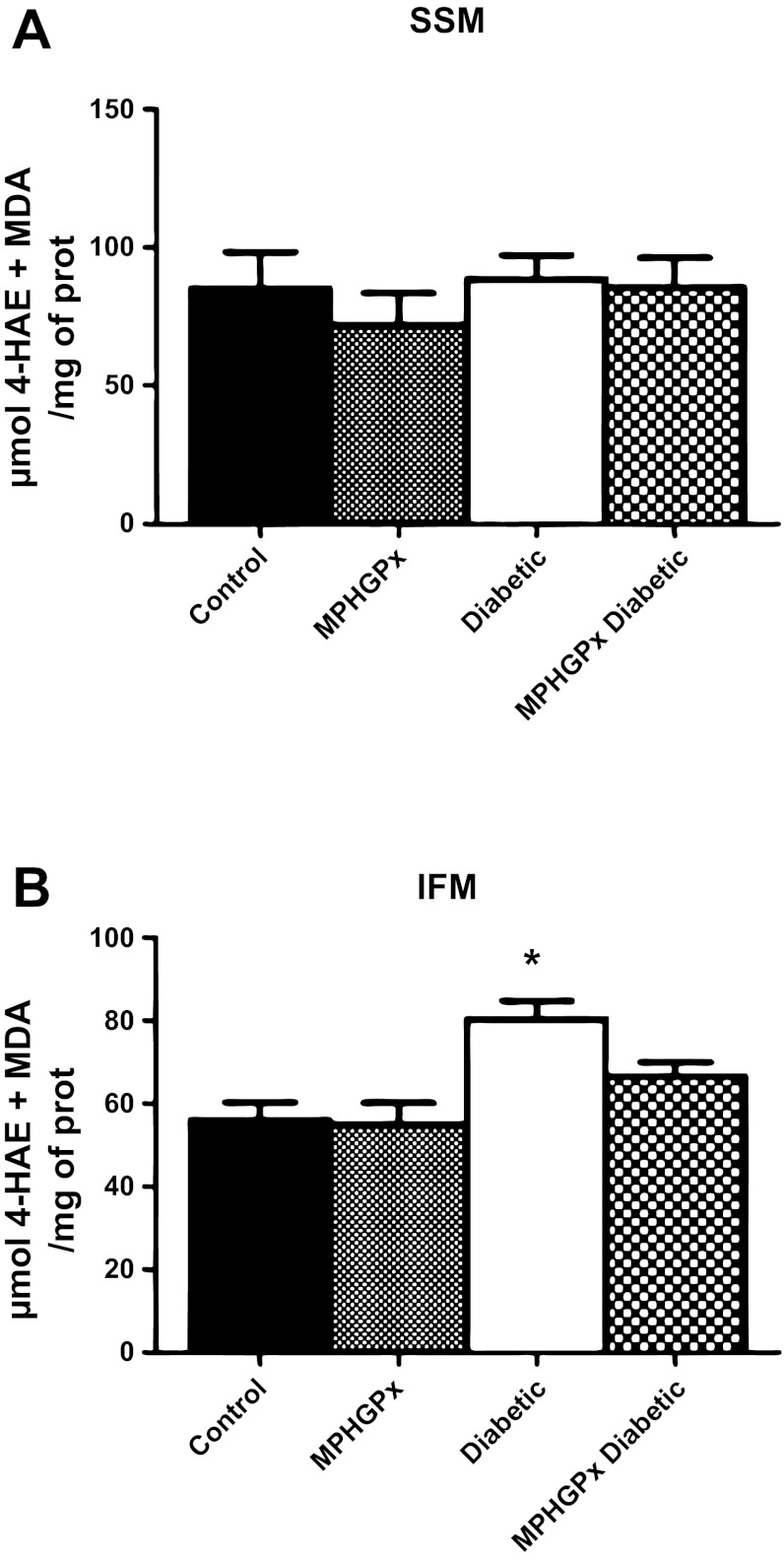
Lipid peroxidation by-products. Oxidative damage to lipids was assessed in SSM (A) and IFM (B) by measuring lipid peroxidation by-products malondialdehyde (MDA) and 4-hydroxyalkenals (4-HAE) using a colorimetric assay and compared against a standard curve of known 4-HAE and MDA concentrations. Values for lipid peroxidation by-products are expressed as means ± SE. *P < 0.05 for diabetic vs. all groups.
Reactive Oxygen Species and Oxidative Stress
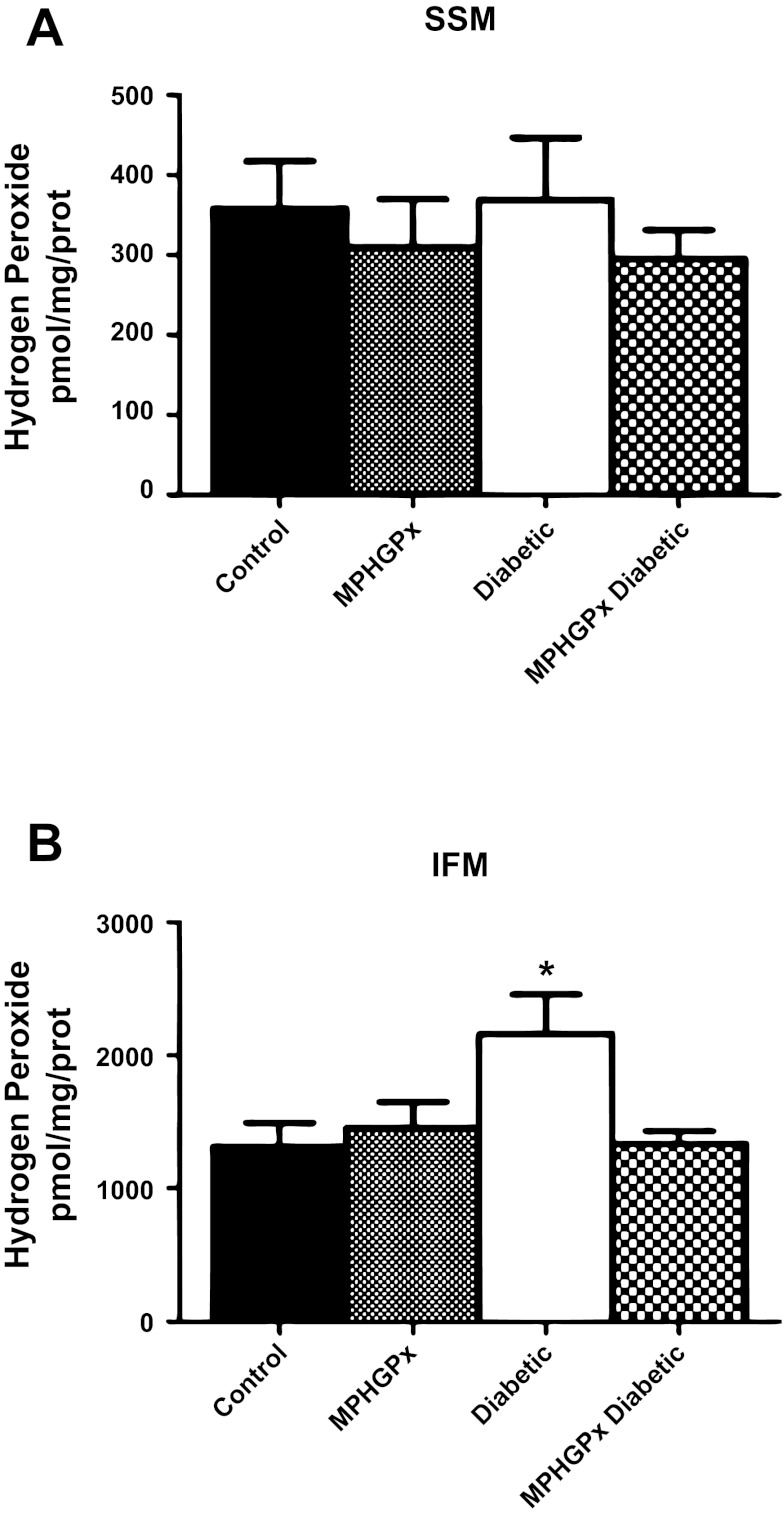
H2O2 can be directly scavenged by mPHGPx. Glutamate/malate-driven H2O2 production was significantly increased in the diabetic IFM compared with control IFM (Fig. 3B, P < 0.05), with no significant changes in diabetic SSM compared with control SSM (Fig. 3A). Overexpression of mPHGPx in the diabetic heart attenuated H2O2 production in the IFM (Fig. 3B, P < 0.05), with no significant changes in the SSM. In addition, lipid peroxidation by-products 4-HAE and MDA were significantly increased in diabetic IFM compared with control IFM (Fig. 4B, P < 0.05), with no significant changes in diabetic SSM compared with control SSM (Fig. 4A, P < 0.05). MPHGPx overexpression decreased the accumulation of lipid peroxidation by-products in diabetic IFM back to values similar to that of control IFM (Fig. 4B, P < 0.05), suggesting the ability of mPHGPx to attenuate ROS-induced damage to lipids within the IFM.
Fig. 3.
H2O2 production. H2O2 production in SSM (A) and IFM (B) was determined using the oxidation of the fluorogenic indicator amplex red in the presence of horseradish peroxidase. H2O2 production was initiated in mitochondria using glutamate-malate as substrates. Standard curves were obtained by adding known amounts of H2O2 to the assay medium in the presence of the substrates amplex red and horseradish peroxidase. Final values were calculated as pmol/mg of protein. Values are expressed as means ± SE. *P < 0.05 for diabetic vs. all groups.
Proteomic Analyses
Because the IFM displayed the greatest dysregulation following diabetic insult, proteomic analyses were focused on this specific subpopulation. To determine how mPHGPx overexpression impacts the proteomic makeup of IFM during a Type 1 diabetic insult, iTRAQ labeling followed by MALDI-TOF/TOF mass spectrometry was performed. Only those proteins that were statistically significant as a result of diabetes mellitus in control or mPHGPx overexpressed animals were reported.
Oxidative Phosphorylation Proteins
As previously reported, multiple mitochondrial respiratory chain proteins were significantly decreased in the IFM by diabetes mellitus (2). The greatest influence was on the NADH dehydrogenase complex (complex I) with five proteins significantly decreased. To a lesser extent, the other respiratory chain complexes were also decreased including two proteins from complex II, three proteins from complex III, 1 protein from complex IV, and two proteins from the ATP synthase (online Supplemental Table S1). Overexpression of mPHGPx in diabetic IFM lead to the preservation of nine OXPHOS proteins significantly decreased in the diabetic IFM including those from complex I, II, III, and V (online Supplemental Table S1). Furthermore, mPHGPx diabetic IFM displayed enhanced protein content of nine OXPHOS proteins that did not display decreased abundance in the diabetic IFM (online supplemental Table S1). These results indicate that mPHGPx has the ability to preserve mitochondrial respiratory chain proteins during a Type 1 diabetic insult.
Lipid Metabolism Proteins
Three mitochondrial proteins involved in lipid metabolism (acyl carrier protein, hydroxyacyl-coenzyme A dehydrogenase, and short-chain specific acyl-CoA dehydrogenase) were significantly decreased when comparing control and diabetic IFM (online supplemental Table S1). Interestingly, two of these proteins, as well as other lipid metabolism constituents including enoyl-CoA hydratase, were significantly increased in the mPHGPx diabetic IFM compared with control diabetic suggesting restoration of lipid metabolism constituents in the face of hyperglycemia (online supplemental Table S1).
Proteins of the TCA
Two proteins of the TCA cycle were significantly decreased in diabetic IFM compared with control including isocitrate dehydrogenase 3 (online supplemental Table S1). Conversely, mPHGPx diabetic IFM displayed increases in isocitrate dehydrogenase 3 (3.5-fold) compared with control diabetic. MPHGPx preserved or elevated four additional TCA cycle proteins that were not influenced by diabetes mellitus. These proteins included aconitate hydratase, citrate synthase, and malate dehydrogenase (online supplemental Table S1).
Transport Proteins
Multiple mitochondrial transport proteins were decreased in diabetic IFM compared with control, including mtHsp70, the key constituent of the PAM complex, essential for all mitochondrial matrix import and to a lesser extent, IMM transport. In contrast, mtHsp70 protein content was preserved in mPHGPx diabetic IFM compared with diabetic control (online supplemental Table S1). The IMM transporter essential for phosphate transport into the mitochondrial matrix, phosphate carrier protein, was decreased in the diabetic IFM and restored in the mPHGPx diabetic IFM. Interestingly, ADP/ATP translocase and Hsp60 protein contents were increased in mPHGPx diabetic IFM relative to diabetic control despite no changes in control versus diabetic (online supplemental Table S1). The outer mitochondrial membrane (OMM) channel, voltage-dependent anion selective channel 1, was also enhanced in mPHGPx diabetic IFM, highlighting the ability of mPHGPx to preserve proteins within the OMM.
Miscellaneous Proteins
MPHGPx diabetic IFM displayed increases in amino acid metabolism protein aspartate aminotransferase as well as ketone metabolism protein succinyl-CoA:3-ketoacid-coenzyme A transferase 1 compared with diabetic IFM (online supplemental Table S1). d-β-Hydroxybutyrate dehydrogenase was significantly decreased in the diabetic IFM compared with control and increased by 2.45-fold in the mPHGPx diabetic IFM (online supplemental Table S1). The antioxidant protein peroxiredoxin-5, which is capable of reducing H2O2 and alkyl hydroperoxides, was significantly decreased within the diabetic IFM compared with control. MPHGPx diabetic IFM showed a trend for increase in peroxiredoxin-5 when compared with diabetic IFM; however, the result was not statistically significant.
Proteomic Loss/Rescue
To begin to understand the impact of mPHGPx overexpression on diabetic IFM, we separated proteins significantly changing into three categories: 1) IFM proteins decreased as a result of diabetes mellitus; 2) IFM proteins upregulated by mPHGPx overexpression as a result of diabetes mellitus; and 3) proteins that were decreased by diabetes mellitus in the IFM, yet restored with mPHGPx overexpression (loss/rescue). Analyses revealed a total of 24 proteins statistically decreased within the diabetic IFM compared with control. Nine of the 24 proteins that were decreased within the diabetic IFM were not restored by mPHGPx overexpression (Fig. 5A). In contrast, the 15 remaining proteins (∼62.5% of the total) were significantly decreased in the diabetic IFM and preserved with mPHGPx overexpression. An additional 20 proteins were identified as being increased with mPHGPx overexpression despite not being influenced by diabetes mellitus insult (Fig. 5A). These results indicate that the majority of proteins negatively impacted by Type 1 diabetes mellitus were preserved with mPHGPx overexpression.
Fig. 5.
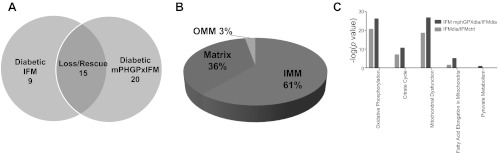
Proteomic alterations in diabetic and mPHPGx diabetic IFM. A: total number of proteins decreased solely in the diabetic IFM (left), proteins decreased in diabetic IFM and preserved within mPHGPx diabetic IFM (middle), and proteins increased solely within the mPHGPx diabetic IFM (right). B: breakdown in approximate percentages of protein contents lost in diabetic IFM and subsequently preserved in the mPHGPx diabetic IFM. C: mitochondrial canonical pathways identifyed using Ingenuity Pathway Analysis (IPA) software most negatively impacted by diabetic insult and subsequently restored by mPHGPx overexpression. Oxidative phosphorylation, tricarboxylic acid cycle (citric acid cycle), fatty acid metabolism, and fatty acid elongation were negatively affected (light gray shade) by diabetes mellitus. Conversely, these pathways as well as pyruvate metabolism were preserved (dark gray shade) in diabetic IFM with the overexpression of mPHGPx. Diab., diabetic; Ctrl, control.
Mitochondrial Subcompartment Proteomes
Because mitochondria are double-membrane organelles, they contain various subcompartments including the OMM, IMS, IMM, and matrix. To determine the submitochondrial location most impacted by mPHGPx overexpression in diabetic IFM, we analyzed the percentage of proteins preserved within the various compartments. As shown in Fig. 5B, proteomic protection via mPHGPx overexpression predominantly occurred within the mitochondrial IMM (61%), followed by the mitochondrial matrix (36%), and then the OMM (3%).
Canonical Pathways
Utilizing IPA software-fed iTRAQ proteomics data (all proteins indicated as being significantly altered), we attempted to predict canonical pathways within the mitochondrion that were most negatively impacted by diabetic insult and subsequently restored by mPHGPx overexpression. As expected, OXPHOS, TCA, FAO, and fatty acid elongation pathways were the most negatively impacted by diabetes mellitus within IFM (green bars) (Fig. 5C). Interestingly, mPHGPx overexpression within diabetic IFM not only corrected, but enhanced, many of the proteins involved in these four mitochondrial processes (red bars) (Fig. 5C). Additionally, processes shown to be influenced by mPHGPx overexpression included proteins involved in pyruvate metabolism. The results suggest that mPHGPx overexpression is able to preserve and even potentially enhance essential mitochondrial processes negatively influenced by diabetes mellitus in IFM. A list of all of the proteins involved in the differentially regulated networks is included in the online supplemental Table S2.
Common Molecular Networks
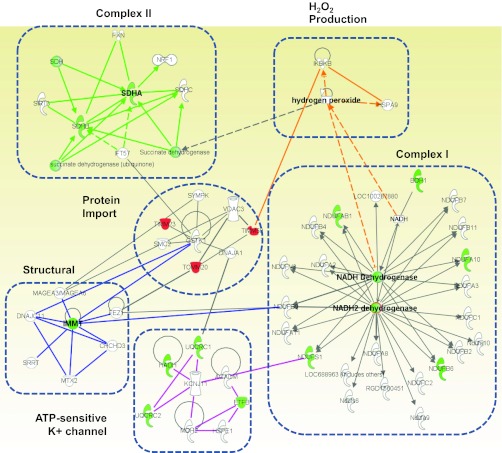
IPA software was able to identify essential mitochondrial molecular networks that were damaged within diabetic IFM and preserved with the overexpression of mPHGPx in the diabetic IFM. Of the six networks identified, five were of IMM origin including ETC complexes I and II, ATP-sensitive K+ channel, the structural protein mitofilin, and the mitochondrial protein import network (Fig. 6). Finally, a H2O2 network was also identified and shown to be preserved in the mPHGPx diabetic IFM (Fig. 6), which is of interest in that mPHGPx is capable of directly scavenging H2O2.
Fig. 6.
Mitochondrial protein networks. Identification using IPA software, of mitochondrial proteomic networks negatively influenced in diabetic IFM and preserved within mPHGPx diabetic IFM. Six networks, including electron transport chain (ETC) complexes I and II, H2O2 production, structure, ATP-sensitive K+ channel, and mitochondrial protein import were identified. Mitochondrial protein import network is the central node linking all other networks. Green indicates proteins preserved within mPHGPx diabetic IFM. Red indicates proteins positively enhanced in mPHGPx diabetic IFM. White indicates proteins unchanged by diabetes mellitus but are part of their respective networks.
Posttranslational Modifications
Utilizing MudPIT technology to examine PTMs, we identified 68 peptide sequences of 44 different proteins that were modified through oxidations, acetylations, or deamidations within the diabetic IFM, which were reversed in the diabetic mPHGPx transgenic (online supplemental Table S3). All proteins analyzed were within the IMM or matrix. Nine proteins were no longer oxidized in the mPHGPx diabetic IFM including 3 from the ETC, 2 from the TCA cycle, and 2 FAO proteins (online supplemental Table S3). Interestingly, 16 IMM proteins and 21 matrix proteins originally identified no longer displayed deamidations within the mPHGPx diabetic IFM group, including an essential mitochondrial import protein mtHsp70 (online supplemental Table S3).
Mitochondrial Protein Import
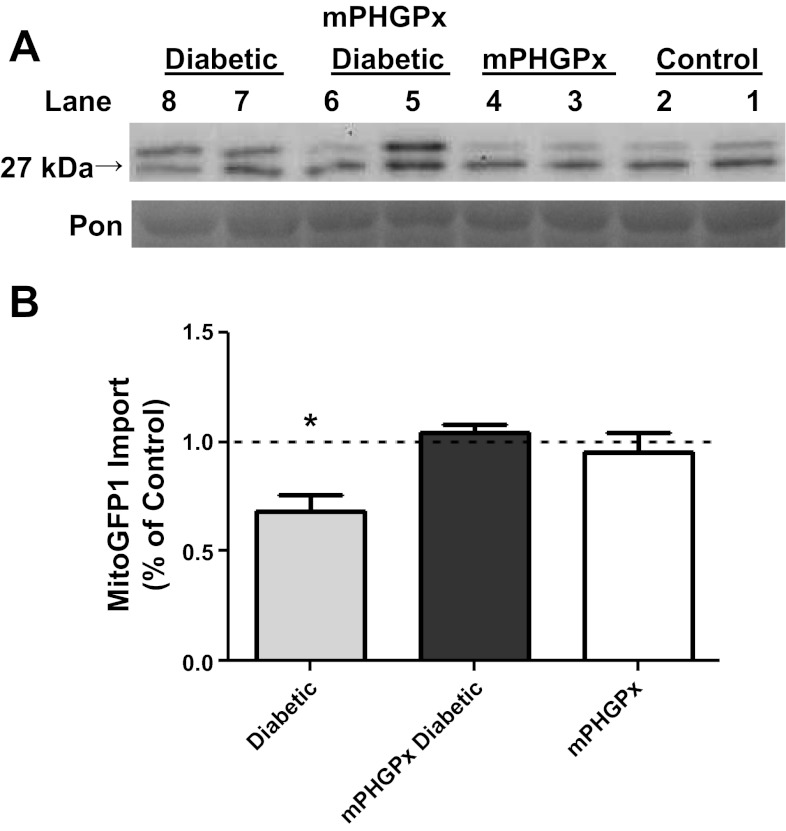
In vitro mitochondrial protein import was evaluated by quantifying the amount of mature MitoGFP1 protein imported into isolated IFM as previously described (2). Western blot analyses highlighted increasing amount of MitoGFP1 at 1 and 2 min, respectively, of the mature 27-kDA band in all IFM groups (Fig. 7A). MitoGFP1 protein import was significantly decreased in the diabetic IFM compared with control, which is in agreement with our previous findings (Fig. 7B) (2). Furthermore, MitoGFP1 import was restored back to control levels in the mPHGPx diabetic indicating preservation of mitochondrial protein import with mPHGPx overexpression.
Fig. 7.
Mitochondrial protein import. Effect of Type 1 diabetes mellitus and mPHGPx overexpression on MitoGFP1 import in IFM. A: representative Western blots from IFM protein import assay. Control for protein loading was confirmed by Ponceau (Pon) staining. B: graphical representation of mitochondrial protein import performed in control diabetic, mPHGPx, and mPHGPx diabetic. The relative amount of imported MitoGFP1 was determined by densitometry. Dashed line denotes control levels. Import efficiency was based off of percent control at the 1- and 2-min time points. Both time point percentages were then averaged to show total import efficiency. Values are presented as means ± SE; *P < 0.05 for Diabetic vs. all groups. N = 5 per group.
Protein Import Machinery
To determine the impact of mPHGPx overexpression on key mitochondrial import constituents, we evaluated the protein abundance of OMM translocase Tom20, IMM translocases Tim23 and Tim50, and essential PAM complex constituent mtHsp70 (Fig. 8). Tom20 protein content was significantly increased in the mPHGPx diabetic IFM compared with diabetic IFM (Fig. 8A). mtHsp70 and Tim23 protein contents were significantly decreased in the diabetic IFM groups compared with the control groups yet were increased in the mPHGPx diabetic groups compared with the diabetic groups (Fig. 8, C and D), which is in agreement with our iTRAQ data examining mtHsp70 (online supplemental Table S1). These results indicate that mPHGPx has the ability to preserve and enhance nuclear-encoded mitochondrial protein import constituents that may be of benefit for nuclear-encoded mitochondrial protein import.
Fig. 8.
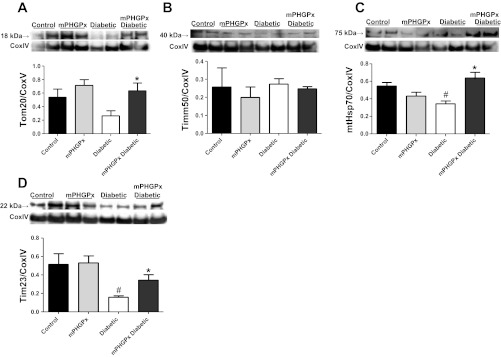
Mitochondrial protein import constituents. Western blot analysis of protein import constituents in IFM. Key proteins involved in mitochondrial protein import were assessed using Western blot analysis. Representative Western blot and densitometry analysis for Tom20 (A), Tim50 (B), mtHsp70 (C), and Tim23 (D) from control, mPHGPx, diabetic, and mPHGPx diabetic. Control for protein loading was confirmed by CoxIV staining. Values are presented as means ± SE; *P < 0.05 for mPHGPx Diabetic vs. Diabetic; #P < 0.05 for Control vs. Diabetic. N = 4 per group.
mtHsp70-Tim44 Complex
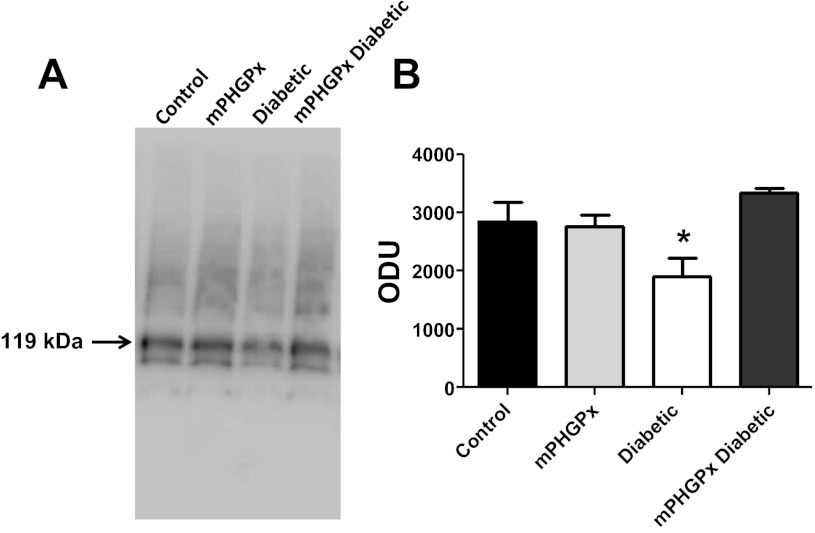
During moments of import into the mitochondrial matrix, mtHsp70 will complex with the inner membrane anchor protein Tim44 to form the functional element of the PAM complex, which provides a critical driving force for nuclear-encoded mitochondrial protein import. To examine mtHsp70 ability to complex with Tim44, we performed BN-PAGE analyses (Fig. 9). Similar to the Western blot results, diabetic IFM displayed a significant decrease in mtHsp70-Tim44 protein complexing compared with all other groups (Fig. 9). Interestingly, mPHGPx diabetic IFM mtHsp70-Tim44 complexing was restored to control levels (Fig. 9). These results suggest that diabetes mellitus may compromise formation of the mitochondrial protein import motor in IFM. Furthermore, overexpression of mPHGPx enables enhanced mtHsp70-Tim44 complexing, such that formation of this critical import component is restored to back to control levels.
Fig. 9.
mtHsp70 protein complexing. Effect of Type 1 diabetes mellitus and mPHGPx overexpression on mtHsp70-Tim44 complexing in isolated IFM from control, diabetic, mPHGPx, and mPHGPx diabetic. IFM were subjected to BN-PAGE, and protein complexes were probed for mtHSP70. A: complex of mtHsp70 and Tim44 exists at 119 kDa. B: mtHsp70 content was analyzed through densitometry. *P < 0.05 for Diabetic vs. all groups. N = 4 per group.
DISCUSSION
Because many functional processes exist within the IMM (oxidative phosphorylation, ATP synthesis, protein import), preservation of this critical locale is essential for a properly functioning mitochondrion, which is of particular relevance during pathological insults such as diabetes mellitus. The goal of the current study was to determine whether overexpression of an antioxidant enzyme that could provide protection to the IMM locale (mPHGPx) would lead to preservation of these essential mitochondrial processes, and ultimately, cardiac function during diabetic insult. The following key findings resulted from these studies: 1) overexpression of mPHGPx reversed many proteomic decrements observed in diabetic IFM; 2) overexpression of mPHGPx preserved nuclear-encoded mitochondrial protein import in diabetic IFM, which was associated with increases in import constituents, as well as preservation of essential PAM complex interactions; and 3) overexpression of mPHGPx restored contractile and mitochondrial functional deficits associated with the diabetic phenotype. Taken together, these findings suggest a positive benefit of targeting a mitochondrial antioxidant enzyme to the IMM locale.
Previous results from our laboratory indicate that mitochondrial functional and proteomic alterations resulting from Type 1 diabetic insult are confined primarily to the IFM, with SSM being minimally impacted. As a result, we focused our proteomic studies on the IFM subpopulation. It is important to point out that mPHGPx overexpression was provided to both SSM and IFM. Because mitochondrial dysfunction was not observed in the SSM subpopulation of mPHGPx transgenic mice, it would suggest that overexpression of mPHGPx in the SSM was not detrimental from a functional context. Proteomic analyses of diabetic IFM confirmed previous findings highlighting decreases in essential mitochondrial proteins governing processes such as OXPHOS, TCA cycle, and transport (2). A rather compelling finding from the proteomic analysis was the observation that mPHGPx was able to preserve a significant portion of the proteins decreased in the diabetic IFM (62.5%) from the mitochondrial processes highlighted above, which was associated with preservation of the functional processes. The majority of these proteins preserved or elevated within the diabetic mPHGPx IFM resided within the IMM. Because mPHGPx exists predominantly at contact sites between the OMM and IMM, one could conclude that its primary locale enables proximal scavenging of IMM and/or OMM lipid hydroperoxides (37, 38). However, it is important to point out that nearly one-third of the proteins protected or elevated within the diabetic mPHGPx IFM resided within the matrix. Because mPHGPx does not reside primarily in this submitochondrial region, the data were somewhat unexpected. MPHGPx contains a mitochondrial bipartite targeting signal, characteristic of IMS proteins (43). Nevertheless, studies have indicated dynamic expression patterns suggestive of movement from one submitochondrial locale to another (23). ETC complexes I and III, sites of electron leakage, were highly preserved within the mPHGPx diabetic IFM potentially decreasing total ROS production and subsequent damage to matrix proteins (11). PTM data support this notion by highlighting decreases in oxidations and deamidations in IMM and matrix proteins from mPHGPx diabetic IFM. Furthermore, mPHGPx is capable of scavenging H2O2, potentially decreasing the oxidative environment seen within the mitochondrion and highlighting another possible mechanism for proteomic protection seen within the matrix of mPHGPX diabetic IFM (52). However, future studies are required for elucidating specific mechanisms accounting for mPHGPx protection to matrix proteins. It should also be pointed out that changes in mitochondrial content, which were not assessed in the current study, may have contributed to alterations in the IFM proteomes observed with mPHGPx overexpression, though enhanced biogenesis would need to be confined to the IFM subpopulation.
Analysis of the processes influenced by enhanced mPHGPx in diabetic IFM identified several molecular networks located within the IMM, including OXPHOS (complexes I and II), the ATP-sensitive K+ channel, mitochondrial structure (mitofilin), and nuclear-encoded mitochondrial protein import. Our results are in agreement with previous work, confirming ETC complexes I and II as potential loci of diabetes mellitus dysfunction (14, 32, 33). Of particular interest was the observation that mPHGPx overexpression corrected a relatively unexplored structural network via preservation of the mitochondrial scaffolding protein mitofilin. IPA analyses identified a central linkage connecting other common networks, namely, the mitochondrial protein import network. Currently, there are an estimated 1,500 proteins in the human mitochondrion, with only 13 transcribed and translated in the organelle itself (1, 7, 40). The vast majority of proteins (>99%) are nuclear encoded and imported into the mitochondrion through a complex mechanism of translocation (8). Mitochondrial protein import is absolutely essential for numerous processes because many proteins involved in mitochondrial morphology/structure and function rely on the import of constituent proteins central for these functions. Thus it is not surprising that preservation of mitochondrial protein import provided a central linkage positively impacting common structural and functional networks negatively impacted during Type 1 diabetes mellitus.
Data from the current study indicate that a key mitochondrial import protein and component of the PAM complex, mtHsp70, was decreased in the diabetic IFM and preserved with mPHGPx overexpression. Furthermore, mtHsp70 content within the PAM complex was decreased in the diabetic IFM and restored with mPHGPx overexpression indicating preservation in a functional context. Functionally, mtHsp70 assists in the import of preproteins into various mitochondrial subcompartments (4, 46, 53). Our hypothesis for the current study was centered on preservation of IMM integrity, which would have impact on the formation of import machinery of which mtHsp70 is a component and the protein import process as a whole. Indeed, in vitro import of mitochondrial matrix-targeted mitoGFP1 protein was significantly decreased in the diabetic IFM compared with control IFM, yet enhanced with mPHGPx overexpression, indicating restoration of matrix import. Key import constituents Tom20 and Tim23 were significantly increased within diabetic mPHGPx IFM compared with diabetic IFM, which may have resulted from preservation of those proteins in the membranes (decreased protein loss or PTM) and/or enhanced protein import itself. A number of reports have identified mitochondrial import decrements when one or more of these essential translocases are disturbed. Mutations to or inactivation of mtHsp70 was shown to inhibit unfolding, translocation, and subsequent folding of mitochondrial preproteins imported into yeast mitochondria (3, 17, 31). Similar findings have been reported by others examining mitochondrial protein import in a model of skeletal muscle disuse through unilateral peroneal nerve denervation (48). Interestingly, in vitro import of TCA cycle protein malate dehydrogenase 2 was decreased coinciding with significant decreases to mtHsp70, Tim23, and Tom20 protein contents (48). Research from our laboratory using rat neonatal cardiac myocytes infected with an adenoviral vector overexpressing mtHsp70 indicated enhanced protein content of mitochondrial proteins following simulated hypoxia/reoxygenation insult compared with control (55). Recently, Gevorkyan-Airepetov et al. (20) mutated the binding region of Tim23 and Tim50, destabilizing the interaction between the translocases. In vitro import of both IMM and matrix-bound proteins were inhibited in the mutant yeast indicating the importance of Tim50 for the transport of numerous proteins within these mitochondrial locales. Overall, mPHGPx overexpression was able to preserve key translocases of the mitochondrial import process, which lead to restoration of mitochondrial protein import within the diabetic IFM subpopulation.
Perspectives and Significance
Diabetes mellitus incidence continues to rise globally while cardiovascular complications remain the leading cause of mortality and morbidity among diabetic patients. Research from our laboratory and others supports the contention that the mitochondrion is centrally involved in the pathological progression of the diabetic heart. As a result, therapeutic interventions targeting the mitochondrion offer the potential for protecting this subcellular locus with the promise of lessening cardiac contractile dysfunction associated with the pathology. Analyses of cardiac mitochondria indicate that the IMM may be particularly susceptible to functional and proteomic alterations associated with the diabetic heart, which may be the result of effects on membrane-associated processes. The data presented in the current paper indicates for the first time that mitochondrially directed enhancement of a phospholipid-targeted antioxidant enzyme (mPHGPx) designed to lessen IMM-associated functional and proteomic loss leads to mitochondrial, and ultimately, cardiac protection following diabetic insult. This effect is correlated with the preservation of essential mitochondrial functional networks. These findings support a paradigm that mechanistically links ROS and oxidative stress to cardiac mitochondrial dysfunction in the diabetic heart. Furthermore, the findings provide the rationale for designing future targeted mitochondrial therapeutics, which take into consideration specific submitochondrial regions at risk, such as the IMM.
GRANTS
This work was supported by the National Institutes of Diabetes and Digestive and Kidney Diseases Grant DP2DK-083095 (to J. M. Hollander). This work was also supported by Grant-In-Aid 0855484D from the American Heart Association (to J. M. Hollander). W. Baseler is a recipient of a National Institutes of Health Predoctoral Fellowship T32HL-090610 and American Heart Association Predoctoral Fellowship 10PRE3420006. E. Dabkowski is a recipient of an American Heart Association Predoctoral Fellowship 0815406D. T. Croston is a recipient of NIH Predoctoral Fellowship T32HL-090610. D. Shepherd is a recipient of NIH Predoctoral Fellowship T32HL-090610. C. Nichols is a recipient of a National Science Foundation IGERT-REN Fellowship DGE-1144676. Animal imaging experiments were supported in part by NIH Grant S10 RR-026378. Ingenuity Pathway Analyses were supported by a WV-INBRE Grant 8 P20 GM103434-12.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: W.A.B. and J.M.H. conception and design of research; W.A.B., E.R.D., D.T., C.E.N., D.L.S., T.L.C., M.J.P., T.T.R., S.E.L., and D.M.S. performed experiments; W.A.B., E.R.D., R.J., D.T., C.E.N., D.L.S., T.L.C., M.J.P., T.T.R., and J.M.H. analyzed data; W.A.B., E.R.D., R.J., D.T., M.J.P., and J.M.H. interpreted results of experiments; W.A.B., R.J., D.T., and J.M.H. prepared figures; W.A.B. and J.M.H. drafted manuscript; W.A.B. and J.M.H. edited and revised manuscript; W.A.B., E.R.D., R.J., D.T., C.E.N., D.L.S., T.L.C., M.J.P., T.T.R., and J.M.H. approved final version of manuscript.
Supplementary Material
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981 [DOI] [PubMed] [Google Scholar]
- 2.Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, Razunguzwa TT, Hollander JM. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol 300: R186–R200, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker D, Krayl M, Strub A, Li Y, Mayer MP, Voos W. Impaired interdomain communication in mitochondrial Hsp70 results in the loss of inward-directed translocation force. J Biol Chem 284: 2934–2946, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol 20: 1227–1232, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254, 1976 [DOI] [PubMed] [Google Scholar]
- 6.Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, Ganesan B, Weimer BC, Abel ED. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58: 1986–1997, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet 38: 576–582, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138: 628–644, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. I. Kinetics of oxygen utilization. J Biol Chem 217: 383–393, 1955 [PubMed] [Google Scholar]
- 10.Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. VI. The effects of adenosine diphosphate on azide-treated mitochondria. J Biol Chem 221: 477–489, 1956 [PubMed] [Google Scholar]
- 11.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem 278: 36027–36031, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Dabkowski ER. Quantitative proteomic analysis of distinct mitochondrial subpopulations in diabetic myocardium. FASEB J 22, 2008 [Google Scholar]
- 13.Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, Hollander JM. Mitochondrial dysfunction in the Type 2 diabetic heart is associated with alterations in spatially-distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol 299: H529–H540, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, Callery PS, Hollander JM. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol 296: H359–H369, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med 45: 855–865, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Feniouk BA, Suzuki T, Yoshida M. Regulatory interplay between proton motive force, ADP, phosphate, and subunit epsilon in bacterial ATP synthase. J Biol Chem 282: 764–772, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol 123: 109–117, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia MJ, McNamara PM, Gordon T, Kannel WB. Morbidity and mortality in diabetics in the Framingham population. Sixteen year follow-up study. Diabetes 23: 105–111, 1974 [DOI] [PubMed] [Google Scholar]
- 19.Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, Rehling P, Meisinger C, Ryan MT, Wiedemann N, Greenberg ML, Pfanner N. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol 19: 2133–2139, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gevorkyan-Airapetov L, Zohary K, Popov-Celeketic D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J Biol Chem 284: 4865–4872, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Godeas C, Sandri G, Panfili E. Distribution of phospholipid hydroperoxide glutathione peroxidase (PHGPx) in rat testis mitochondria. Biochim Biophys Acta 1191: 147–150, 1994 [DOI] [PubMed] [Google Scholar]
- 22.Hamblin M, Friedman DB, Hill S, Caprioli RM, Smith HM, Hill MF. Alterations in the diabetic myocardial proteome coupled with increased myocardial oxidative stress underlies diabetic cardiomyopathy. J Mol Cell Cardiol 42: 884–895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haraguchi CM, Mabuchi T, Hirata S, Shoda T, Yamada AT, Hoshi K, Yokota S. Spatiotemporal changes of levels of a moonlighting protein, phospholipid hydroperoxide glutathione peroxidase, in subcellular compartments during spermatogenesis in the rat testis. Biol Reprod 69: 885–895, 2003 [DOI] [PubMed] [Google Scholar]
- 24.He H, Giordano FJ, Hilal-Dandan R, Choi DJ, Rockman HA, McDonough PM, Bluhm WF, Meyer M, Sayen MR, Swanson E, Dillmann WH. Overexpression of the rat sarcoplasmic reticulum Ca2+ ATPase gene in the heart of transgenic mice accelerates calcium transients and cardiac relaxation. J Clin Invest 100: 380–389, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol 264: 476–483, 1996 [DOI] [PubMed] [Google Scholar]
- 26.Hollander JM, Lin KM, Scott BT, Dillmann WH. Overexpression of PHGPx and HSP60/10 protects against ischemia/reoxygenation injury. Free Radic Biol Med 35: 742–751, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Hollander JM, Martin JL, Belke DD, Scott BT, Swanson E, Krishnamoorthy V, Dillmann WH. Overexpression of wild-type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation 110: 3544–3552, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem 275: 22387–22394, 2000 [DOI] [PubMed] [Google Scholar]
- 29.Judge S, Jang YM, Smith A, Hagen T, Leeuwenburgh C. Age-associated increases in oxidative stress and antioxidant enzyme activities in cardiac interfibrillar mitochondria: implications for the mitochondrial theory of aging. FASEB J 19: 419–421, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Jullig M, Hickey AJ, Middleditch MJ, Crossman DJ, Lee SC, Cooper GJ. Characterization of proteomic changes in cardiac mitochondria in streptozotocin-diabetic rats using iTRAQ isobaric tags. Proteomics Clin Appl 1: 565–576, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143, 1990 [DOI] [PubMed] [Google Scholar]
- 32.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950, 2002 [DOI] [PubMed] [Google Scholar]
- 33.King KL, Young ME, Kerner J, Huang H, O'Shea KM, Alexson SE, Hoppel CL, Stanley WC. Diabetes or peroxisome proliferator-activated receptor alpha agonist increases mitochondrial thioesterase I activity in heart. J Lipid Res 48: 1511–1517, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685, 1970 [DOI] [PubMed] [Google Scholar]
- 35.Lesnefsky EJ, Chen Q, Slabe TJ, Stoll MS, Minkler PE, Hassan MO, Tandler B, Hoppel CL. Ischemia, rather than reperfusion, inhibits respiration through cytochrome oxidase in the isolated, perfused rabbit heart: role of cardiolipin. Am J Physiol Heart Circ Physiol 287: H258–H267, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Lin D. Multidimensional protein identification technology as an effective tool for proteomics. Am Genomic/Proteomic Technol 1: 38–46, 2001 [Google Scholar]
- 37.Lu J, Holmgren A. Selenoproteins. J Biol Chem 284: 723–727, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Nam S, Nakamuta N, Kurohmaru M, Hayashi Y. Cloning and sequencing of the mouse cDNA encoding a phospholipid hydroperoxide glutathione peroxidase. Gene 198: 245–249, 1997 [DOI] [PubMed] [Google Scholar]
- 39.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem 252: 8731–8739, 1977 [PubMed] [Google Scholar]
- 40.Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, Bork P, Prokisch H, Steinmetz LM. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet 2: e170, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem 278: 52873–52880, 2003 [DOI] [PubMed] [Google Scholar]
- 42.Pullman ME, Penefsky HS, Datta A, Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J Biol Chem 235: 3322–3329, 1960 [PubMed] [Google Scholar]
- 43.Pushpa-Rekha TR, Burdsall AL, Oleksa LM, Chisolm GM, Driscoll DM. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J Biol Chem 270: 26993–26999, 1995 [DOI] [PubMed] [Google Scholar]
- 44.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes 54: 8–14, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Rosca MG, Okere IA, Sharma N, Stanley WC, Recchia FA, Hoppel CL. Altered expression of the adenine nucleotide translocase isoforms and decreased ATP synthase activity in skeletal muscle mitochondria in heart failure. J Mol Cell Cardiol 46: 927–935, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Schwarz E, Seytter T, Guiard B, Neupert W. Targeting of cytochrome b2 into the mitochondrial intermembrane space: specific recognition of the sorting signal. EMBO J 12: 2295–2302, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen X, Zheng S, Thongboonkerd V, Xu M, Pierce WM, Jr, Klein JB, Epstein PN. Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am J Physiol Endocrinol Metab 287: E896–E905, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Singh K, Hood DA. Effect of denervation-induced muscle disuse on mitochondrial protein import. Am J Physiol Cell Physiol 300: C138–C145, 2011 [DOI] [PubMed] [Google Scholar]
- 49.Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol 80: 783–806, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol 264: 484–509, 1996 [DOI] [PubMed] [Google Scholar]
- 51.Turko IV, Murad F. Quantitative protein profiling in heart mitochondria from diabetic rats. J Biol Chem 278: 35844–35849, 2003 [DOI] [PubMed] [Google Scholar]
- 52.Ursini F, Maiorino M, Brigelius-Flohe R, Aumann KD, Roveri A, Schomburg D, Flohe L. Diversity of glutathione peroxidases. Methods Enzymol 252: 38–53, 1995 [DOI] [PubMed] [Google Scholar]
- 53.Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta 1422: 235–254, 1999 [DOI] [PubMed] [Google Scholar]
- 54.Williamson CL, Dabkowski ER, Baseler WA, Croston TL, Alway SE, Hollander JM. Enhanced apoptotic propensity in diabetic cardiac mitochondria: influence of subcellular spatial location. Am J Physiol Heart Circ Physiol 298: H633–H642, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson CL, Dabkowski ER, Dillmann WH, Hollander JM. Mitochondria protection from hypoxia/reoxygenation injury with mitochondria heat shock protein 70 overexpression. Am J Physiol Heart Circ Physiol 294: H249–H256, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Ye G, Metreveli NS, Donthi RV, Xia S, Xu M, Carlson EC, Epstein PN. Catalase protects cardiomyocyte function in models of type 1 and type 2 diabetes. Diabetes 53: 1336–1343, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem 253: 162–168, 1997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.