Abstract
The effect of differential signalling by IL-6 and leukaemia inhibitory factor (LIF) which signal by gp130 homodimerisation or LIFRβ/gp130 heterodimerisation on survival and hypertrophy was studied in neonatal rat cardiomyocytes. Both LIF and IL-6 [in the absence of soluble IL-6 receptor (sIL-6Rα)] activated Erk1/2, JNK1/2, p38-MAPK and PI3K signalling peaking at 20 min and induced cytoprotection against simulated ischemia-reperfusion injury which was blocked by the MEK1/2 inhibitor PD98059 but not the p38-MAPK inhibitor SB203580. In the absence of sIL-6R, IL-6 did not induce STAT1/3 phosphorylation, whereas IL-6/sIL-6R and LIF induced STAT1 and STAT3 phosphorylation. Furthermore, IL-6/sIL-6R induced phosphorylation of STAT1 Tyr701 and STAT3 Tyr705 were enhanced by SB203580. IL-6 and pheneylephrine (PE), but not LIF, induced cardiomyocyte iNOS expression and nitric oxide (NO) production. IL-6, LIF and PE induced cardiomyocyte hypertrophy, but with phenotypic differences in ANF and SERCA2 expression and myofilament organisation with IL-6 more resembling PE than LIF. Transfection of cardiomyocytes with full length or truncated chimaeric gp130 cytoplasmic domain/Erythropoietin receptor (EpoR) extracellular domain fusion constructs showed that the membrane proximal Box 1 and Box 2 containing region of gp130 was necessary and sufficient for MAPK and PI3K activation; hypertrophy; SERCA2 expression and iNOS/NO induction in the absence of JAK/STAT activation. In conclusion, IL-6 can signal in cardiomyocytes independent of sIL-6R and STAT1/3 and furthermore, that Erk1/2 and PI3K activation by IL-6 are both necessary and sufficient for induced cardioprotection. In addition, p38-MAPK may act as a negative feedback regulator of JAK/STAT activation in cardiomyocytes.
Keywords: Interleukin-6, Glycoprotein130, Cardiomyocyte
Highlights
► IL-6 signals independently of soluble IL-6 receptor in cardiomyocytes. ► IL-6 activates MAPK and PI-3-kinase in the absence of JAK/STAT activation. ► IL-6 induces hypertrophy and cytoprotection in the absence of JAK/STAT activation. ► IL-6 induced Erk-1 and 2 MAPK activation is necessary and sufficient for cytoprotection.
1. Introduction
Interleukin-6 (IL-6) is a pleiotropic cytokine of the IL-6 family which is expressed in many cell types and regulates many physiological and pathophysiological processes including immune system regulation, inflammation, wound healing and cell survival [1]. In the heart, IL-6 is expressed by integral tissue cellular components such as cardiomyocytes, fibroblasts and vascular endothelial and smooth muscle cells as well as interstitial macrophages [2,3]. IL-6 receptors consist of a membrane-tethered gp80 (cognate) or soluble form of the α-ligand-binding domain (IL-6Rα or sIL-6Rα). IL-6Rs are expressed primarily in myocardial interstitial cells such as fibroblasts and monocytes, rather than cardiomyocytes [3], but are upregulated in cardiomyocytes in response to activation by pro-inflammatory signals such as angiotensin II, endothelin-1 and noradrenalin as well as mechanical stretch [3–5]. However, recent evidence suggests that a cognate IL-6R is expressed in adult rat cardiomyocytes [6]. IL-6 plays an important role in myocardial remodelling whilst other IL-6 family cytokines such as leukaemia inhibitory factor (LIF) and cardiotrophin-1 (CT-1) also elicit survival and hypertrophy signalling via LIFRβ in cardiomyocytes [7,8].
IL-6 family cytokines include IL-6, LIF, CT-1, cilliary neurotrophic factor (CNTF), oncostatin M (OSM), IL-11, IL-27, erythropoietin (EPO) and others. They utilise type I cytokine receptors which consist of a ligand binding α-receptor subunit and a signal transducing β-receptor subunit containing a cytoplasmic signalling domain. They are type I membrane proteins with an N-terminal extracellular domain and a single membrane-spanning domain. IL-6 family cytokines utilise the common transmembrane glycoprotein component gp130 which forms all or part of the signal transducing domain depending on whether the ligand/receptor induces gp130 homodimerisation (i.e. In the case of IL-6, IL-11) or a gp130/β-receptor heterodimer (i.e. In the case of LIF, CT-1 or OSM). Only formation of the gp130 homo- or heterodimer can induce signalling. IL-6 can induce gp130-dependent signalling via two modes, either classically by binding to the cognate gp80 IL-6R or via trans-signalling which utilises the sIL-6R [1,5]. IL-6 trans-signalling thus allows IL-6 to signal in tissues/cells which do not necessarily express cognate IL-6R. The receptors themselves do not contain intrinsic kinase activity, but rather dimerisation activates constitutively bound Janus family kinases, JAK1, JAK2 or Tyk2 which then phosphorylate tyrosine residues in the distal cytoplasmic domain of gp130 which in turn act as docking sites for the SH2 domains of the Signal Transducer and Activator of Transcription (STAT) family of transcription factors (typically STATs 1, 3 and 5). Subsequent phosphorylation of STATs then induces STAT dimerisation and translocation to the nucleus where they activate gene transcription. Thus JAK/STAT activation is a classical hallmark of gp130-dependent cytokine signalling [1,9]. However, IL-6 family cytokines also activate a full complement of mitogen- and stress-activated protein kinases (MAPKs/SAPKs), including Erk1/2, JNK1/2 and p38-MAPK and also the phospho-inositide-3-kinase (PI3K)/Akt (protein kinase B: PKB) axis [1,10]. MAPK activation is thought to occur primarily through recruitment of the protein tyrosine phosphatase SHP2 to Tyr757 in the central region of gp130 which then recruits Grb2/Shc/Ras/Mek1/2/Erk1/2 [11]. However, JAK2 can also activate Erk1/2 and PI3K/Akt independently of SHP2 [12,13].
The relative contributions of Erk1/2, PI3K/Akt and JAK/STAT signalling to cardiomyocyte hypertrophy and survival is complex. Erk1/2 appears to play an important role in regulating hypertrophic gene expression and aspects of myofilament remodelling, whereas JAK/STAT activation may be more important in myofilament organisation [7,10,14]. PI3K/Akt plays an important role in adaptive hypertrophy [15,16]. Erk1/2 and PI3K play a critical role in cardiomyocyte survival, since activation by cardiac cytokines IL-6, LIF and CT-1 protects cardiomyocytes against apoptosis induced by a variety of stressors, including the chemotherapeutic agent doxorubicin, angiotensin II and myocardial injury [8,17–19]. In this study we have explored differences in signalling between IL-6 and LIF, these two cytokines representing the two principal different modes of gp130 recruitment, i.e. IL-6-induced gp130 homodimerisation and LIF-induced gp130/LIFRβ heterodimerisation. Despite the reported lack of basal cognate IL-6R expression in cardiomyocytes, we found that IL-6 can induce a full complement of MAPK and PI3K activation even in the absence of sIL-6R and STAT1/STAT3 activation. This was in contrast to LIF which activated MAPK/PI3K and JAK/STAT1/3. In the case of IL-6, JAK-STAT1/3 activation only occurred in the presence of exogenously added sIL-6R. Furthermore, Erk1/2 activation was both necessary and sufficient for IL-6-induced cytoprotection.
2. Materials and methods
2.1. Materials
Recombinant murine IL-6 and LIF (I9646; L51558: endotoxin < 1 EU/μg) and recombinant human erythropoietin (rhEPO: E5627) were from Sigma, UK. Recombinant soluble IL-6 receptor (sIL-6R) was a gift from Professor Peter Heinrich. Antibodies to phospho-STAT1 (Tyr701); phospho-STAT3 (Tyr705); pan-STAT1; pan-STAT3; phospho-ERK1/2; phospho-p38; phospho-JNK and phospho-PKB/Akt (Ser473) were from Cell Signalling, UK. Antibodies to pan-p42-MAPK, pan-Erk1/2; pan-p38 and pan-JNK were from Santa Cruz Biotech, USA. Antibodies to iNOS (NOS2) were from BD-Transduction Labs, UK. Antibodies to FLAG-epitope and α-actinin were from Sigma, UK. ALEXA-fluor 568 goat-anti-mouse antibody and ToPro were from Molecular Probes, UK. General purpose reagents were from Sigma-Aldrich, UK and Merck-BDH, UK unless otherwise stated. Plasmid and RNA preparation reagents were from Qiagen, UK. Collagenase was from Worthington Biochemicals, USA. Pancreatin was from Gibco-BRL. Gelatin solution was from Sigma, UK. Integrin-targeting peptide (peptide 6) was a gift from Dr Stephen Hart, Institute of Child Health, UK and has been described previously [19]. Lipofectin™ reagent was from Invitrogen, UK. Creatine phosphokinase (CPK) assay kit was from Boehringer-Mannheim, Germany. PD98059 and SB203580 were from Calbiochem, UK.
2.2. Isolated neonatal rat cardiomyocyte culture and treatment
All animal experiments were performed in accordance with European Commission and UK Home Office guidelines. Neonatal rat ventricular cardiomyocytes (NRVMs/CMs) were isolated from 1–2 day-old Sprague–Dawley rats as previously described [20,21] by collagenase/pancreatin digestion. Cells were plated at a density of approximately 2 × 106 cells per well on gelatin coated 6-well plates (NUNC) in medium containing 4:1 DMEM:M199 supplemented with 5% foetal calf serum; 10% horse serum and 1% penicillin/streptomycin (Gibco/BRL) for 24 h and then transferred to maintenance medium (serum-free DMEM:M199 plus antibiotics). Cells were maintained for 2 days in maintenance medium (3 days after plating) prior to treatment. Cells were treated with 10–50 ng/ml sterile IL-6 or LIF reconstituted in 0.1% (w/v) BSA/PBS as appropriate to the experiment for the indicated times. sIL-6R was used at 100 ng/ml. IL-6 was complexed with sIL-6R for 30 min in maintenance medium prior to addition. For inhibitor experiments cells were pretreated with 50 μM PD98059, 10 μM SB203580 or vehicle (0.01% DMSO) for 30 min prior to IL-6 or LIF application.
2.3. Transfections
Full length (Eg) and truncated (ΔB) gp130–EpoR fusion constructs were transfected into cardiomyocytes using a Lipofectin-Integrin targeting peptide–DNA (LID) protocol as previously described [19,22]. Full length (Eg) and truncated (ΔB) gp130–EpoR fusion constructs [23] were sub-cloned into the pcDNA3.1 vector (Invitrogen, UK). They were transiently expressed in CMs by overnight transfection using LID followed by 72 h in serum free medium as previously described [19,22]. Subsequently CMs were treated with 100 ng/ml recombinant human EPO (rhEPO) in serum free medium for 30 min (for harvesting for phospho-immunoblots) or 24 h (for RNA isolation; for hypertrophy; confocal analysis).
2.4. Simulated ischemia/reperfusion (sI/R)
sI/R was carried out using a modified Krebs buffer/anaerobic environment protocol as described previously [18,21] and is a modification of the method described by Esumi et al. [24]. Briefly, cells in 6-well tissue culture plates were exposed to 1.0 ml per well of sI buffer consisting of modified Krebs–Henseleit buffer containing (in mM): 137 NaCl; 12 KCl; 0.49 MgCl2; 1.8 CaCl2; 4 HEPES; 10 2-deoxyglucose; 20 Na lactate; pH 6.8. Cells were then exposed to a hypoxic environment by placing the plates into BBL GasPak anaerobic pouches (Becton Dickinson, UK) for 6 h. Simulated reperfusion was achieved by removing the plates from hypoxia and replacing the buffer with 1.0 ml of maintenance medium. The supernatant was then removed after 3 h for analysis of creatine phosphokinase (CPK) release (as a measure of cell injury).
2.5. Immunocytochemistry and confocal fluorescence microscopy
Cardiomyocytes were plated on gelatin-coated chamber slides in 1.0 ml of medium. Following treatment samples were fixed in 4% paraformaldehyde for 10 min at room temperature (RT). Slides were aspirated and left overnight in PBS at 4 °C. Slides were washed for 3 × 5 min in PBS at RT then blocked for 1 h in PBS; 2% BSA; 2% horse serum; 0.1% Triton x-100 at RT. Primary antibody was added at 1:800 dilution in blocking buffer and incubated for 1 h at RT. Slides were washed for 3 × 5 min in PBS; 0.1% Triton x-100 (PBST). FITC conjugated anti-mouse secondary antibody (Sigma) was added at 1:200 dilution in blocking buffer For 2 h at RT. Slides were washed for 3 × 5 min in PBST, aspirated, mounting medium added and coverslipped. Immunofluorescence confocal microscopy was performed using a Zeiss LSM 510 microscope. Where appropriate, the DNA stain ToPro was added to mounting medium for visualisation of nuclei.
2.6. RT-PCR analysis
Following treatment RNA was extracted from cells using RNeasy (Qiagen). RNA was reverse transcribed in 50 μl reactions using: 10 μl 5 × first strand buffer (Promega); 5 μl 0.1 M DTT; 1 μl oligo dT (10 μg/ml); 1 μl random hexamers; 1 μl dNTPs (10 μg/ml); 28 μl ddH2O; 3 μl total RNA. Samples were mixed, heated to 72 °C for 15 min [21] and cooled to RT. 1 μl RNase inhibitor and 1 μl reverse transcriptase were added. RT was performed at 42 °C for 2–3 h. cDNA was stored at − 20 °C and 1 μl used for PCR reactions. PCR was performed in 50 μl reactions using: 5 μl 10 × buffer (5 mM MgCl2); 1 μl dNTPs (10 mM); 1 μl oligo A; 1 μl oligo B; 0.25 μl Taq polymerase; 40 μl ddH2O; 1.5 μl DNA; 0.25 μl DMSO. Primers used were as follows: GAPDH: (forward) 5′-CACCATCTTCCAGGAGCGAG-3′; (reverse) 5′-ACAGCCTTGGCAGC ACCAGT-3′; ANF: (forward) 5′-GTGAGCTTCCTCCTTTTACTGG-3′; (reverse) 5′-AG GATGGACAGGATTGGAAATCA-3′; SERCA2: (forward) 5′-GACGAGTTTGGGGAAC AGCT-3′; (reverse) 5′-CAGGTGGTGATGACAGCAGG-3′.
3. Results
3.1. Differential JAK/STAT and MAPK/PI3K activation by IL-6 and LIF
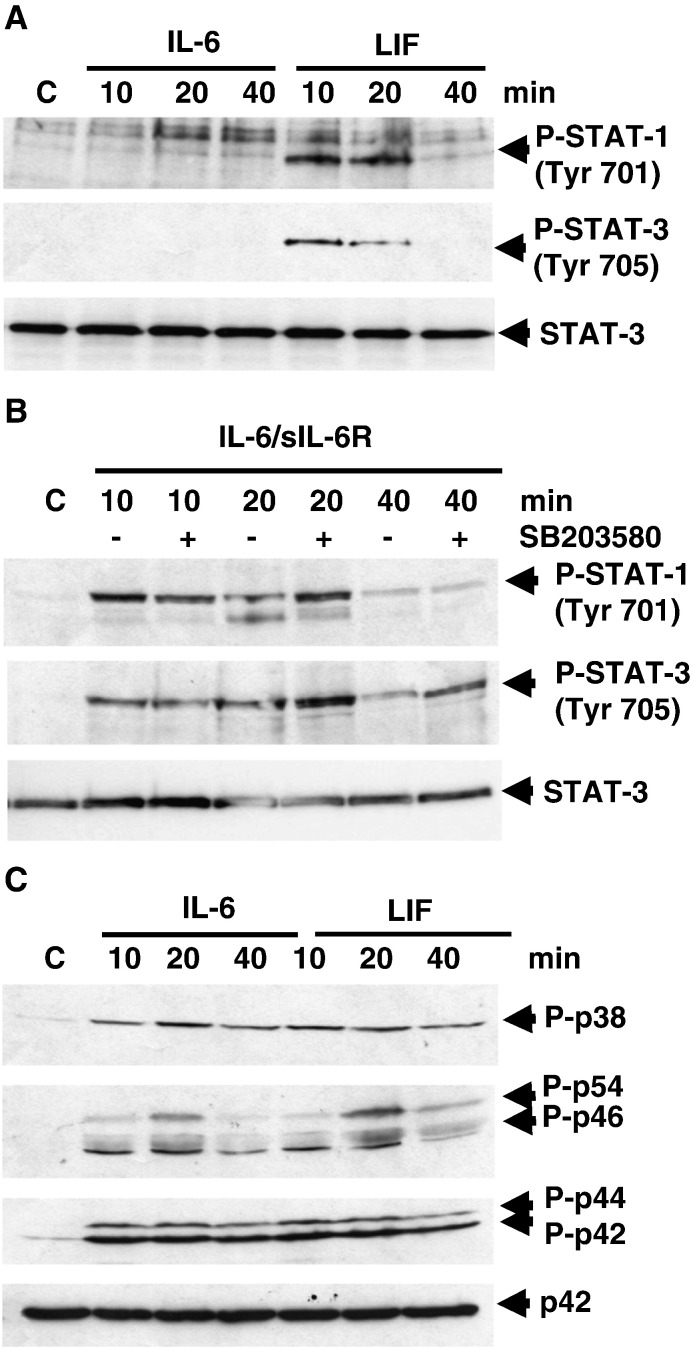
Under basal conditions cardiomyocytes (CMs) express low levels of IL-6R, but express the common IL-6 family cytokine receptor signalling component gp130 and also the ligand-binding domain of the leukaemia inhibitory factor β (LIFRβ). However, IL-6R expression is upregulated in CMs following stress, injury or neurohormonal activation [3,5,25]. We have shown previously that IL-6 induced PI3K- and iNOS-dependent cytoprotection in CMs [18]. Therefore, we determined the requirement for MAPK and JAK/STAT activation by IL-6 and LIF under different conditions. Treatment of CMs with IL-6 or LIF showed that only LIF but not IL-6 induced rapid and transient STAT1 Tyr701 and STAT3 Tyr705 phosphorylation (Fig. 1A). However, treatment of CMs with IL-6 in the presence of exogenously added soluble IL-6R (sIL-6R) was able to reconstitute STAT1 Tyr701 and STAT3 Tyr705 phosphorylation (Fig. 1B). Furthermore, pre-treatment with the p38-MAPK inhibitor SB203580 enhanced STAT1 and STAT3 phosphorylation by IL-6/sIL-6R (Fig. 1B). This is consistent with negative regulation of STAT3 activation by p38-MAPK/MK2-dependent phosphorylation of gp130 Ser782 [26] and suggests that Ser782 is constitutively phosphorylated by p38-MAPK/MK2 in CMs in the absence of sIL-6R.
Fig. 1.
Differential signalling to JAK/STAT by IL-6 and LIF. Neonatal rat cardiomyocytes were treated with IL-6 (with or without sIL-6R) or LIF for the indicated times (min) in serum-free medium and harvested for Western Blot analysis. Blots were probed for: Panels A and B: Phospho-STAT 1 (Tyr701); Phospho-STAT3 (Tyr705)/Total STAT3; Panel C: Phospho-p38-MAPK (Thr180/Tyr182); Phospho-JNK (Thr183/Tyr185); Phospho-Erk1/2 (Thr202/Tyr204)/Total Erk2.
The phosphorylation of STAT1 and STAT3 requires JAK1/2-dependent phosphorylation of critical tyrosine residues within the membrane distal gp130 cytoplasmic domain which are then able to recruit the STATs via their SH2 domains. However, Tyr757 within the membrane proximal region is thought to be required for MAPK activation via recruitment of SHP2. Furthermore, two proximal juxtamembrane regions termed Box 1 and Box 2 are sufficient for JAK1 and JAK2 recruitment. Given that the results above suggest that IL-6 cannot recruit and activate STAT1 and STAT3 in the absence of sIL-6R, we analysed the activation of MAPKs (Erk1/2, p38 and JNK) in response to IL-6 and LIF. Surprisingly, IL-6 (in the absence of sIL-6R) and LIF induced equivalent phosphorylation of Erk, JNK and p38-MAPK (Fig. 1C). Taken together, these results demonstrate that IL-6 can induce a distinct pattern of signalling in the absence of sIL-6R and gp130-dependent JAK/STAT1/3 activation.
3.2. MAPK-dependent protection against simulated ischemia
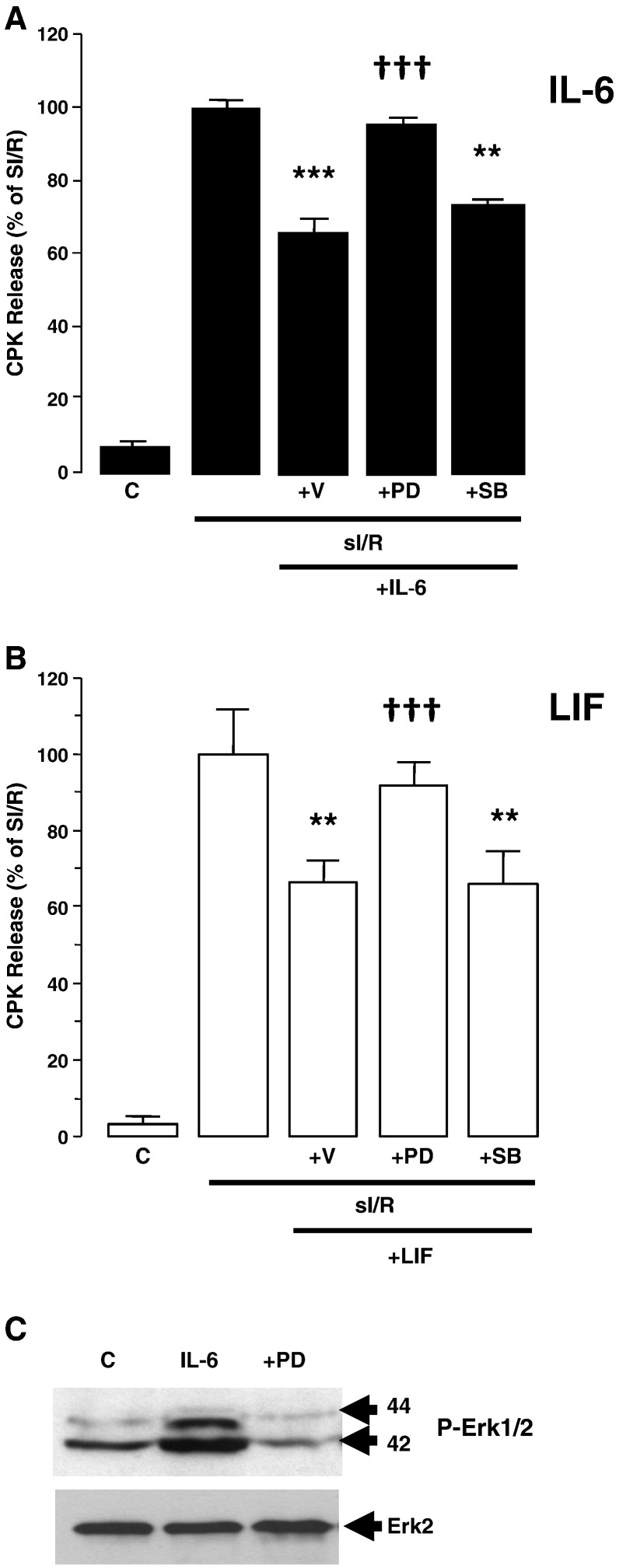
Both gp130- dependent MAPK and JAK/STAT activation have been implicated in cardiomyocyte survival. Since we have previously shown that IL-6 can induce CM survival [18] and IL-6 can induce PI3K/PKB activation independently of JAK/STAT activation, we compared IL-6 and LIF in their ability to protect CMs against simulated ischaemia/reperfusion injury (sI/R) using creatine phosphokinase (CPK) release as an index of injury. Both IL-6 and LIF pre-treatment 24 h before sI/R induced an equivalent reduction in sI/R injury (Fig. 2). To determine whether protection was p42/p44-MAPK (Erk1/2) or p38-MAPK-dependent, IL-6 and LIF pre-treatment were performed in the presence of pathway-selective pharmacological inhibitors. Interestingly, PD98059 (MEK1/2 inhibitor) reversed protection. In contrast, SB203580 (p38-MAPK inhibitor) had no effect. The efficacy of PD98059 to block Erk1/2 phosphorylation is shown in panel C. In keeping with our previous findings [18] the results show that both IL-6 and LIF induce Erk1/2 and PI3K-dependent protection independently of JAK/STAT and that MAPK/PI3K activation are both necessary and sufficient for protection in this context.
Fig. 2.
Erk dependent protection induced by IL-6 and LIF. Neonatal rat cardiomyocytes were treated with IL-6 (50 ng/ml) or LIF (10 ng/ml) in the presence of vehicle (DMSO) or PD98059 (50 μM), or SB203580 (10 μM) for 24 h in serum-free medium and then subjected to simulated ischemia/reoxygenation (sI/R) injury for 2 h. Cell injury was determined by assay of CPK release into the supernatant (Abs at 340 nm) after 3 h of reoxygenation expressed as % of sI/R (set to 100%). Results are expressed as mean ± SEM for triplicate experiments (n = 3) each performed in triplicate. Statistically significant differences are indicated as ***p < 0.001 vs sI/R or ††† vs vehicle Panel A: IL-6 pre-treatment; Panel B: LIF pre-treatment. Panel C: Western blots of Phospho-Erk1/2 (top) and total Erk2 (bottom) demonstrating the efficacy of PD98059 to inhibit phosphorylation of the respective downstream kinases.
3.3. iNOS/nitric oxide induction by IL-6 and LIF
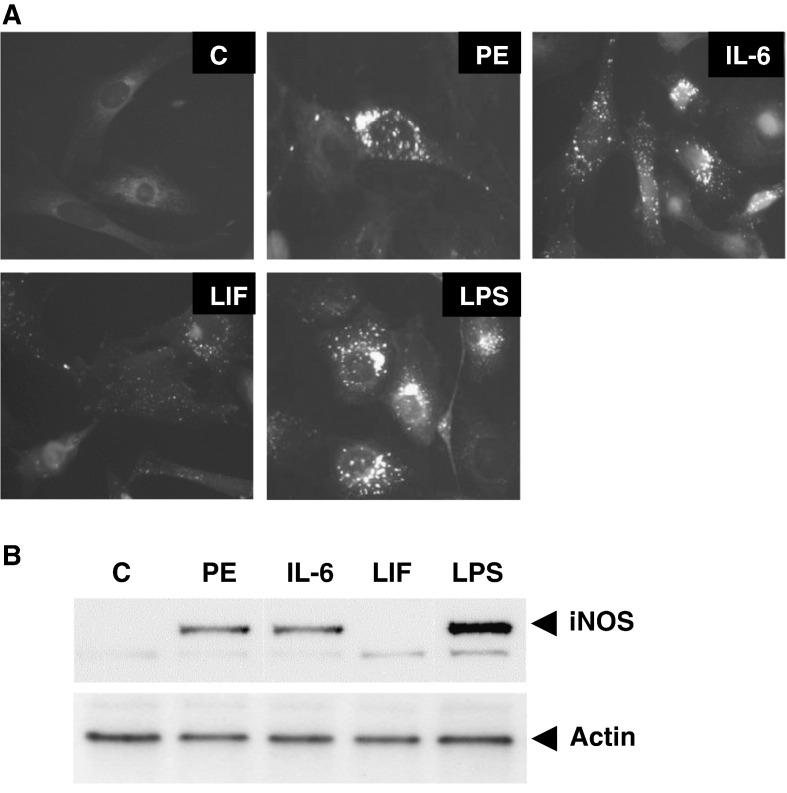
We have previously reported that IL-6-induced protection of CMs is dependent on iNOS-derived nitric oxide (NO) [18]. Therefore, we analysed the expression of iNOS and NO-production in CMs following pre-treatment with the α1-adrenergic receptor (α1-AR) agonist phenylephrine (PE), IL-6 and LIF. The inflammatory (Toll-like receptor) agonist bacterial lipopolysaccharide (LPS) was used as a positive control. Fig. 3 shows that PE and IL-6 but not LIF induced strong NO production as determined using the fluorescent NO probe DAF2-DA. In keeping with this, PE and IL-6 but not LIF, induced upregulation of iNOS expression. We have previously shown that IL-6-induced protection was iNOS-dependent, since when CMs were treated with the selective iNOS inhibitor amino guanidine (AG) prior to sI/R [18], AG blocked IL-6-induced protection.
Fig. 3.
Analysis of nitric oxide/iNOS expression. Panel A: Fluorescence micrographs of neonatal rat cardiomyocytes loaded with DAF-2DA. Cells were treated with Phenylephrine (PE: 10 μM), IL-6 (50 ng/μl), LIF (50 ng/μl) and LPS (10 μM) for 24 h in serum-free medium. Control myocytes (C) were left untreated. Panel B: Western blot containing 60 μg of protein per lane from control and treated cardiomyocytes immunoblotted with anti-iNOS antibody.
3.4. IL-6 and LIF-dependent cardiomyocyte hypertrophy
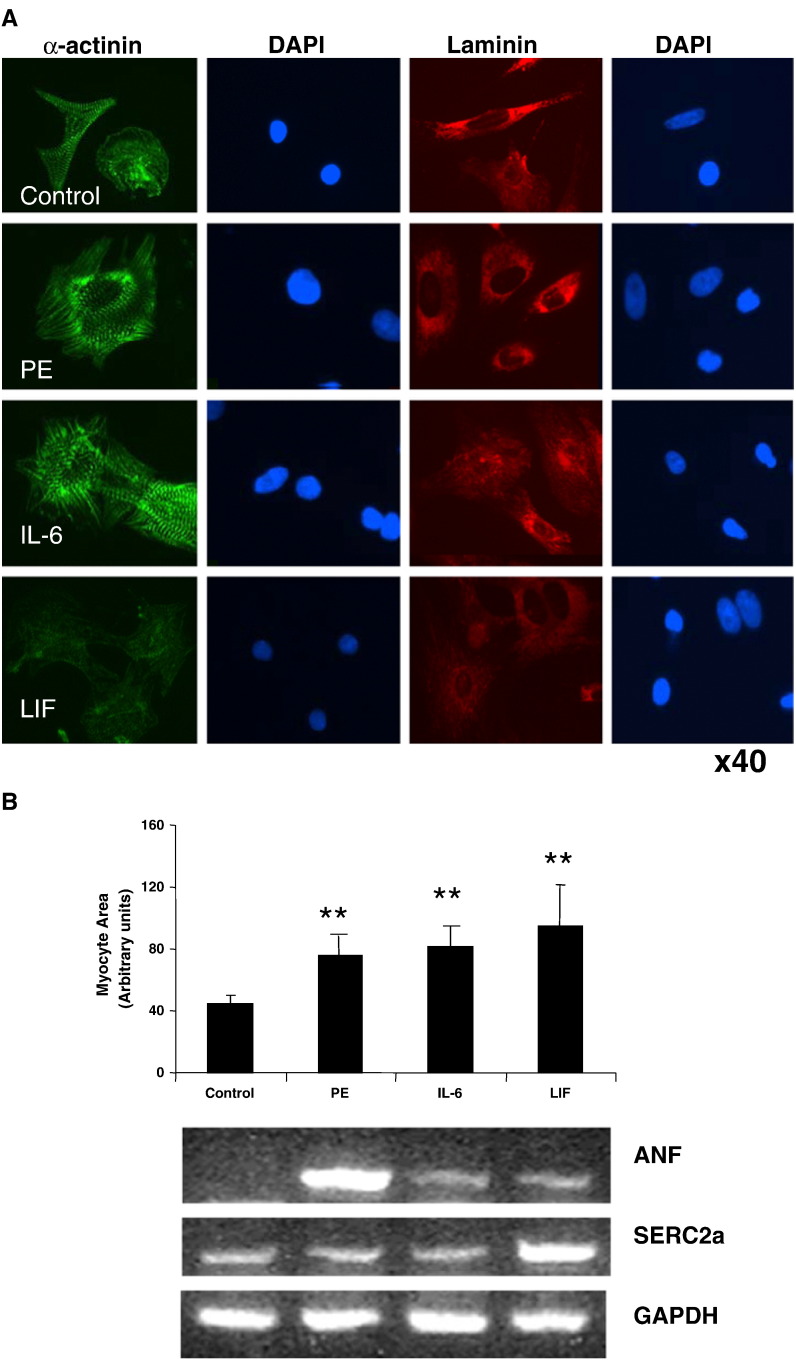
The IL-6 family of gp130-dependent cytokines including IL-6, LIF and CT-1 have been implicated in CM hypertrophy and heart failure. Furthermore, Erk1/2 and PI3K/PKB play a role in adaptive hypertrophy [15,16]. The role of JAK/STAT activation in hypertrophy is less clear, but STAT3 may play a role in gp130-dependent hypertrophy [27–29]. Because IL-6 family cytokines activate the Erk1/2, PI3K/PKB and JAK/STAT axes concomitantly, their individual contribution to hypertrophy is complex. We compared the ability of IL-6, LIF and PE to induce hypertrophy in isolated cultured neonatal rat ventricular cardiomyocytes (NRVMs) as determined by changes in morphological characteristics such as cell size and myofilament organisation, staining with CM-specific markers and gene expression. Fig. 4A shows that IL-6 and LIF both induced CM hypertrophy at 24 h as shown by increased cell size and elaboration of myofilaments as visualised by α-actinin staining. Both IL-6- and LIF-induced hypertrophy were associated with a reorganisation of myofilaments from a linear pattern aligning with the long axis of the cell to a radial pattern accompanied by increased stress fibre formation. Interestingly, the intensity of staining for the Z-disc marker α-actinin and the ECM marker laminin were reduced in LIF-treated cells compared to IL-6 or PE. This suggests that there are phenotypic differences between the IL-6- and LIF-induced hypertrophy.
Fig. 4.
Analysis of hypertrophic cell morphology and gene markers. Panel A: Confocal fluorescence micrographs of neonatal rat ventricular cardiomyocytes stained with α-actinin (green), laminin (red) and DAPI (blue) following treatment with Phenylephrine (10 μM), IL-6 (50 ng/ml), LIF (50 ng/ml) for 24 h in serum-free medium. The upper panel represents untreated myocytes (control). Magnification 40 ×. Panel B: mean cell area ± SEM (n = 200) of untreated (control) and neonatal rat cardiomyocytes treated (as above) for 24 h in serum free medium. Cell area was analysed with using Adobe Photoshop. Statistical analysis was performed using Student's t-test. **p < 0.05 versus control. Representative RT-PCR of untreated (control) and neonatal rat cardiomyocytes treated (as above) for 12 h in serum-free medium. RT-PCR was performed using oligonucleotide primers for the hypertrophic markers, ANF and SERC2a. GAPDH was used as a housekeeping control.
Since IL-6 and LIF appear to induce equivalent but distinct patterns of hypertrophy in CMs, we analysed the expression of hypertrophy-associated gene markers in response to PE, IL-6 and LIF, including atrial natriuretic factor (ANF) and the sarcoplasmic reticulum calcium pump SERCA2a. The results shown in Fig. 4B demonstrate that PE, IL-6 and LIF induced similar increases in myocyte area. In contrast, differential expression of ANF and SERCA2a were observed. Whereas PE induced strong upregulation ANF mRNA, IL-6 and LIF both induced weak expression of ANF. Furthermore, PE and IL-6 did not induce SERCA2 expression, whereas LIF induced strong SERCA2a mRNA expression.
3.5. Requirement for proximal versus distal gp130 regions
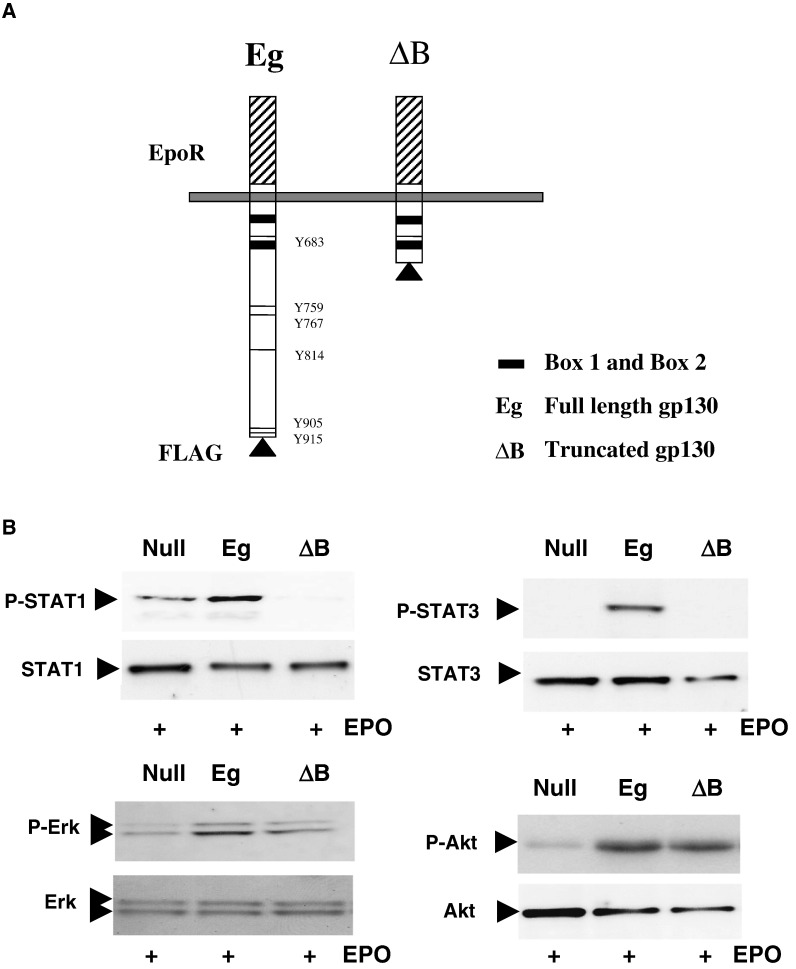
Because IL-6 and LIF induce similar but distinct responses in CMs (and distinct patterns of JAK/STAT activation) we next determined the requirement for the proximal and distal regions of gp130 in these responses. CMs were transfected with fusion constructs consisting of the full length, epitope-tagged cytoplasmic domain of gp130 coupled to the erythropoietin receptor (EpoR) extracellular ligand-binding domain (referred to as Eg) or a truncated gp130 containing only the Box 1 and Box 2 regions (referred to as ΔB) as depicted in Fig. 5A [23]. EpoR is also a type I cytokine receptor which utilises gp130. Furthermore, EpoR has been reported to be expressed in CMs and mediate cardioprotection [30,31]. However, both the presence of EpoR itself and the specific molecular determinants of EpoR-protective mechanisms in CMs have recently been challenged [32–34]. Nevertheless, the possible presence of endogenous EpoR coupled to gp130 in the system is a possible confounder. However, treatment of CMs with EPO alone (100 ng/ml) did not elicit a significant response under control conditions (null vector). Therefore, firstly, we analysed the ability of these constructs to induce activation of STAT1 and STAT3 phosphorylation in response to exogenously added EPO. As expected, both STAT1 and STAT3 phosphorylation were increased in the Eg but not the ΔB expressing cells (Fig. 5B). In fact ΔB appeared to have a slightly dominant negative effect on STAT1 phosphorylation and STAT3 expression. Erk1/2 and PKB(Akt) phosphorylation were intact in ΔB expressing cells, suggesting that sites within Box 1 and Box 2 are sufficient for Erk1/2 and PI3K activation.
Fig. 5.
Analysis of JAK/STAT, MAPK and PI3K Activation by gp130. Panel A: Schematic representation of FLAG-tagged full length (Eg) and truncated (ΔB) gp130–EpoR fusion constructs showing the EpoR extracellular ligand binding domain (hatched), the membrane-spanning region and the cytoplasmic domain. Box 1 and Box 2 JAK binding domains are shown (shaded) and the phosphotyrosine residue docking sites for STATs are indicated. Panel B: Neonatal rat cardiomyocytes were transfected with pcDNA3.1, Eg and ΔB for 72 h in serum-free medium and subsequently stimulated with 100 ng/ml EPO for 30 min and harvested for Western Blot analysis. Blots were probed for Phospho-STAT 1 (Tyr701)/Total STAT1; Phospho-STAT3 (Tyr705)/Total STAT3; Phospho-Erk1/2 (Thr202/Tyr204)/Total Erk1/2; Phospho-Akt (PKB: Ser473)/Total Akt(PKB).
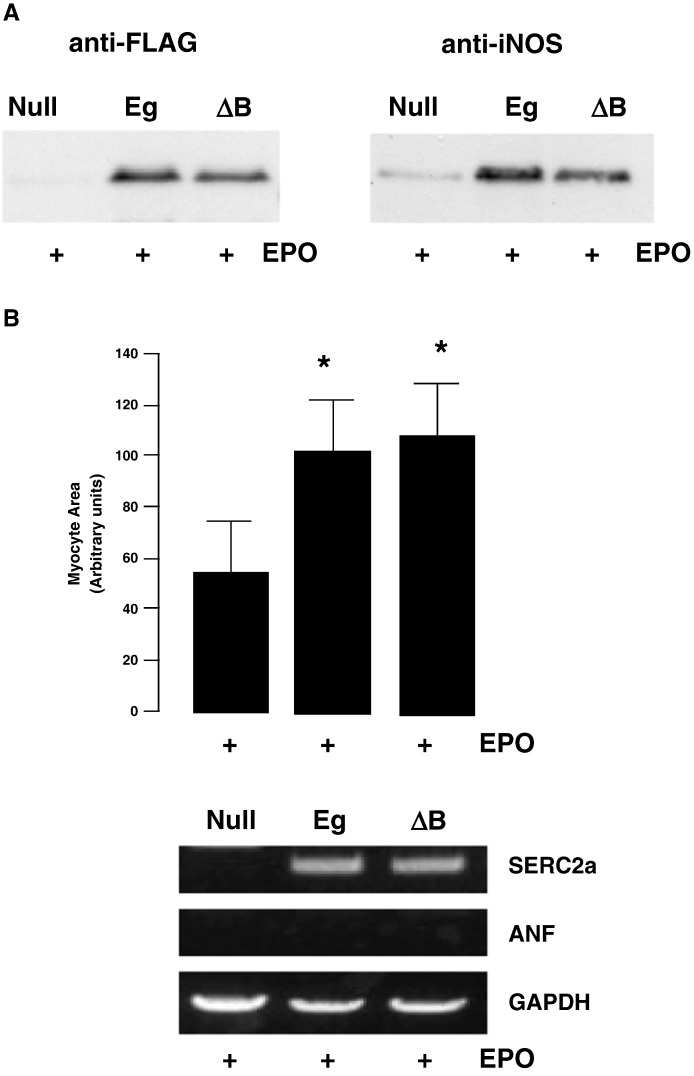
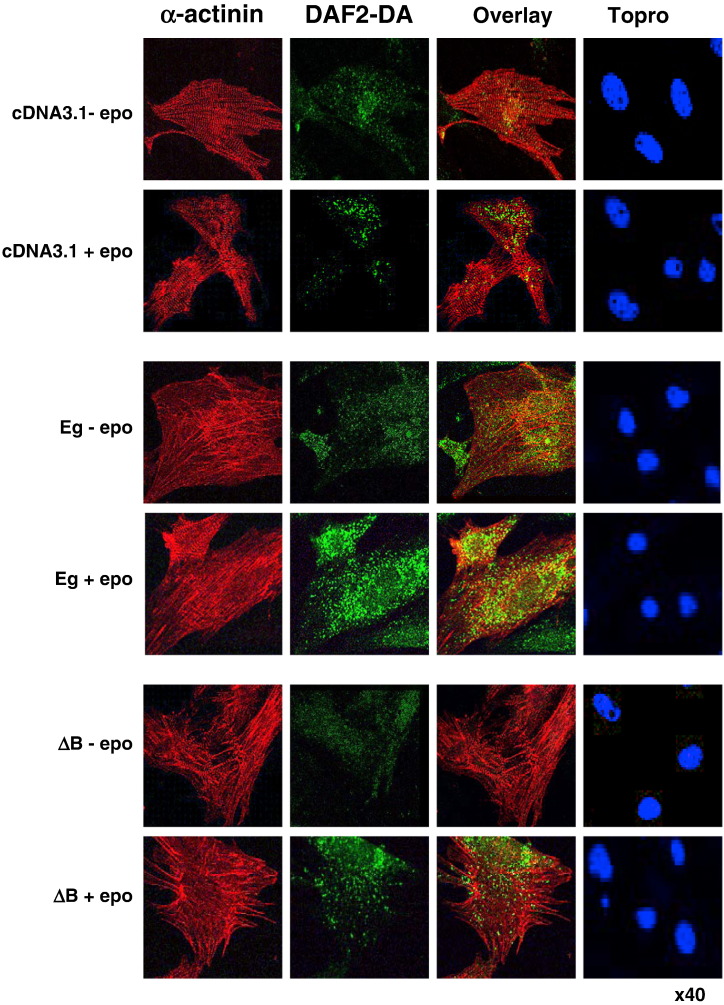
These results confirm the fidelity of the model in the absence of a significant effect of EPO on endogenous pathways. Equivalent expression of Eg and ΔB was confirmed by blotting with anti-FLAG antibodies (Fig. 6A). Interestingly, both Eg and ΔB induced identical increases in myocyte area and SERCA2 mRNA expression (Fig. 6B), indicating that STAT1 and STAT3 phosphorylation are dispensible for gp130-dependent hypertrophy. We further analysed the NO production in CMs expressing Eg and ΔB. Fig. 7 shows that both Eg and ΔB induced similar increases in NO production as determined by confocal analysis of DAF2-DA-loaded CMs. This was also associated with similar induction of iNOS expression in Eg and ΔB expressing CMs (Fig. 6A).
Fig. 6.
Analysis of gp130-dependent hypertrophy. Panel A: Western blot analysis of neonatal rat cardiomyocytes transfected with pcDNA3.1 (empty vector), Eg or ΔB for 72 h in serum-free medium and subsequently stimulated with 100 ng/ml EPO. Cells were harvested for Western blotting 24 h after EPO treatment. Blots were probed with anti-FLAG or anti-iNOS antibodies. Panel B: Mean cell area ± SEM (n = 100) of neonatal rat cardiomyocytes transfected with pcDNA3.1 (empty vector), Eg or ΔB for 72 h in serum-free medium and subsequently stimulated with 100 ng/ml EPO. Cells were fixed and micrographed after 24 h of EPO treatment. Cell area was analysed using Adobe Photoshop. Statistical analysis was performed using a student's t-test. *p < 0.05 versus pcDNA3.1. Below is shown representative RT-PCR of transfected neonatal rat cardiomyocytes which have been cultured for 72 h in serum-free medium and subsequently stimulated with EPO for 6 h. RNA was extracted and RT-PCR was performed using oligonucleotide primers for hypertrophic markers ANF and SERC2a. GAPDH was used as a housekeeping control.
Fig. 7.
Analysis of gp130-dependent changes in hypertrophic cell morphology and NO production. Confocal fluorescence micrographs of neonatal rat ventricular cardiomyocytes stained with α-actinin (red), DAF2-DA (green) and ToPro (blue) following transfection with pcDNA3.1 (empty vector), Eg or ΔB for 72 h in serum-free medium and subsequently stimulated with 100 ng/ml EPO. Cells were loaded with DAF2-DA and/or fixed for staining and confocal analysis 24 h after EPO treatment.
4. Discussion
These studies show that STAT1 and STAT3 phosphorylation are dispensible for the induction of hypertrophy and iNOS-dependent survival of cardiomyocytes. We have identified a minimal IL-6 signalling modality in CMs which only requires the membrane proximal region of gp130. Nevertheless, it remains to be determined how IL-6, or indeed the minimal gp130ΔB construct signals to Erk1/2 and PI3K/Akt. Signalling of IL-6 via gp130 would be expected to be associated with JAK/STAT activation, since JAKs are constitutively associated with the gp130β chain [11,35]. The activation of the Erk/MAPK and PI3K axes in this study appeared to be maximal despite the absence of STAT1/3 phosphorylation. It is possible that MAPK/PI3K activation has a lower threshold. Activation of the Erk1/2 cascade via gp130 is thought to require recruitment of the SH2 containing cytolplasmic protein tyrosine phosphatase SHP2. Phosphorylation of the membrane proximal Tyr757 is necessary and sufficient for SHP2 recruitment. Tyr phosphorylation of SHP2 leads to activation of the Ras/Erk1/2 module through recruitment of the Grb2/Sos docking protein/RasGTP exchange factor complex [11,35]. SHP2 can also form a complex with the scaffolding proteins Gab1/2 and the p85 subunit of PI3K, thus leading to activation of the PI3K pathway and ERK5 [36]. Also, activated STAT5A and B can form a complex between Gab2/Grb1/2 and the p85 subunit of PI3K leading to PI3K activation, which is independent of SHP2 [12]. Mutation of Tyr757 to Phe has been shown to block Erk1/2 activation [37]. However, this paradigm does not explain our results unless JAK-dependent Tyr757 phosphorylation and SHP2 recruitment versus distal Tyr phosphorylation and STAT1/3 recruitment are mutually independent events. Even so, the gp130ΔB construct is truncated N-terminal to Tyr757 [23]. However, it has been reported that JAK2 can also activate Erk1/2 and PI3K/Akt independently of SHP2 [12,13]. This latter mechanism could explain our results if IL-6 binds to a cognate receptor in CMs (possibly at low levels of expression) that can activate Erk/MAPK and PI3K and which has a different threshold for Erk/MAPK/PI3K activation than STAT3 activation compared to IL-6/sIL-6R.
With respect to differing thresholds for MAPK/PI3K versus JAK/STAT activation, this could also be achieved, at least in part, by negative feedback regulation of JAK/STAT activation. Indeed our results suggest that signalling crosstalk exists between p38-MAPK and STAT1/3 activation. Activation of the p38-MAPK/MK2 axis by stress or other receptors basally represses STAT1/3 phosphorylation via Ser782 phosphorylation of gp130 monomers which leads to gp130 internalisation and degradation [26]. Thus the absence of STAT1/3 phosphorylation may be due to low endogenous levels of the cIL-6Rα, whereas stabilisation of gp130 homodimers in the presence of sIL-6R is able to overcome this.
There is further evidence that a balance exists between JAK/STAT and Erk1/2 activation and that Erk1/2 activation may also act to prevent sustained STAT3 activation [11]. STAT3 activation in itself appears to play an important, although complex role in cardiomyocyte survival and remodelling. For instance, overexpression of the negative regulator of gp130/STAT signalling, suppressor of cytokine signalling 1 (SOCS1) promotes the transition to heart failure in a model of pressure overload [38]. Conversely, CM-restricted knockout of SOCS3 prevents post-MI remodelling [39] but increases SR Ca2 + overload, cardiac dysfunction and arrhythmias [40]. The SHP2/Erk1/2 axis may also negatively regulate gp130/STAT activation because mice lacking gp130Y757 develop various organ defects resulting from over-activation of gp130/STAT1/3, since Tyr757 is also necessary for recruitment of SOCS3. In addition, STAT1 and STAT3 may have opposing roles in CM survival, since STAT1 has been shown to promote CM apoptosis [41,42].
Since it is well established that Erk1/2 and PI3K/Akt can cooperate to mediate CM survival [18,21], it is not surprising that our results show that activation of these pathways can protect CMs in the absence of JAK/STAT activation. However, our results do not preclude a protective role for STAT3 in other contexts and likewise do not preclude a role for STAT3 in myocardial hypertrophy and myofilament remodelling in vivo in response to complex stimuli comprising biomechanical stresses and neurohumoral and/or cytokine activation. For instance, STAT3 overexpression alone induces CM hypertrophy in vitro and in vivo [9,27,28]. STAT3 has also been shown to have positive effects on CM survival which may be independent of its function as a transcription factor (i.e. non-genomic) and act at the mitochondrial level [43]. Furthermore, recent evidence suggests that pharmacological inhibition of STAT3 but not STAT1 reduced collagen deposition and cardiac fibrosis in a model of renal artery ligation in rats. This was accompanied by reduced cardiac hypertrophy, reduced re-expression of foetal gene markers and improved cardiac function [44]. This suggests that STAT3 may play an important role in cardiac remodelling driven by IL-6/STAT3 activation in cardiac fibroblasts. Interestingly, inhibition of p38-MAPK showed similar effects, showing that p38-MAPK activation is also pro-fibrotic.
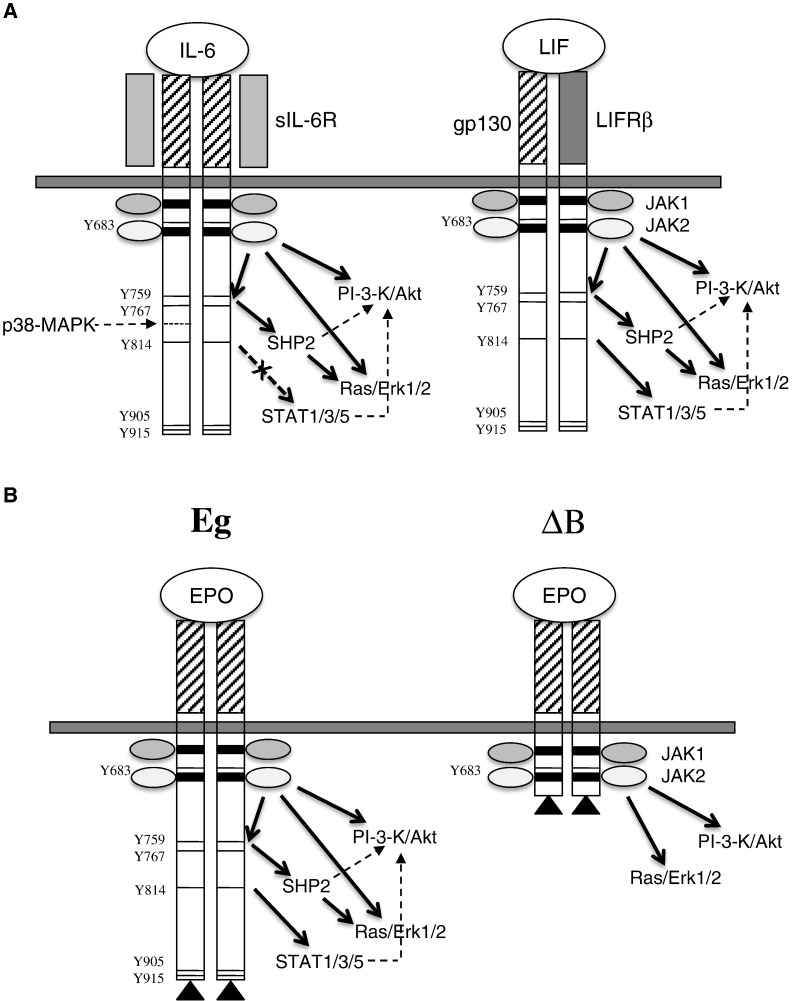
There may also be important compartmental considerations in relation to the role of JAK/STAT signalling in myocardial remodelling. For instance, Lindner et al. 45] found that STAT3 knockout mice showed reduced cardiac function and increased collagen deposition in a model of cocksackie virus-induced myocarditis. Importantly, this was a cardiomyocyte-restricted knockout. Therefore, they conclude that STAT3 signalling in CMs protects against fibrosis. Furthermore, cardiomyocyte-specific knockout of STAT3 induces age-dependent heart failure [46]. Therefore activation of STAT3 in cardiomyocytes versus myofibroblasts may have opposite effects on fibrosis and CM hypertrophy. In support of our results, Szabo-Fresnais et al. recently reported that IL-6 activates Erk1/2 and PI3K signalling in adult rat cardiomyocytes in the absence of JAK/STAT activation via a cognate receptor [6]. Therefore, it seems highly likely that CMs do express a cognate receptor of some form, but how differential activation of Erk1/2 MAPK and PI3K independent of JAK/STAT is achieved remains to be determined. A diagram depicting the possible modes of IL-6/LIF receptor/gp130 activation is shown in Fig. 8.
Fig. 8.
Modes of gp130 dimerisation and signalling. Panel A: Schematic representation of IL-6R-induced gp130 homo-dimerisation or LIF-induced gp130 hetero-dimerisation showing the extracellular ligand binding domain (hatched/stippled), the membrane-spanning region and the cytoplasmic domain. Box 1 and Box 2 JAK binding domains are shown (shaded) and the phosphotyrosine residue docking sites for STATs are indicated. The differential activation of PI3K/Akt; Ras/Erk1/2 or STAT1, 3, 5 are indicated by arrows. Panel B: Schematic representation of FLAG-tagged full length (Eg) and truncated (ΔB) gp130–EpoR fusion constructs showing the EpoR extracellular ligand binding domain (hatched), the membrane-spanning region and the cytoplasmic domain. Box 1 and Box 2 JAK binding domains are shown (shaded) and the phosphotyrosine residue docking sites for STATs are indicated. The differential activation of PI3K/Akt; Ras/Erk1/2 or STAT1, 3, 5 are indicated by arrows.
In conclusion, our results show that IL-6 has important pro-survival effects in cardiomyocytes which are mediated by Erk1/2 and PI3K/Akt activation and which are independent of gp130-dependent JAK/STAT activation. Furthermore, Erk1/2 and PI3K/Akt activation appear to be necessary and sufficient for cardiomyocyte hypertrophy induced by IL-6, or by a minimal gp130 construct containing only the membrane-proximal Box 1 and Box 2 regions.
Conflict of interest statement
None declared.
Funding
This work was supported by British Heart Foundation grants FS/95/039 (AP); PG/96/164; PG/98/077 (NS) and PG/2001/066 (AF).
Acknowledgements
Soluble IL-6 receptor was kindly provided by Professor Peter Heinrich. Gp130–EpoR fusion constructs were kindly provided by Professor Lutz Graeve.
References
- 1.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Muller-Newen G., Schaper F. The Biochemical Journal. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plenz G., Eschert H., Erren M., Wichter T., Bohm M., Flesch M. Journal of the American College of Cardiology. 2002;39:1508–1512. doi: 10.1016/s0735-1097(02)01791-6. [DOI] [PubMed] [Google Scholar]
- 3.Briest W., Rassler B., Deten A., Leicht M., Morwinski R., Neichel D. Pflügers Archiv. 2003;446:437–446. doi: 10.1007/s00424-003-1043-x. [DOI] [PubMed] [Google Scholar]
- 4.Saito S., Aikawa R., Shiojima I., Nagai R., Yazaki Y., Komuro I. FEBS Letters. 1999;456:103–107. doi: 10.1016/s0014-5793(99)00936-9. [DOI] [PubMed] [Google Scholar]
- 5.Coles B., Fielding C.A., Rose-John S., Scheller J., Jones S.A., O'Donnell V.B. The American Journal of Pathology. 2007;171:315–325. doi: 10.2353/ajpath.2007.061078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo-Fresnais N., Lefebvre F., Germain A., Fischmeister R., Pomerance M. Cellular Signalling. 2010;22:1143–1152. doi: 10.1016/j.cellsig.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Wollert K.C., Taga T., Saito M., Narazaki M., Kishimoto T., Glembotski C.C. The Journal of Biological Chemistry. 1996;271:9535–9545. doi: 10.1074/jbc.271.16.9535. [DOI] [PubMed] [Google Scholar]
- 8.Sheng Z., Pennica D., Wood W.I., Chien K.R. Development. 1996;122:419–428. doi: 10.1242/dev.122.2.419. [DOI] [PubMed] [Google Scholar]
- 9.Haghikia A., Stapel B., Hoch M., Hilfiker-Kleiner D. Heart Failure Reviews. 2011;16:35–47. doi: 10.1007/s10741-010-9170-x. [DOI] [PubMed] [Google Scholar]
- 10.Kodama H., Fukuda K., Pan J., Sano M., Takahashi T., Kato T. American Journal of Physiology. Heart and Circulatory Physiology. 2000;279:H1635–H1644. doi: 10.1152/ajpheart.2000.279.4.H1635. [DOI] [PubMed] [Google Scholar]
- 11.Fischer P., Hilfiker-Kleiner D. British Journal of Pharmacology. 2008;153(Suppl. 1):S414–S427. doi: 10.1038/bjp.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyga R., Pecquet C., Harir N., Gu H., Dhennin-Duthille I., Regnier A. The Biochemical Journal. 2005;390:359–366. doi: 10.1042/BJ20041523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada O., Ozaki K., Akiyama M., Kawauchi K. Molecular Cancer Therapeutics. 2012;11:1112–1121. doi: 10.1158/1535-7163.MCT-11-0850. [DOI] [PubMed] [Google Scholar]
- 14.Wollert K.C., Studer R., Doerfer K., Schieffer E., Holubarsch C., Just H. Circulation. 1997;95:1910–1917. doi: 10.1161/01.cir.95.7.1910. [DOI] [PubMed] [Google Scholar]
- 15.Aoyagi T., Matsui T. Current Pharmaceutical Design. 2011;17:1818–1824. doi: 10.2174/138161211796390976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weeks K.L., Gao X., Du X.J., Boey E.J., Matsumoto A., Bernardo B.C. Circulation. Heart Failure. 2012;5:523–534. doi: 10.1161/CIRCHEARTFAILURE.112.966622. [DOI] [PubMed] [Google Scholar]
- 17.Negoro S., Oh H., Tone E., Kunisada K., Fujio Y., Walsh K. Circulation. 2001;103:555–561. doi: 10.1161/01.cir.103.4.555. [DOI] [PubMed] [Google Scholar]
- 18.Smart N., Mojet M.H., Latchman D.S., Marber M.S., Duchen M.R., Heads R.J. Cardiovascular Research. 2006;69:164–177. doi: 10.1016/j.cardiores.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Hart S.L., Arancibia-Carcamo C.V., Wolfert M.A., Mailhos C., O'Reilly N.J., Ali R.R. Human Gene Therapy. 1998;9:575–585. doi: 10.1089/hum.1998.9.4-575. [DOI] [PubMed] [Google Scholar]
- 20.Pennica D., King K.L., Shaw K.J., Luis E., Rullamas J., Luoh S.M. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:1142–1146. doi: 10.1073/pnas.92.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punn A., Mockridge J.W., Farooqui S., Marber M.S., Heads R.J. The Biochemical Journal. 2000;350(Pt 3):891–899. [PMC free article] [PubMed] [Google Scholar]
- 22.Obasanjo-Blackshire K., Mesquita R., Jabr R.I., Molkentin J.D., Hart S.L., Marber M.S. Cardiovascular Research. 2006;71:672–683. doi: 10.1016/j.cardiores.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Gerhartz C., Heesel B., Sasse J., Hemmann U., Landgraf C., Schneider-Mergener J. The Journal of Biological Chemistry. 1996;271:12991–12998. doi: 10.1074/jbc.271.22.12991. [DOI] [PubMed] [Google Scholar]
- 24.Esumi K., Nishida M., Shaw D., Smith T.W., Marsh J.D. The American Journal of Physiology. 1991;260:H1743–H1752. doi: 10.1152/ajpheart.1991.260.6.H1743. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs M., Hilfiker A., Kaminski K., Hilfiker-Kleiner D., Guener Z., Klein G. The FASEB Journal. 2003;17:2118–2120. doi: 10.1096/fj.03-0331fje. [DOI] [PubMed] [Google Scholar]
- 26.Radtke S., Wuller S., Yang X.P., Lippok B.E., Mutze B., Mais C. Journal of Cell Science. 2010;123:947–959. doi: 10.1242/jcs.065326. [DOI] [PubMed] [Google Scholar]
- 27.Hirota H., Chen J., Betz U.A., Rajewsky K., Gu Y., Ross J., Jr. Cell. 1999;97:189–198. doi: 10.1016/s0092-8674(00)80729-1. [DOI] [PubMed] [Google Scholar]
- 28.Kunisada K., Negoro S., Tone E., Funamoto M., Osugi T., Yamada S. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:315–319. doi: 10.1073/pnas.97.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto T., Takeishi Y., Takahashi H., Shishido T., Arimoto T., Tomoike H. Basic Research in Cardiology. 2004;99:328–337. doi: 10.1007/s00395-004-0482-7. [DOI] [PubMed] [Google Scholar]
- 30.Wright G.L., Hanlon P., Amin K., Steenbergen C., Murphy E., Arcasoy M.O. The FASEB Journal. 2004;18:1031–1033. doi: 10.1096/fj.03-1289fje. [DOI] [PubMed] [Google Scholar]
- 31.Tada H., Kagaya Y., Takeda M., Ohta J., Asaumi Y., Satoh K. Cardiovascular Research. 2006;71:466–477. doi: 10.1016/j.cardiores.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Santhanam A.V., d'Uscio L.V., Katusic Z.S. Advances in Pharmacology. 2010;60:257–285. doi: 10.1016/B978-0-12-385061-4.00009-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanellakis P., Pomilio G., Agrotis A., Gao X., Du X.J., Curtis D. British Journal of Pharmacology. 2010;160:2085–2096. doi: 10.1111/j.1476-5381.2010.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinclair A.M., Coxon A., McCaffery I., Kaufman S., Paweletz K., Liu L. Blood. 2010;115:4264–4272. doi: 10.1182/blood-2009-10-248666. [DOI] [PubMed] [Google Scholar]
- 35.Haan C., Kreis S., Margue C., Behrmann I. Biochemical Pharmacology. 2006;72:1538–1546. doi: 10.1016/j.bcp.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Nakaoka Y., Nishida K., Fujio Y., Izumi M., Terai K., Oshima Y. Circulation Research. 2003;93:221–229. doi: 10.1161/01.RES.0000085562.48906.4A. [DOI] [PubMed] [Google Scholar]
- 37.Jenkins B.J., Grail D., Nheu T., Najdovska M., Wang B., Waring P. Nature Medicine. 2005;11:845–852. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- 38.Cittadini A., Monti M.G., Iaccarino G., Castiello M.C., Baldi A., Bossone E. Cardiovascular Research. 2012 doi: 10.1093/cvr/cvs261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oba T., Yasukawa H., Hoshijima M., Sasaki K., Futamata N., Fukui D. Journal of the American College of Cardiology. 2012;59:838–852. doi: 10.1016/j.jacc.2011.10.887. [DOI] [PubMed] [Google Scholar]
- 40.Yajima T., Murofushi Y., Zhou H., Park S., Housman J., Zhong Z.H. Circulation. 2011;124:2690–2701. doi: 10.1161/CIRCULATIONAHA.111.028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephanou A., Scarabelli T.M., Townsend P.A., Bell R., Yellon D., Knight R.A. The FASEB Journal. 2002;16:1841–1843. doi: 10.1096/fj.02-0150fje. [DOI] [PubMed] [Google Scholar]
- 42.McCormick J., Suleman N., Scarabelli T.M., Knight R.A., Latchman D.S., Stephanou A. Journal of Cellular and Molecular Medicine. 2012;16:386–393. doi: 10.1111/j.1582-4934.2011.01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boengler K., Hilfiker-Kleiner D., Heusch G., Schulz R. Basic Research in Cardiology. 2010;105:771–785. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mir S.A., Chatterjee A., Mitra A., Pathak K., Mahata S.K., Sarkar S. The Journal of Biological Chemistry. 2012;287:2666–2677. doi: 10.1074/jbc.M111.246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindner D., Hilbrandt M., Marggraf K., Becher P.M., Hilfiker-Kleiner D., Klingel K. Cardiology Research and Practice. 2012;2012:437623. doi: 10.1155/2012/437623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilfiker-Kleiner D., Hilfiker A., Fuchs M., Kaminski K., Schaefer A., Schieffer B. Circulation Research. 2004;95:187–195. doi: 10.1161/01.RES.0000134921.50377.61. [DOI] [PubMed] [Google Scholar]