Abstract
Msi1-like (MSIL) proteins, which are eukaryote-specific and contain a series of WD40 repeats, have pleiotropic roles in chromatin assembly, DNA damage repair, and regulation of nutrient/stress-sensing signaling pathways. In the fungal kingdom, the functions of MSIL proteins have been studied most intensively in the budding yeast model Saccharomyces cerevisiae, an ascomycete. Yet their functions are largely unknown in other fungi. Recently, an MSIL protein, Msl1, was discovered and functionally characterized in the pathogenic yeast Cryptococcus neoformans, a basidiomycete. Interestingly, MSIL proteins appear to have redundant and unique roles in both fungi, suggesting that MSIL proteins may have evolutionarily divergent roles in different parts of the fungal kingdom. In this review, we will describe the current findings regarding the role of MSIL proteins in fungi and discuss future directions for research on this topic.
Keywords: Chromatin assembly factor, Histone, Msi1, Retinoblastoma, WD40
The Msi1-like (MSIL) proteins belong to a subgroup of the large family of WD40 repeat proteins. The WD40 repeat domain consists of about 40 amino acid-conserved motifs terminating in a Trp-Asp (WD) dipeptide. WD40 repeat domains form a β-propeller structure that mediates a plethora of protein-protein interactions [1]. The fact that MSIL proteins have been discovered in diverse eukaryotes, ranging from yeasts to humans, but not in prokaryotes indicates that they have eukaryote-specific functions [2]. Among plants, thale cress (Arabidopsis thaliana) contains five MSIL proteins (MSI1-5), and rice (Oryza sativa) has three MSIL proteins (OsRBAP1-3). In vertebrates, two copies of MSIL proteins are usually found, such as RbAp48 and RbAp46 in humans, mice, chickens, and zebra fish. In fungi, one or two copies of MSIL proteins are usually found. A review by Hennig et al. [2] provides detailed information on the phylogenetic diversification of MSIL proteins.
MSIL proteins have multifunctional roles in histone binding, nucleosome/chromatin assembly and modification, DNA damage repairs, and regulation of signaling pathways. Because MSIL proteins have no catalytic functions, their roles are mainly mediated as adaptors or regulators. Although MSIL proteins have crucial roles in basic cellular functions and genome architecture in mammalian cells, they do not appear to be essential for cell growth in fungi. There are some exceptions, however, including one of the two MSIL proteins in Schizosaccharomyces pombe, Mis16, which is essential for viability because it is required for kinetochore assembly during mitosis [3].
In humans, two MSIL proteins, RbAp46/48 (retinoblastoma protein [pRB]-associated proteins 46/48), alone are able to interact directly with helix 1 of histone H4 not yet assembled to the chromatin [4]. By interacting with two larger components, an MSIL protein forms a heterotrimeric complex called chromatin assembly factor 1 (CAF-1), which assists in assembling the tetramers of two histone H3-H4 dimers on newly synthesized DNA to form nucleosomes. In addition, the MSIL proteins interact with histone modification enzymes, such as histone deacetylases (HDACs), acetyltransferases, and methyltransferases (see review [2]). Additionally, the mammalian MSIL proteins serve as a co-repressor for the pRB, a well-known tumor suppressor, by repressing the transcription of E2F target genes [5]. pRB can carry out transcriptional repression by recruiting HDACs [6-11].
MSIL proteins also have been studied extensively in plants, particularly Arabidopsis thaliana. Among the five Arabidopsis MSIL proteins [12-14], MSI2 and MSI3 are related genes, although their functions are not yet known. MSI4 and MSI5 are also related genes, and MSI4 is an allelic gene of FVE, which functions in the autonomous floral inductive pathway [15, 16] responsible for suppressing the floral repressor Flowering Locus C (FLC) [17]. Therefore, fve mutants exhibit delayed flowering behavior by increasing FLC expression [17]. Like other MSIL proteins, Arabidopsis MSI1 can be a part of several protein complexes. In particular, MSI1 is a subunit of the fertilization independent seed (FIS)-Polycomb repressive complex 2 (PRC2) complex consisting of FIS1 (MEDEA), FIS2, and FIS3 (FIE), which controls seed development and parental imprinting [18]. Additionally, MSI1 physically interacts with the CUL4-DDB1 complex, which regulates embryo formation and has a role in maintaining parental imprinting of MEDEA [19]. Thus, MSI1 is also associated with Arabidopsis CAF-1 [20, 21] and the retinoblastoma-related protein RBR1 [12, 22]. Furthermore, transcriptome profiling of msi1-cs (cosuppression line) plants reveals that abscisic acid-responsive genes (osmotic and salt stress-related genes) are upregulated in msi1-cs, suggesting a novel role of MSI1 for negative regulation in response to drought stress [23].
Despite their pleiotropic roles in multicellular, complex eukaryotes, knowledge of fungal MSIL proteins has largely stemmed from studies of MSIL proteins in the budding yeast model Saccharomyces cerevisiae, an ascomycete. Information about the functions of MSIL proteins in other fungi is quite limited, except for a recent publication about the role of MSIL protein Msl1 in the human fungal pathogen Cryptococcus neoformans, a basidiomycete [24]. Interestingly, this report showed that the MSIL protein in C. neoformans also has pleiotropic roles and yet appears to have a rather different regulatory mechanism from that of the MSIL proteins in S. cerevisiae. This finding raises the possibility that the functions and regulatory mechanisms of MSIL proteins are diverse throughout the fungal kingdom despite their evolutionary conservation. Several excellent reviews of MSIL proteins or WD40-repeat proteins are available [1, 2, 25-29]. In this review, we focus mainly on describing the functions of MSIL proteins in model yeasts and pathogenic fungi to discuss their potential as novel therapeutic targets.
FUNGAL MSIL PROTEINS AS COMPONENTS FOR THE CAF-1 AND HISTONE-MODIFYING COMPLEX
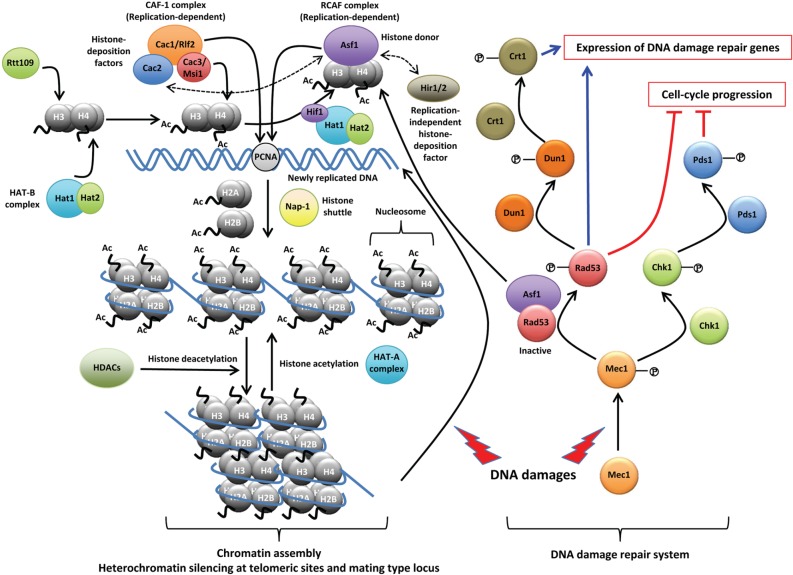
The primary function of two MSIL proteins, Cac3 (also known as Msi1) and Hat2, in the non-pathogenic model yeast S. cerevisiae is the assembly and modification of histones (Fig. 1) [30]. During cell division, newly replicated DNA is rapidly assembled into the chromatin, which consists of repeating units of nucleosomes. A single nucleosome is deposited with a tetramer of histone H3 and H4 followed by the addition of two dimers of the histones H2A and H2B. S. cerevisiae contains three CAF-1 proteins, designated Cac1 (also known as Rlf2; Rap1p localization factor 2), Cac2, and Cac3/Msi1, which correspond to the human CAF-1 proteins p150, p60, and p48, respectively. CAF-1 helps in the binding of histone H3 and H4 onto newly synthesized DNA during chromosome replication or onto recently repaired DNA sites after DNA damage by directly interacting with proliferating cell nuclear antigen, which is a DNA polymerase processivity factor [31-33]. Therefore, deletion of any of the CAC genes causes increased susceptibility to DNA damaging agents, such as UV irradiation, methyl methanesulfonate (MMS), and hydroxyurea (HU) [34]. Despite the seemingly essential role of the CAF-1 complex, deletion of any or all of these genes does not cause lethality, indicating that other chromatin assembly factor(s) exists [31, 35]. In fact, another chaperone for histones H3 and H4 is the replication-coupling assembly factor (RCAF) complex, which consists of anti-silencing function 1 (Asf1) and acetylated H3 and H4 histones [36]. It is known that RCAF and CAF-1 act synergistically for chromatin assembly by interacting with newly synthesized DNA during both DNA replication and DNA damage repair [36]. Expectedly, the asf1Δ cac1Δ double mutant grows much more poorly than does either mutant alone [36]. After DNA damages are repaired, CAF-1 and Asf1 work together to recover the normal chromatin structure at the repair sites and deactivate the checkpoint proteins to stop the double-strand break response [37].
Fig. 1.
The proposed roles of Msi1-like (MSIL) proteins in chromatin assembly and DNA damage repair. In Saccharomyces cerevisiae, two MSIL proteins, histone acetyltransferase 2 (Hat2) and Cac3/Msi1, are primarily involved in nucleosome assembly and chromatin assembly. As a component of the histone acetyltransferase B (HAT-B) complex, Hat2 assists Hat1, which binds to and acetylates newly synthesized histone H4. Then, the HAT-B complex transfers the histone H3/H4 tetramer into the nucleus. In the nucleus, the HAT-B complex forms a NuB4 complex, which delivers the H3/H4 tetramer to anti-silencing function 1 (Asf1) by interaction with the Hat1p-interacting factor 1 (Hif1) histone chaperone. The histone H3 in the delivered H3/H4 tetramer is acetylated by another nuclear histone, acetyltransferase Rtt109, associated with Asf1. As a histone chaperone, Cac3/Msi1 is the smallest subunit of the chromatin assembly factor 1 (CAF-1) complex, which additionally contains Cac1/Rlf2 (the largest subunit) and Cac2. CAF-1 is involved in chromatin assembly as a histone-deposition factor during DNA replication as well as DNA damage repair. The replication-coupled assembly factor (RCAF), composed of Asf1 and an acetylated tetramer of H3/H4, serves as a histone donor and also plays overlapping roles with CAF-1 in chromatin assembly during DNA replication. Histon regulatory (Hir) 1 and Hir2 are also histone-deposition factors, which act independently from DNA replication. Asf1 can interact with the Cac2 of CAF-1 and Hir1/2. CAF-1 assembles histone H3/H4 onto newly replicated DNA by targeting proliferating cell nuclear antigen. Two dimers of histones H2A and H2B are transferred by the nucleasome assembly protein 1 (Nap-1) which shuttles between the cytoplasm and the nucleus. Histone deacetylation by histone deacetylases (HDACs) facilitates aggregation of the nucleosome and results in chromatin assembly. This complex, compact structure of chromatin provides the basis for heterochromatin gene silencing at telomeric regions and mating type locus. When DNA damage occurs, such as double-strand DNA breaks, histones in the chromatin structure are dissociated and a series of DNA-repair signaling cascades, including Mec1, Rad53, Chk1, Pds1, Dun1, and Crt1, are activated to induce the expression of genes involved in DNA damage repair and to block the progression of cell-cycle. Notably, Asf1 physically interacts with Rad53 under normal conditions, and upon DNA damage, Asf1 and Rad53 phosphorylated by Mec1 are released, serving as a histone donor and activator of the DNA damage repair system, respectively. PCNA, proliferating cell nuclear antigen. For more detailed information, please refer to the following review [30].
In addition, CAF-1 is required to maintain heterochromatin gene silencing at telomeric sites and at the mating type locus (Fig. 1) [38]. For this position-dependent gene silencing, Asf1 also has been implicated because double deletion of CAC1 and ASF1 synergistically reduces heterochromatin gene silencing [36]. Asf1 requires interaction with histone regulatory (Hir) proteins, which are involved in nucleosome formation and heterochromatin gene silencing [39]. Therefore, loss of both the functional CAF-1 complex and the Hir proteins leads to increased chromosome missegregation, increased mutation in kinetochore protein genes, and altered centromeric chromatin structures [40].
Compared with CAC1 and CAC2, whose expression is differentially regulated during cell cycle progress, CAC3 expression is largely constant during the cell cycle [41]. In CAF-1, Cac1 physically interacts with Cac2 and Cac3 separately, whereas Cac2 and Cac3 do not interact with each other [42]. Interestingly, Cac1 is usually co-purified with Cac2 and Cac3, but Cac3 is not always purified with Cac1 or Cac2, suggesting that Cac3 has other cellular roles or interaction partners [43]. Cac1 was independently identified as one of the Cac3/Msi1-interacting proteins through yeast two-hybrid screening with Cac3 as the bait protein [44]. Interaction between Cac1 and Cac3 does not require Cac2 [44]. Furthermore, Cac3 localizes to both the nucleus and the cytoplasm, whereas Cac1 primarily localizes to the nucleus [45].
S. cerevisiae contains a second MSIL protein, Hat2, which is a core component of histone acetyltransferases (HATs). Hat2 is the regulatory subunit of the yeast HAT-B (type B HAT) complex composed of Hat1 and Hat2 in the cytoplasm that acetylates newly synthesized histones. In the nucleus, the HAT-B complex further interacts with Hat1p-interacting factor 1 (Hif1), causing the Nub4 (nuclear type B HAT; Hat1-Hat2-Hif1) complex to mediate the DNA repair-linked chromatin reassembly process (Fig. 1) [46, 47]. In contrast, another HAT family, the HAT-A (type A HAT) complex, localizes in the nucleus, where the complex acetylates histones in a chromatin context to facilitate transcriptional activation [48]. Hat1 is the catalytic subunit of the HAT-B complex and acetylates H4 on lysine residues 5 and 12 [49]. Hat1, which is not an MSIL protein, is involved in histone deposition, nucleosome formation, and chromatin assembly during the progression of DNA replication, and it is required for histone modifications during DNA repair [50, 51]. Hat2 has an adaptor role between the Hat1 and Hif1 proteins and is responsible for the high affinity binding of Hat1 to histone H4, while Hif1p is a histone chaperone [47]. In the absence of either Hat1 or Hat2, no growth deficiencies have been reported [51]. Hat1 and Hat2 are involved in telomeric silencing [50]. Notably, however, in transcriptome analysis, hat1Δ and hat2Δ mutants exhibit distinct transcriptional patterns in yeast cells [52]. In addition, deletion of HAT2, but not HAT1, expands the chronological lifespan of yeast [52], providing further support for the suggestion that Hat1 and Hat2 do not have completely overlapping functions in the cell. Recently, the genetic interaction of the Hat1-Hat2 complex with origin-recognition complex, which is required for the initiation of chromosomal DNA replication, has been identified [53].
Besides studies in S. cerevisiae, a single study by Yang et al. [24] has investigated the role of an MSIL protein in C. neoformans, which is a basidiomycetous fungus that causes fatal meningoencephalitis in both immunocompromised and immunocompetent individuals. A single gene (CNAG_03297.2) called MSI1-like gene 1 (MSL1) was identified as the closest Cryptococcus ortholog to both the yeast Msi1 and Hat2. Similar to the yeast Cac3/Msi1, Msl1 also appears to promote resistance to DNA damaging agents, such as UV irradiation, HU, and MMS, indicating that Msl1 may also have a role as a component of the CAF-1-like complex in C. neoformans (Fig. 1) [24]. As further evidence in support of this idea, Msl1 localizes to both the cytoplasm and nucleus but is more enriched in the nucleus [24]. Furthermore, the Cac2 ortholog of the CAF-1 complex was identified in C. neoformans, and its deletion mutant (cac2Δ) shows hypersensitivity to genotoxic agents such as the msl1Δ mutant [24]. Despite the presence of the conserved Msl1 (Cac3 ortholog) and Cac2, rather unique features have been discovered in the CAF-1 components in C. neoformans. First, MSL1 does not functionally complement the yeast CAC3 (unpublished data by Y.-S. Bahn). Second, Cryptococcus does not contain an obvious Cac1 ortholog, which is a key CAF-1 protein that interacts with both Cac2 and Msi1/Cac3. It is conceivable that Cryptococcus may have a structurally distinct Rlf2/Cac1-like protein, which might have unique interfaces that interact separately with Msl1 and Cac2. This may explain why Msl1 cannot be substituted for Msi1 in S. cerevisiae. To investigate the function of CAF-1 in Cryptococcus, an Rlf2/Cac1-like protein must be identified and functionally characterized with relation to Cac2 and Msl1 in C. neoformans.
FUNGAL MSIL PROTEINS AS REGULATORS OF THE NUTRIENT- AND STRESS-SENSING SIGNALING PATHWAYS
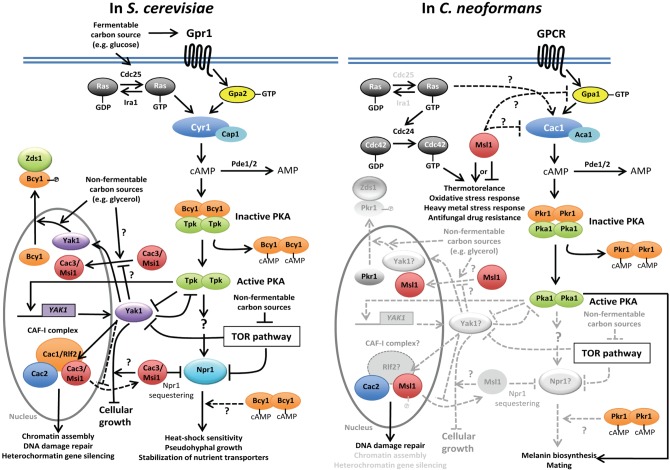
Between the two MSIL proteins, Msi1/Cac3 appears to have additional cellular roles besides its function in histone and chromatin assembly and modification. In fact, Msi1 was first identified as a negative regulator of the Ras/cAMP pathway in S. cerevisiae because it was screened as multicopy suppressors of the ira1Δ mutation [54]. The Ras/cAMP pathway is a nutrient-sensing signaling pathway required for the growth, stress response, and differentiation of S. cerevisiae (Fig. 2) [55]. Under certain environmental stresses, the Cyr1/Cdc35 adenylyl cyclase produces cAMP, which binds to Bcy1, the regulatory subunit of protein kinase A (PKA), and releases the catalytic subunit PKA (Tpk1/Tpk2/Tpk3) from Bcy1. The released Tpk proteins regulate downstream transcription factors to respond to environmental stresses [56]. The cAMP pathway is negatively controlled by feedback regulation from phosphodiesterase (Pde) 1 and Pde2 [57, 58]. Upstream of Cyr1, two bifurcated signaling branches exist: a heterotrimeric GTP-binding protein (G-protein) and small G-protein Ras. Ras1 and Ras2 are activated by Cdc25 guanine nucleotide exchange factor, which enhances the replacement of GDP with GTP [59]. Ira1 and Ira2, which are GTPase-activating proteins, negatively control Ras activation, in contrast to Cdc25 [60]. Alternatively, a G-protein coupled receptor (GPCR) associated with a heterotrimeric G-protein complex (Gα, Gβ, and Gγ) receives signals, such as glucose addition, and undergoes conformational changes with the release of the active GTP-binding form of the Gα subunit [61, 62]. Activated Ras or Gα subunit activates Cyr1/Cdc35 adenylyl cyclase directly and subsequently controls PKA (Fig. 2) [55, 63].
Fig. 2.
The proposed roles of Msi1-like (MSIL) proteins as regulators of the nutrient- and stress-sensing signaling pathways in fungi. In Saccharomyces cerevisiae, Cac3/Msi1 likely acts downstream of protein kinase A (PKA) in the presence of functional Bcy1. In response to certain environmental cues (such as glucose addition), adenylyl cyclase (Cyr1/Cdc35) is activated by either activated GTP-bound Ras proteins or activated GTP-bound Gpa2 Ga subunit through the Gpr1 G-protein coupled receptor (GPCR) to produce cAMP, which is a secondary messenger. Synthesized cAMP either is degraded to AMP by phosphodiesterases (Pde1/2) as a negative feedback regulation or binds to the regulatory subunit (Bcy1) of PKA, which normally suppresses the activation of the catalytic subunits (Tpk1/2/3) of PKA. Released and activated Tpks, which have redundant and unique roles, phosphorylate or activate diverse target genes. Downstream of PKA, nitrogen permease reactivator 1 (Npr1) activates downstream effector proteins, mediating heat shock sensitivity or stabilization of nutrient transporters, which otherwise are degraded by ubiquitination. Cac3/Msi1 normally sequesters Npr1 and inhibits its activation. At this step, it is unclear how Bcy1 acts on the Cac3/Msi1 (depicted as dotted arrow). Activated PKA also phosphorylates and inhibits yet another kinase 1 (Yak1), which antagonizes the Ras/cAMP signaling, although the YAK1 transcription itself is also activated by PKA, perhaps as an autoregulatory loop. Interestingly, Yak1 stabilizes the interaction between Cac1 and Cac3/Msi1 in the nucleus without affecting other CAF-1 interactions. Under non-fermentable carbon sources, Yak1 inhibits the nuclear localization of Cac3/Msi1 without phosphorylation, whereas Yak1 phosphorylates a nuclear-localized Bcy1 and facilitates its export to the cytoplasm. Conversely, Msi1 can also control Yak1. Overexpression of YAK1 confers growth arrest only in the presence of Msi1. In Cryptococcus neoformans, however, the regulatory mechanism of the Msl1 MSIL protein in terms of the Ras and cAMP/PKA pathways is rather different from that in S. cerevisiae. First, the Ras signaling pathway is less functionally connected to the cAMP pathway and instead governs thermotolerance and other stress-related phenotypes by controlling an additional GTPase protein Cdc42 and a Cdc24 guanine nucleotide exchange factor protein. All of the Msl1-related phenotypes appear to be independent from the Ras-signaling pathways. Second, there is no evidence showing that Msl1 acts downstream of PKA in C. neoformans. In fact, the negative role of Msl1 in melanin and mating efficiency is abolished by the loss of Cac1, indicating that Msl1 may act on Cac1 or its upstream factors (depicted dotted lines). In C. neoformans, Yak1-like or Npr1-like kinases have not been identified or functionally characterized. Therefore, physical and functional relationships between Msl1 and other PKA downstream kinases remain to be elucidated in future studies (in gray symbols and dotted arrows). CAF-1, chromatin assembly factor 1. Zds1, zillion different screens 1.
The multicopy expression of MSI1 inhibits the phenotypes of ira1 mutation-mediated Ras over-activation, such as heat-shock sensitivity [54]. Interestingly, overexpression of human RbAp48 can also suppress the over-activated Ras/cAMP pathway in S. cerevisiae [64]. The mechanism of how Msi1 regulates the Ras/cAMP pathway, however, has not been completely elucidated (Fig. 2). The step at which Msi1 works in the cAMP-signaling pathway was originally proposed to be upstream of PKA because MSI1 overexpression suppresses heat-shock sensitive phenotypes caused by the ira1Δ or RAS2G19V mutation, but not by the bcy1Δ mutation [54]. Furthermore, the overexpression of MSI1 decreases the intracellular cAMP levels induced by glucose addition, suggesting that Msi1 may regulate Ras1, like Ira1 or Cyr1 [54]. Due to the similar β-propeller structures, Ruggieri et al. [54] proposed that Msi1 is similar to the β-subunit of the G-protein. Nevertheless, the msl1Δ deletion mutant does not show any cAMP-related phenotypes, suggesting that other MSI1-like genes may exist [44, 54]. The role of Msi1/Cac3 as a negative regulator of the Ras/cAMP-signaling pathway is independent from the function of the CAF-1 complex because overexpression of MSI1/CAC3 can still suppress the overactive phenotypes of the RAS2G19V mutant in the cac1Δ or cac2Δ mutant backgrounds [45]. Furthermore, functional Gpr1 (GPCR)/Gpa1 (Gα-subunit) signaling is dispensable for phenotypic recovery of the RAS2G19V mutant by MSI1/CAC3 overexpression [45].
Nevertheless, a series of studies have been reported to show that Msi1 may work downstream of Cyr1. Overexpression of MSI1 does not significantly affect intracellular cAMP levels or PKA activity in itself [44]. In addition, the heat-shock sensitivity caused by the deletion of two phosphodiesterase genes, PDE1 and PDE2, can be suppressed by the overexpression of MSI1 even in the presence of increased intracellular cAMP levels, but not in the bcy1Δ mutant [44]. These data suggest that Msi1 may work downstream of PKA, but in a Bcy1-dependent manner. In efforts to identify the Cac3/Msi1-interacting proteins, Johnston et al. [45] performed yeast two-hybrid screening with CAC3/MSI1 as the bait and identified the nitrogen permease reactivator 1 (NPR1) gene, which encodes a Ser/Thr protein kinase. It appears that the Npr1 kinase, which is one of the positive PKA targets, is normally sequestered and inactivated by Msi1. Upon release from Msi1-mediated inhibition, the activated Npr1 can phosphorylate and stabilize a number of nutrient transporters, such as Gap1 (a general amino acid permease) [65] and Mep2 (an ammonium permease) [66], which otherwise is normally degraded by the ubiquitin-mediated proteasome (Fig. 2) [44, 45]. In a supportive finding, the phenotypic restoration caused by MSI1 overexpression in the RAS2G19V mutant is inhibited by the overexpression of NPR1, and ubiquitin overexpression can suppress the RAS2G19V mutant [45]. Another piece of supporting evidence for this working model is that the expression of the target genes downstream of the Ras/cAMP pathway, such as TPS1 and HSP104, is not affected by MSI1 overexpression, which indicates that Msi1 may negatively regulate the Ras/cAMP pathway by controlling protein degradation, possibly through Npr1 and ubiquitination rather than through transcriptional regulation of target genes.
In addition, Msi1 has mutual interdependence with yet another kinase 1 (Yak1), which is a dual-specificity tyrosine-regulated protein kinase that antagonizes the Ras/cAMP pathway of S. cerevisiae (Fig. 2) [42]. YAK1 was initially identified as a gene whose deletion rescues the growth defect caused by the loss of the cAMP-dependent PKA function or Ras function [67]. In other reports, it was proposed that PKA directly phosphorylates the Yak1 kinase in vitro [68] and indirectly activates transcription of the YAK1 gene, suggesting that Yak1 antagonizes the Ras/cAMP signaling with an autoregulatory loop [69]. Yak1 has additional functions in regulating several cellular processes. In the absence of YAK1, strong pseudohyphal growth induced by either constitutive over-activation of Ras or catalytic subunits of PKA appeared to be blocked [70]. Indeed, the yak1Δ mutant shows 50- to 100-fold enhanced sensitivity in response to heat-shock, which was also observed in strains with low PKA activity [71]. This heat shock sensitivity is suppressed by overexpression of MSI1 or deletion of NPR1, which implies that Npr1 sequestration by Msi1 regulates the downstream targets of the Ras/cAMP pathway in a Yak1-dependent manner [42]. Additionally, the interaction between Msi1 and Cac1 for the formation of the CAF-1 complex requires Yak1 [42]. Therefore, it is possible that the deletion of YAK1 could release Msi1 from CAF-1, which subsequently sequesters Npr1 to inactivate the Ras/cAMP signaling (Fig. 2). However, the interaction of either Cac1-Cac2 or Msi1-Npr1 occurs in a Yak1-independent manner. Yak1 affects the subcellular location of Msi1 as well as the interaction of Msi1 with Cac1. Msi1 is normally localized in both the cytoplasm and nucleus when the cells were grown with a fermentable carbon source (e.g., glucose), and this localization is Yak1-independent. Under the growth condition with non-fermentable carbon sources (e.g., glycerol or ethanol), however, Msi1 accumulates in the nucleus, and this nuclear accumulation is blocked by overexpression of YAK1 [42]. In contrast to the Yak1-dependent regulation of Msi1, Yak1 itself also appears to be controlled by Msi1. Yak1 negatively regulates the cell growth of S. cerevisiae, and the overproduction of YAK1 thereby arrests the cell cycle. The cell growth defect caused by an excess of YAK1 is diminished in the msi1Δ mutant, whereas deletion of CAC1 does not restore the growth defect by YAK1 overexpression; this indicates that Yak1 has a role in cell growth in an Msi1-dependent manner but in a CAF-1-independent manner [42]. Therefore, Msi1 and Yak1 mutually control each other.
The functions and regulatory mechanisms of Msl1 in the Ras and cAMP/PKA signaling pathways of C. neoformans are somewhat different from those of the yeast Msi1 (Fig. 2). In fact, the Ras and cAMP/PKA-signaling pathways are not strongly connected in C. neoformans, as they control distinct cellular features. The cAMP/PKA pathway positively controls two major virulence factors of C. neoformans, the polysaccharide cell surface capsule and antioxidant melanin pigment and sexual differentiation (see reviews [72, 73]). In contrast, the Ras-signaling pathway does not significantly control capsule and melanin biosynthesis, but it affects sexual differentiation (see reviews [72, 73]). Instead, the Ras-signaling pathway has a major role in controlling growth at high temperatures, including the host physiological temperature (37℃) [74]. In a supportive finding, transcriptome profiles between ras1Δ and cAMP-signaling mutants have been found to be quite different in C. neoformans [75]. Msl1 mainly acts independently from the Ras signaling pathway in C. neoformans. Msl1 has pleiotropic roles in diverse stress responses. Similar to the ras1Δ mutant, the msl1Δ mutant shows increased susceptibility to high temperature and membrane destabilizers, such as sodium dodecyl sulfate, and the msl1Δ ras1Δ mutant shows even greater susceptibility [24]. In contrast, Msl1 is involved in other stress responses, such as oxidative and heavy metal stress responses, unlike Ras2. Therefore, it is likely that the Msl1 and Ras signaling pathways mainly control the stress response of C. neoformans independently. In contrast to the Ras-signaling pathway, the cAMP/PKA pathway is partly related to the Msl1-signaling pathway. Msl1 appears to negatively regulate the cAMP-signaling pathway in melanin production and sexual reproduction. Distinct from yeast Msi1, however, deletion of the adenylyl cyclase gene (CAC1) can completely abolish the effect of MSL1 deletion, indicating that Msl1 may act upstream of Cac1 in the cAMP pathway [24]. Strangely, capsule production, which is known to be positively controlled by the cAMP pathway, is not affected by the msl1Δ mutation [24]. In conclusion, the role of the Msi1/Cac3-like protein as regulators of the nutrition- and stress-signaling pathways appears to be divergent among fungi.
FUNGAL MSIL PROTEINS AS DEVELOPMENTAL REGULATORS
In plants, MSIL proteins are known to repress genes involved in the developmental process and sexual reproduction. In Drosophila, the MSIL protein p55 is required for the repression of developmentally regulated dE2F2/RBF-target genes but not for proliferation-regulated E2F target genes [76]. Furthermore, expression of p55 is considerably higher in actively differentiating cells than in non-differentiating ones [77]. In fungi, also, several lines of evidence strongly suggest that MSIL proteins could be involved in the developmental process and morphological differentiation. In S. cerevisiae, expression of MSI1 is greatly increased in meiotic division during sporulation [78], although the physiological meaning of this finding is not known. Msi1 serves as a co-repressor for pRB and helps recruit Rpd3 HDAC to pRB [79]. pRB controls differentiation as well as proliferation by repressing the target genes of the E2F transcription factor [79]. The role of Msi1 in the transcriptional repression of pRB is independent of its function as a component of the CAF-1 complex. In C. neoformans, deletion of MSL1 significantly increases mating efficiency [24]. The repressing role of Msl1 in mating requires the functional components in the Ras or cAMP pathway because deletion of genes in these signaling pathways abolishes the enhanced mating effect by the msl1Δ mutation [24]. Taken together, the role of MSIL proteins in the developmental program and differentiation appears to be highly conserved in fungi.
MSIL PROTEINS AS TRANSCRIPTIONAL ACTIVATORS IN THE PRESENCE OF NON-FERMENTABLE CARBON SOURCES
Pratt and colleagues reported an additional interesting function of Msi1: a cryptic transcriptional activation activity in non-fermentable carbon sources in S. cerevisiae [42]. Surprisingly, Msi1 fused to the Gal4 DNA-binding domain exhibits one-hybrid activity (the ability of transcriptional activation) only in the presence of non-fermentable carbon sources (e.g., glycerol, ethanol, acetate, lactase, etc.), but not in the presence of fermentable carbon sources (e.g., glucose, fructose, etc.) [42]. This activity requres only Yak1 and not CAF-1 and Ras/cAMP/Npr1-signaling, whereas Yak1 alone is not sufficient for this Msi1 function. During growth of S. cerevisia on non-fermentable carbon sources, the total protein accumulation of Msi1 (not by transcriptional induction) and its nuclear localization both increase, whereas Msi1 localizes to both the cytoplasm and the nucleus during growth on glucose [42]. The accumulation of Msi1 protein does not require Yak1. Instead, Yak1 inhibits nuclear localization of Msi1, although Yak1 does not phosphorylate Msi1 [42]. The in vivo target genes regulated by Msi1 and other unknown co-factors under non-fermentable carbon conditions have not been discovered. Furthermore, it is still not known whether this carbon-source-regulated function of the MSIL protein is conserved in C. neoformans and other fungi.
MSIL PROTEINS IN OTHER FUNGAL PATHOGENS
So far, the functions and regulatory mechanisms of MSIL proteins in fungi have been studied only in non-pathogenic S. cerevisiae and pathogenic C. neoformans. Most of the animal and plant fungal pathogens appear to have at least one MSIL protein, containing four to six intact WD40 repeats (Fig. 3). In S. cerevisiae, Msi1 has seven WD40 repeats (six intact WD40), and the N-terminal and C-terminal domains have separate functions. The first four WD40 repeats are required for sequestration of Npr1 and suppression of Ras/cAMP signaling, whereas the last three WD40 repeats are required for interaction with Cac1 of CAF-1 and for the cryptic ability to activate transcription in the presence of non-fermentable carbon sources (Fig. 3) [42]. Particularly, the latter functions of the Msi1 C-terminus require Yak1 [42]. Most MSIL-like proteins share certain features in common, such as the length of the amino acid sequences and the locus of the conserved WD40 repeats. Generally, the C-terminal WD40 domain structure is more conserved than the N-terminal WD40 domain structure, where less conserved WD40 motifs are often discovered. In addition, C. glabrata XP_448393 and A. gossypii NP_984338 have an extra-extended N-terminus compared with the others. This finding may indicate that the CAF-1 function of MSIL proteins could be more conserved among other fungi than their Npr1-sequestration function for suppressing Ras/cAMP signaling.
Fig. 3.
The conserved WD40 repeat domains in pathogenic fungal Msi1-like (MSIL) proteins. The domain architecture of the MSIL proteins with the conserved WD40 repeats is shown by the multiple sequence alignment and sequence comparison of the MSIL proteins. Each fungal MSIL protein sequence was retrieved by protein blast search (blastp) using the Saccharomyces cerevisiae Msi1 protein sequence as the query in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Phanerochaete chrysosporium database JGI (http://genome.jgi-psf.org/Phchr1/Phchr1.info.html). The purple box is the complete conserved WD40 domain containing the WD, FD, or YD terminal dipeptide. The grey box is the WD40-like domain without the conserved terminal WD dipeptide. The pink box is the last WD40 domain that contains the conserved Trp or random amino acid (WX) as a terminal motif. aa, amino acids; WD, Trp-Asp; FD, Phe-Asp; YD, Tyr-Asp; Npr1, nitrogen permease reactivator 1; Yak1, yet another kinase 1; S. cerevisiae, Saccharomyces cerevisiae; S. pombe, Schizosaccharomyces pombe; H. sapiens, Homo sapiens; A. clavatus, Aspergillus clavatus; A. fumigatus, Aspergillus fumigatus; A. terreus, Aspergillus terreus; C. albicans, Candida albicans; C. glabrate, Candida glabrate; C. lusitaniae, Candida lusitaniae; C. immitis, Coccidiodes immitis; C. neoformans, Cryptococcus neoformans; E. cuniculi, Encephalitozoon cuniculi; A. gossypii, Ashbya gossypii; C. gloeospiriodes, Colletotrichum gloeospiriods; G. graminicola, Glomerella graminicola; G. zeae, Gibberella zeae; M. oryzae, Mangnaporthe oryzae; P. chrysospotium, Phanerochaete chrysosporium; S. sclerotiorum, Sclerotinia sclerotiorum; U. maydis, Ustilago maydis; Z. tritici, Zymoseptoria tritici.
When the structural organization of the WD40 repeats of MSIL proteins in fungal pathogens were further compared, the following common features were discovered. First, all of the MSIL proteins in the fungal pathogens contain a starting WD40-like domain without a conserved terminal WD dipeptide at the N-terminus, unlike the S. cerevisiae Msi1 and human MSIL proteins (Fig. 3). Second, the terminal WD40 domains always contain the conserved Trp or random amino acid (WX) motif (Fig. 3). The MSIL proteins in fungal pathogens show a greater than 40% sequence identity to C. neoformans Msl1 (AEX_58661). Therefore, it seems that the general structural features of MSIL proteins in fungal pathogens are more similar to C. neoformans Msl1 than to S. cerevisiae Msi1. This structural similarity suggests a potential role for other MSIL proteins in fungal pathogenicity, based on the role of Msl1 in the virulence of C. neoformans.
POTENTIAL FOR MSIL PROTEINS AS NOVEL ANTIFUNGAL DRUG TARGETS
The potential to exploit MSIL proteins as novel antifungal drug targets has been suggested by studies in C. neoformans [24]. A strain with a MSL1 deletion exhibits attenuated virulence [24]. Interestingly, however, the msl1Δ mutant was found to enhance azole resistance, suggesting that a potential inhibitor of Msl1 or its related signaling components may have antagonistic effects with azole drugs. Using direct inhibitors of MSIL proteins as antifungal drugs is not likely to be a good approach because MSIL proteins are evolutionarily conserved throughout eukaryotes. Instead, however, fungal-specific regulators downstream of MSIL or the manipulation of specific interactions between the MSIL protein and its binding partner could be alternative therapeutic methods. For example, if there is an Rlf2/Cac1-like protein in the CAF-1 complex of C. neoformans, it could be quite divergent from its counterpart in yeast or humans.
Transcriptome analysis performed in C. neoformans uncovered several Msl1-regulated genes, which could potentially contribute to the virulence of the pathogen [24]. In line with a previous finding that the expression of some target genes of cAMP-signaling is not affected by msi1 mutation in S. cerevisiae [44, 45], only a small number of genes (17 genes) are differentially regulated by msl1 mutation in C. neoformans. This fact indicates that an MSIL protein alone may not be sufficient to negatively regulate the Ras/cAMP-signaling pathway at full scale. Interestingly, a significant portion of Msl1-regulated genes are stress-related, including Hsp12 (small heat shock proteins) and Hsp78 (an oligomeric mitochondrial matrix chaperone), further supporting the role of Msl1 in stress response and adaptation in C. neoformans. Because MSIL proteins appear to play physiological roles by interacting with other proteins, such as the CAF-1 components, pRB, Npr1, and Yak1 in yeast, Msl1-interacting proteins must be screened in C. neoformans and other pathogens. For example, an ortholog for one of the yeast Msi1-interacting kinases, Npr1, does not seem to be found in the C. neoformans genome, suggesting the possibility that novel MSIL-interacting proteins may exist in fungal pathogens.
CONCLUSION AND FUTURE RESEARCH DIRECTIONS
MSIL proteins with WD40 repeats have pleiotropic roles as regulators of stress-signaling pathways such as Ras/cAMP and other pathways, as a component of the chromatin-assembly factor and as a developmental regulator in pathogenic and non-pathogenic model yeasts. In particular, in pathogenic fungi such as C. neoformans, MSIL proteins govern the production of virulence factors and in vivo virulence, suggesting that MSIL-related signaling pathways could be developed as novel antifungal drug targets. Despite their evolutionary conservation and multi-functionality, the functions of MSIL in other pathogenic yeasts and filamentous fungi, such as Candida albicans, Candida glabrata, and Aspergillus fumigatus, remain completely unknown. Therefore, the following questions need to be further addressed in future studies.
1. Are such pleiotropic functions of MSIL proteins conserved in other ascomycete fungi, such as S. pombe and Neurospora crassa, similar to those in S. cerevisiae?
2. How divergent among fungi is the chromatin assembly factor, functionally and structurally?
3. Are Msi1-interacting proteins discovered in S. cerevisiae, such as the CAF-1 complex, Npr1, and Yak1, evolutionarily and functionally conserved in C. neoformans and other fungi?
4. Is the role of MSIL proteins in virulence conserved in other pathogenic fungi?
5. If MSIL proteins are conserved in other pathogenic fungi, are they functionally connected to the Ras and/or cAMP/PKA signaling pathways?
6. Is the role of MSIL proteins in development and morphological differentiation conserved in filamentous fungi?
7. Is the function of MSIL proteins in sensing and responding to non-fermentable carbon sources conserved in C. neoformans and other fungi?
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MEST) (Nos. 2010-0029117 and 2008-0061963).
References
- 1.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 2.Hennig L, Bouveret R, Gruissem W. MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 2005;15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 5.Ross JF, Liu X, Dynlacht BD. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- 6.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 8.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci U S A. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iavarone A, Massagué J. E2F and histone deacetylase mediate transforming growth factor beta repression of cdc25A during keratinocyte cell cycle arrest. Mol Cell Biol. 1999;19:916–922. doi: 10.1128/mcb.19.1.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiegler P, De Luca A, Bagella L, Giordano A. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 1998;58:5049–5052. [PubMed] [Google Scholar]
- 12.Ach RA, Taranto P, Gruissem W. A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell. 1997;9:1595–1606. doi: 10.1105/tpc.9.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenzior AL, Folk WR. AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Lett. 1998;440:425–429. doi: 10.1016/s0014-5793(98)01500-2. [DOI] [PubMed] [Google Scholar]
- 14.Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- 15.Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- 17.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci U S A. 2000;97:10637–10642. doi: 10.1073/pnas.170292997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumbliauskas E, Lechner E, Jaciubek M, Berr A, Pazhouhandeh M, Alioua M, Cognat V, Brukhin V, Koncz C, Grossniklaus U, et al. The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. Embo J. 2011;30:731–743. doi: 10.1038/emboj.2010.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaya H, Shibahara KI, Taoka KI, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 21.Exner V, Taranto P, Schönrock N, Gruissem W, Hennig L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development. 2006;133:4163–4172. doi: 10.1242/dev.02599. [DOI] [PubMed] [Google Scholar]
- 22.Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandre C, Möller-Steinbach Y, Schönrock N, Gruissem W, Hennig L. Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant. 2009;2:675–687. doi: 10.1093/mp/ssp012. [DOI] [PubMed] [Google Scholar]
- 24.Yang DH, Maeng S, Strain AK, Floyd A, Nielsen K, Heitman J, Bahn YS. Pleiotropic roles of the Msi1-like protein Msl1 in Cryptococcus neoformans. Eukaryot Cell. 2012;11:1482–1495. doi: 10.1128/EC.00261-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 26.Smith TF. Diversity of WD-repeat proteins. Subcell Biochem. 2008;48:20–30. doi: 10.1007/978-0-387-09595-0_3. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 32.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 33.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 34.Game JC, Kaufman PD. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 36.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 37.Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 40.Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3277. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pratt ZL, Drehman BJ, Miller ME, Johnston SD. Mutual interdependence of MSI1 (CAC3) and YAK1 in Saccharomyces cerevisiae. J Mol Biol. 2007;368:30–43. doi: 10.1016/j.jmb.2007.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marheineke K, Krude T. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Démolis N, Jacquet M, Michaeli T. MSI1 suppresses hyperactive RAS via the cAMP-dependent protein kinase and independently of chromatin assembly factor-1. Curr Genet. 2000;38:60–70. doi: 10.1007/s002940000133. [DOI] [PubMed] [Google Scholar]
- 45.Johnston SD, Enomoto S, Schneper L, McClellan MC, Twu F, Montgomery ND, Haney SA, Broach JR, Berman J. CAC3 (MSI1) suppression of RAS2G19V is independent of chromatin assembly factor I and mediated by NPR1. Mol Cell Biol. 2001;21:1784–1794. doi: 10.1128/MCB.21.5.1784-1794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ge Z, Wang H, Parthun MR. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem. 2011;286:16790–16799. doi: 10.1074/jbc.M110.216846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poveda A, Pamblanco M, Tafrov S, Tordera V, Sternglanz R, Sendra R. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 48.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 50.Kelly TJ, Qin S, Gottschling DE, Parthun MR. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol Cell Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 52.Rosaleny LE, Antúnez O, Ruiz-García AB, Pérez-Ortín JE, Tordera V. Yeast HAT1 and HAT2 deletions have different life-span and transcriptome phenotypes. FEBS Lett. 2005;579:4063–4068. doi: 10.1016/j.febslet.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 53.Suter B, Pogoutse O, Guo X, Krogan N, Lewis P, Greenblatt JF, Rine J, Emili A. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007;5:38. doi: 10.1186/1741-7007-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 56.Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sass P, Field J, Nikawa J, Toda T, Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson RB, Tatchell K. SRA5 encodes the low-Km cyclic AMP phosphodiesterase of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:505–510. doi: 10.1128/mcb.8.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson LC, Gibbs JB, Marshall MS, Sigal IS, Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987;235:1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- 61.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 62.Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 63.Fedor-Chaiken M, Deschenes RJ, Broach JR. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990;61:329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- 64.Qian YW, Wang YC, Hollingsworth RE, Jr, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 65.Vandenbol M, Jauniaux JC, Grenson M. The Saccharomyces cerevisiae NPR1 gene required for the activity of ammonia-sensitive amino acid permeases encodes a protein kinase homologue. Mol Gen Genet. 1990;222:393–399. doi: 10.1007/BF00633845. [DOI] [PubMed] [Google Scholar]
- 66.Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- 68.Garrett S, Menold MM, Broach JR. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol Cell Biol. 1991;11:4045–4052. doi: 10.1128/mcb.11.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Smith MM, Mymryk JS. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol Biol Cell. 2001;12:699–710. doi: 10.1091/mbc.12.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartley AD, Ward MP, Garrett S. The Yak1 protein kinase of Saccharomyces cerevisiae moderates thermotolerance and inhibits growth by an Sch9 protein kinase-independent mechanism. Genetics. 1994;136:465–474. doi: 10.1093/genetics/136.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, Cardenas ME. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 73.Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–380. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 75.Maeng S, Ko YJ, Kim GB, Jung KW, Floyd A, Heitman J, Bahn YS. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans. Eukaryot Cell. 2010;9:360–378. doi: 10.1128/EC.00309-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor-Harding B, Binné UK, Korenjak M, Brehm A, Dyson NJ. p55, the Drosophila ortholog of RbAp46/RbAp48, is required for the repression of dE2F2/RBF-regulated genes. Mol Cell Biol. 2004;24:9124–9136. doi: 10.1128/MCB.24.20.9124-9136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyler JK, Bulger M, Kamakaka RT, Kobayashi R, Kadonaga JT. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 79.Kennedy BK, Liu OW, Dick FA, Dyson N, Harlow E, Vidal M. Histone deacetylase-dependent transcriptional repression by pRB in yeast occurs independently of interaction through the LXCXE binding cleft. Proc Natl Acad Sci U S A. 2001;98:8720–8725. doi: 10.1073/pnas.151240898. [DOI] [PMC free article] [PubMed] [Google Scholar]