Abstract
Bisphenol-A (BPA), a ubiquitous environmental endocrine disrupting chemical, is a component of polycarbonate plastic and epoxy resins. Because of its estrogenic properties, there is increasing concern relative to risks from exposures during critical periods of early organ differentiation. Prenatal BPA treatment in sheep results in low birth weight, hypergonadotropism, and ovarian cycle disruptions. This study tested the hypothesis that gestational exposure to bisphenol A, at an environmentally relevant dose, induces early perturbations in the ovarian transcriptome (mRNA and microRNA). Pregnant Suffolk ewes were treated with bisphenol A (0.5 mg/kg, sc, daily, produced ∼2.6 ng/mL of unconjugated BPA in umbilical arterial samples of BPA treated fetuses approaching median levels of BPA measured in maternal circulation) from days 30 to 90 of gestation. Expression of steroidogenic enzymes, steroid/gonadotropin receptors, key ovarian regulators, and microRNA biogenesis components were measured by RT-PCR using RNA derived from fetal ovaries collected on gestational days 65 and 90. An age-dependent effect was evident in most steroidogenic enzymes, steroid receptors, and key ovarian regulators. Prenatal BPA increased Cyp19 and 5α-reductase expression in day 65, but not day 90, ovaries. Fetal ovarian microRNA expression was altered by prenatal BPA with 45 down-regulated (>1.5-fold) at day 65 and 11 down-regulated at day 90 of gestation. These included microRNAs targeting Sry-related high-mobility-group box (SOX) family genes, kit ligand, and insulin-related genes. The results of this study demonstrate that exposure to BPA at an environmentally relevant dose alters fetal ovarian steroidogenic gene and microRNA expression of relevance to gonadal differentiation, folliculogenesis, and insulin homeostasis.
The differentiation of organ systems depends on precise exposure to steroid hormones at specific times during development. Therefore, abnormal exposure to compounds (native or environmental steroids) during critical periods of development can interfere with normal homeostasis and lead to long-term deleterious effects at the reproductive and metabolic levels. In humans, inappropriate exposure to excess native or environmental steroids can occur early during development from disease states, use of anabolic steroids, continued use of contraceptive pills (1, 2), or unintended exposure to environmental steroid mimics (3). The current increase in the prevalence of some common reproductive and metabolic diseases (4) may stem from inadvertent exposure during development to endocrine disrupting chemicals (EDCs), which can adversely influence the developmental trajectory of target tissue differentiation.
EDCs are a broad group of substances that include persistent pollutants, chemicals used in plant or animal food production, and plant-derived substances as well as industrial compounds used in consumer products (3). Bisphenol A (BPA), one such EDC, is a component of polycarbonate plastic and epoxy resins, which is used in the manufacture of protective coatings of food cans, dental sealants, and polyvinyl chloride tubes (5). It has also been detected in dust, air, and water (6–8). Exposure of humans to BPA is nearly universal. BPA has been found in maternal circulation (9, 10), amniotic and placental fluids (11–13), breast milk (14), and the urine of infants (15). It has estrogenic (16, 17) as well as antiandrogenic properties (18, 19). BPA has been found to inhibit transcriptional activity stimulated by thyroid hormone (20), act as a glucocorticoid receptor agonist (21), and impair insulin homeostasis (22). The multitude of targets raises concerns relative to risks from BPA exposures during critical periods of early differentiation.
At the reproductive level, prenatal BPA treatment of sheep induces effects similar to prenatal T treatment (23). Our studies with the native steroid T, an aromatizable androgen, found that exposure to excess T during fetal development leads to a reproductive and metabolic phenotype that resembles that of women with polycystic ovary syndrome (24). These disruptions are manifested at the neuroendocrine, ovarian, and metabolic level, culminating in oligo-/anovulation and insulin resistance. The ovarian changes appear to involve early changes in microRNA (miRNA) (25), posttranscriptional regulators that induce translational repression and gene silencing (26). Subtractive approaches using prenatal dihydrotestosterone, a nonaromatizable androgen as a programming agent (27, 28), or cotreatment of T with antiandrogen flutamide (25) have demonstrated that androgens as well as estrogens are involved in mediating the effects of prenatal T excess at the ovarian level.
Prenatal BPA treatment may cause similar disruptions in ovarian transcriptome during early ovarian differentiation because of the following: 1) prenatal BPA treatment, similar to T treatment (23), induces low birth weight, hypergonadotropism, and cycle disruptions (29); 2) effects of excess prenatal T on ovarian gene expression are in part facilitated via estrogenic actions (25); and 3) BPA is viewed as an estrogen mimic (16, 17). Therefore, the aim of this study was to gain a comprehensive understanding of early developmental changes in the ovarian transcriptome, determine the impact of gestational exposure to an environmentally relevant dose of BPA on these developmental changes, specifically the expression of steroidogenic enzymes, and steroid hormone receptors as well as key ovarian regulators, in addition to identify changes in fetal ovarian miRNA expression impacted by gestational exposure to BPA.
Materials and Methods
Animals, prenatal treatments, and tissue collection
All procedures were approved by the University Animal Care and Use Committee at the University of Michigan. Suffolk ewes (2–3 years old) were purchased locally and group fed 0.5 kg of shelled corn and 1.0–1.5 kg of alfalfa hay per ewe per day (2.31 Mcal/kg of digestible energy). The diet meets the nutrient requirements for sheep defined by the National Research Council. Breeder animals assigned to generate control, and BPA-treated fetuses were blocked by maternal weight, body score, and animal providers. Preventive abortion treatment involved administration of aureomycin crumbles (chlortetracycline: 250 mg/ewe per day). The day of mating was determined by visual confirmation of paint markings left on the rumps of ewes by the raddled rams. Gestational BPA treatment consisted of daily sc injections of 0.5 mg/kg·day of BPA (purity ≥ 99%, catalog number 239658; Aldrich Chemical Co, Milwaukee, Wisconsin) in corn oil from days 30 through 90 of gestation (term ∼147 days). Control (C) mothers received vehicle. For sample and tissue procurement, all dams were anesthetized with 20–30 mL of pentobarbital iv (Nembutol Na solution, 50 mg/mL; Abbott Laboratories, Chicago, Illinois) and maintained with isofluorane under general anesthesia (1%–2%; Halocarbon Laboratories, River Edge, New Jersey).
To determine the internal dose of BPA achieved using the current treatment regimen (0.5 mg/kg daily), umbilical arterial samples (fetal output; n = 3–4/group) were collected from both C and prenatal BPA-treated groups at fetal day 90. The uterus was exposed through a midline incision and the uterine wall incised. Dams were administered a barbiturate overdose (Fatal Plus; Vortech Pharmaceuticals, Dearborn, Michigan) and fetuses removed for tissue harvest. Fetal ovaries were removed from gestational day 65 and gestational day 90 C and BPA-treated fetuses, weighed, washed with PBS, quick frozen, and stored at −80°C until processing. One ovary from 1 female offspring of each dam (the mother was the experimental unit) from gestational day 65 and gestational day 90 control (n = 4 and 5, respectively) and prenatal BPA-treated (n = 5, both ages) animals were used for gene expression studies. The number of fetuses per dam did not differ between treatment groups at either gestational age.
BPA measurements
In our previous study, 5 mg/kg·day BPA produced approximately 40 ng/mL free BPA in maternal blood of sheep (29), levels approximating twice the highest level found in pregnant US women (10). The dose used in the current study, 0.5 mg/kg·day BPA was aimed to produce maternal blood levels of BPA (∼5 ng/mL) approaching the median level of BPA measured in maternal circulation of US women (10). BPA levels in samples were quantified using a HPLC coupled with an API 5500 electrospray triple-quadrupole mass spectrometer (Applied Biosystems, Foster City, California). The method is based on that described earlier (10). In brief, analyte separation and detection were carried out using an Agilent 1100 series HPLC interfaced with an Applied Biosystems API 5500 electrospray triple-quadrupole mass spectrometer. Ten microliters of the extract were injected onto an analytical column (Betasil C18, 100 × 2.1 mm column; Thermo Electron Corp, Waltham, Massachusetts), which was connected to a Javelin guard column (Betasil C18, 20 × 2.1 mm). The tandem mass spectrometry was operated in the electrospray negative ion mode. Instrumental parameters were optimized to transmit the [M-H]ion before fragmentation to 1 or more product ions. Data were acquired using multiple reaction monitoring for the transitions of 227 greater than 212 for BPA, and 239 greater than 224 for 13C12-BPA.
Quality assurance and quality control parameters included validation of the method by spiking 13C12-BPA into the sample matrices and passing through the entire analytical procedure to calculate recoveries of BPA through the analytical method. A procedural blank, containing milli-Q water in place of plasma, was analyzed with the samples to check for interferences or laboratory contamination. The limits of detection were 0.10 and 0.05 ng/mL for 13C12-BPA and BPA, respectively, and these were calculated as twice that of the valid lowest acceptable calibration standard. Reported concentrations were corrected for the recoveries of surrogate standard (isotopic dilution method). An external calibration curve was prepared by injecting 10 μL of 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 50, and 100 ng/mL standards, and the regression coefficient was 0.99.
Quantitative RT-PCR
Isolation of fetal ovarian RNA was performed with Trizol and assessed for quality using the Agilent Bioanalyzer Nano Chip (Agilent Technologies, Santa Clara, California). High quality of RNA was assessed by 18S and 28S peaks and RNA integrity numbers (>8.0). Total RNA (250 ng) isolated from whole ovarian fetal tissue was reverse transcribed (miScript system; QIAGEN, Valencia, California) and cDNA amplified by quantitative RT-PCR (Applied Biosystems 7900HT system) using forward and reverse primers designed to ovine or bovine sequences (see Table 1 and Supplemental Material, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org) and Power Sybr Green (Applied Biosystems, Carlsbad, California).
Table 1.
List of miRNAs Differentially Expressed Between Day 65 and Day 90 in Control Group
| miRNA | ddCT | Fold Change | P Value |
|---|---|---|---|
| hsa-mir-608 | 13.547 | 11972.194 | .006 |
| hsa-mir-217 | 10.417 | 1367.002 | .023 |
| hsa-mir-641 | 6.267 | 77.013 | .029 |
| hsa-mir-1539 | 3.760 | 13.546 | .044 |
| hsa-mir-515-5p | 2.094 | 4.270 | .031 |
| hsa-mir-361-3p | 1.778 | 3.430 | .040 |
| hsa-let-7b | 1.438 | 2.710 | .048 |
| hsa-mir-491-5p | 1.409 | 2.655 | .028 |
| hsa-mir-518a-3p | −1.591 | −3.012 | .032 |
| hsa-mir-514 | −1.876 | −3.670 | .021 |
| hsa-mir-564 | −1.893 | −3.715 | .015 |
| hsa-mir-363 | −1.909 | −3.755 | .028 |
| hsa-mir-885-5p | −2.076 | −4.217 | .042 |
| hsa-mir-519e | −2.084 | −4.239 | .010 |
| hsa-mir-302b | −2.302 | −4.931 | .024 |
| hsa-mir-124 | −3.035 | −8.195 | .037 |
| hsa-mir-508-3p | −3.068 | −8.387 | .026 |
| hsa-mir-371-5p | −3.387 | −10.460 | .008 |
| hsa-mir-1205 | −3.971 | −15.677 | .042 |
| hsa-mir-375 | −4.566 | −23.682 | .040 |
| hsa-mir-875-5p | −5.992 | −63.642 | .043 |
Abbreviation: ddCT, δδCT. The comparison with which the P value applies refers to the 2 gestational ages within the control group.
The focus was on genes coding enzymes in steroidogenic pathway [steroidogenic acute regulatory protein (StAR), cholesterol side chain cleavage enzyme (P450scc/Cyp11a1), 3β1-hydroxysteroid dehydrogenase (HSD), 3β2HSD, 17βHSD, cytochrome P450 17α hydroxylase/17,20 lyase (Cyp17), aromatase (Cyp19), and steroid-5-α-reductase (SRD5A1)]; steroid receptors [androgen receptor (AR), estrogen receptors (ESR1, ESR2), progesterone receptor (PGR)]; gonadotropin receptors (FSHR and LHR); key regulators of ovarian development and function [ferredoxin 1 (FDX1), FDX1 receptor (FDX1R), cyclin D2 (CCND2), IGF-I (IGF1), IGF-I receptor (IGFR1), and growth differentiation factor 9 (GDF9)]; members of the insulin signaling pathway [insulin receptor (IR), insulin receptor substrates 1 and 2 (IRS1 and IRS2), protein kinase B (AKt), phosphatidylinositol-3 kinase (PI3K), mammalian target of rapamycin (mTOR), glucose transport protein 4 (Glut4), and peroxisome proliferator-activated receptor (PPAR)-α; and miRNA processing enzymes [Argonaute 2 (Ago2), Dicer (Dicer1), DiGeorge syndrome critical region 8 (DGCR8), Drosha (Drosha), and rat sarcoma-related nuclear protein (Ran GTPase)]. Standard cycling conditions (50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute) and a dissociation curve to determine product melting temperature to ensure presence of a single amplicon were used in all assays. A relative standard curve was analyzed for each specific gene target. Normalization and data analysis involved comparison of the relative amount of gene product to small RNA U6 (RNU6)-2. RNU6-2 (typically referred to as U6) and 18S expression were examined, and U6 was chosen for normalization based on its consistent level of expression across age and treatment groups (Supplemental Figure 1). We used U6 for both mRNA and miRNA normalization for consistency as done previously (25).
To identify miRNA expressed in fetal ovarian tissue, 50 ng of total RNA was reverse transcribed using the miRCURY LNA universal cDNA synthesis kit, as per the manufacturer's instruction (Exiqon, Vedback, Denmark) from 3 ovaries within each of the treatment groups (gestational day 65 control, n = 3; gestational day 90 control, n = 3; gestational day 65 BPA, n = 3; and gestational day 90 BPA, n = 3). The reverse transcribed miRNA products were diluted 1:110, added to an equal volume of Sybr Green master mix (Exiqon), and loaded onto miRCURY LNA miRNA PCR panels containing a total of 742 miRNA based on human miRBase 16 (Human Panels I and II, V2.M; Exiqon). Quantitative RT-PCR was performed using the ABI 7900HT real-time system (Applied Biosystems) with an initial polymerase activation/denaturation step at 95°C for 10 minutes followed by 40 cycles at 95°C for 10 seconds and 60°C for 60 seconds. After the product amplification, a melting curve analysis was performed to ensure the amplification of a sole miRNA product. The resulting cycle threshold (CT) values were analyzed using the ΔΔCΤ method with the U6 used as the normalizer.
To identify putative mRNA targets, a bioinformatic analysis was conducted on all differentially expressed miRNA using TargetScan 5.1 (www.targetscan.org). These analyses focused on steroid receptors/enzymes, ovarian regulatory genes, insulin signaling molecules, and lipid metabolic hormones and key regulators of gonadal differentiation. A comprehensive literature-based analysis was also undertaken for all differentially expressed miRNA linked to steroid receptor action, steroidogenesis, ovarian function, sexual differentiation, insulin signaling, diabetes, and lipid metabolism, as done previously (25).
Statistical analysis
All mRNA expression data generated by quantitative RT-PCR were log transformed and analyzed by 2-way ANOVA followed by a Bonferroni post hoc test (Prism, version 4; GraphPad Software, La Jolla, California). The differences in BPA concentrations between groups were analyzed using an independent t test after testing for normality and homogeneity of data (SPSS for Windows, release 17.0.0; SPSS Inc, Chicago, Illinois). P < .05 was considered significant.
Results
BPA internal exposure
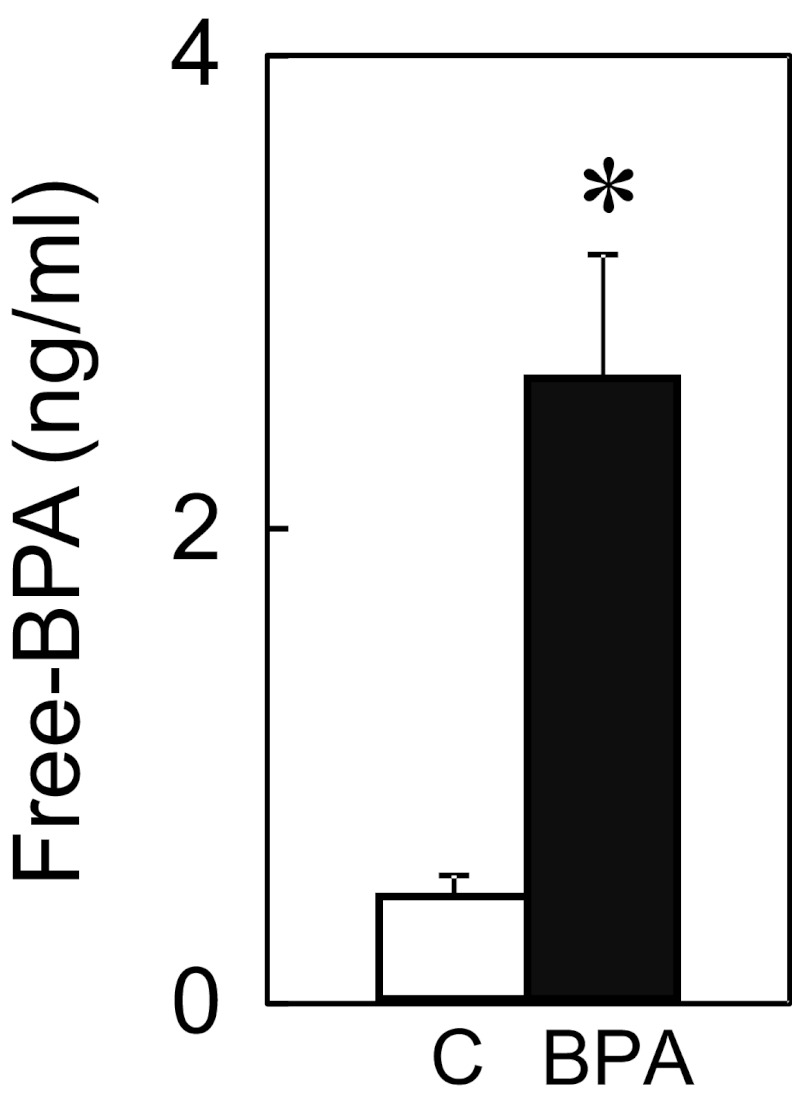
Concentrations of free BPA achieved in umbilical arterial samples from female fetuses were higher in BPA-treated fetuses compared with controls (Figure 1). Free BPA levels averaged 0.43 ± 0.09 and 2.62 ± 0.52 ng/mL in controls and BPA-treated fetuses, respectively (C vs BPA; P < .01) at day 90 of gestation (30).
Figure 1.
Free-BPA concentrations (nanograms per milliliter) in umbilical arterial plasma of female fetuses at gestational day 90. Controls (C; n = 3) are shown as open bars and BPA treated (n = 4) as closed bars. Asterisk above mean ± SEM denotes significant (P < .01) difference between treatment groups.
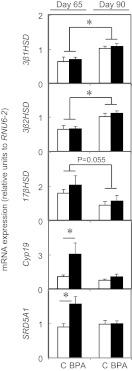
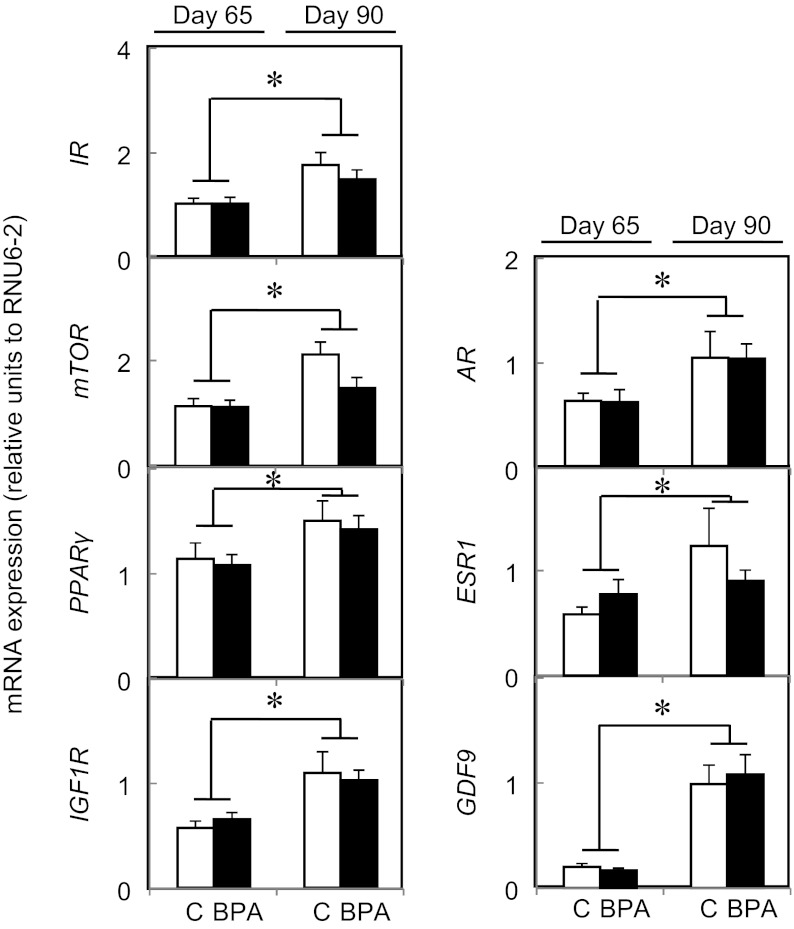
Age-specific changes in expression of key ovarian regulators
Data for those genes that did not show a statistical difference between treatment groups across age were subjected to combined analyses. The results from this combined analyses are shown in Figure 2 and showed that there was a fetal age-dependent increase (P < .005) in 3β1HSD and 3β2HSD and a tendency for decreased expression of 17βHSD (P = .055) from fetal day 65 to 90. A fetal age-related increase in the mRNA expression of AR, ESR1, GDF9, IR, mTOR, PPARα, and IGF1R was also observed between day 65 and day 90 (Figure 3) but not in the rest of the genes studied (see Supplemental Material Table 1 for a complete list).
Figure 2.
Age-dependent gene expression changes in fetal ovarian steroidogenic enzymes (top 3 panels) between day 65 (left graph) and day 90 (right graph) of gestation in control (C; open bars) and BPA-treated (closed bars) females. Results for 3β1HSD, 3β2HSD, and 17βHSD of control and BPA-treated females were combined for testing age effects when there was no treatment effect within age. Treatment effects were observed only with Cyp19 and 5α-reductase expression (2 bottom panels). Asterisks represent statistical differences between ages (P < .005) or treatment groups. Control (n = 4 and 5, respectively, at day 65 and day 90) and prenatal BPA-treated (n = 5, both ages) groups.
Figure 3.
Age-dependent changes in fetal ovarian steroid receptors, growth factors, and insulin-related gene expression between day 65 (left graph) and day 90 (right graph) of gestation in control (open bars) and BPA-treated (closed bars) females. Results of control and BPA-treated females were combined for testing age effects when there was no treatment effect within age. Asterisks represent statistical differences between ages (P < .005). Control (n = 4 and 5, respectively, at day 65 and day 90) and prenatal BPA-treated (n = 5, both ages) groups.
Gestational BPA-induced changes in expression of key ovarian regulators
Genes that showed age-specific treatment effect are shown in Figure 2. Gestational BPA treatment increased the mRNA expression of Cyp19, the enzyme responsible for the conversion of androgens into estrogens, and 5α-reductase, the enzyme responsible for the conversion of T into dihydrotestosterone at fetal day 65 (Figure 2) but not day 90. Prenatal BPA treatment did not alter the mRNA expression of the rest of the genes studied (see Supplemental Table 1 for a complete list) at either time point.
Age-specific changes in expression of miRNA
Comparison of miRNA expression between fetal day 65 and day 90 in the control group indicated that 21 genes were differentially expressed, with 8 being up-regulated and 13 down-regulated in gestational day 90 fetuses (Table 1). Two of the up-regulated miRNAs, miR-608 and miR-217, were minimally expressed in gestational day 65 fetal ovaries, as noted by the approximately 12 000- and approximately 1300-fold increases observed between the gestational day 65 and gestational day 90 tissues, respectively. Comparison of miRNA expression between gestational day 65 and gestational day 90 ovaries from BPA-treated fetuses indicated that 39 miRNAs were differentially expressed, with all but 1 showing up-regulation in day 90 fetuses (Table 2). miR-766 expression on day 90 was one-twelfth the level seen at fetal day 65. Only 3 of the differentially expressed miRNAs were common to both the control and BPA groups. Two of these 3, miR-let-7b and miR-491-5p, were up-regulated in both groups. The third, miR-519e-3p, was up-regulated 13.7-fold in BPA ovaries between gestational day 65 and gestational day 90 but down-regulated (1:4.2) in control ovaries between gestational day 65 and gestational day 90.
Table 2.
List of miRNAs Differentially Expressed Between Day 65 and Day 90 in BPA-Treated Group
| miRNA | ddCT | Fold Change | P Value |
|---|---|---|---|
| hsa-mir-886-3p | 9.132 | 561.028 | .014 |
| hsa-mir-519e-5p | 4.470 | 22.168 | .028 |
| hsa-mir-222-5p | 4.436 | 21.644 | .035 |
| hsa-mir-520d-3p | 4.189 | 18.240 | .036 |
| hsa-mir-1243 | 3.880 | 14.721 | .039 |
| hsa-mir-519e-3p | 3.783 | 13.768 | .042 |
| hsa-mir-371-3p | 3.348 | 10.183 | .033 |
| hsa-mirplus-c1066 | 3.029 | 8.163 | .047 |
| hsa-mir-1911 | 2.896 | 7.443 | .027 |
| hsa-mir-29a | 2.601 | 6.067 | .046 |
| hsa-mir-1256 | 2.207 | 4.618 | .037 |
| hsa-mir-99a | 2.056 | 4.157 | .011 |
| hsa-mir-193b-5p | 1.660 | 3.159 | .014 |
| hsa-mir-125b | 1.659 | 3.157 | .004 |
| hsa-let-7b | 1.640 | 3.118 | .042 |
| hsa-mir-150 | 1.626 | 3.087 | .004 |
| hsa-mir-33b-3p | 1.495 | 2.818 | .008 |
| hsa-mir-423-3p | 1.434 | 2.702 | .037 |
| hsa-mir-423-5p | 1.375 | 2.594 | .017 |
| hsa-mir-328 | 1.374 | 2.592 | .008 |
| hsa-mir-140-3p | 1.340 | 2.532 | .047 |
| hsa-mir-138-2-3p | 1.311 | 2.482 | .036 |
| hsa-mir-491-5p | 1.303 | 2.467 | .000 |
| hsa-mir-30d | 1.300 | 2.463 | .020 |
| hsa-mir-505 | 1.261 | 2.397 | .027 |
| hsa-mir-320a | 1.244 | 2.369 | .003 |
| hsa-mir-378a-5p | 1.191 | 2.283 | .011 |
| hsa-mir-320b | 1.158 | 2.232 | .036 |
| hsa-mir-423-5p | 1.143 | 2.209 | .007 |
| hsa-mir-145 | 1.133 | 2.192 | .016 |
| hsa-mir-92b | 1.058 | 2.082 | .029 |
| hsa-mir-891b | 1.034 | 2.048 | .023 |
| hsa-mir-502-5p | 1.004 | 2.006 | .045 |
| hsa-mir-500 | 1.003 | 2.004 | .028 |
| hsa-mir-214 | 0.757 | 1.690 | .024 |
| hsa-mir-193a-5p | 0.727 | 1.656 | .029 |
| hsa-mir-339-5p | 0.659 | 1.579 | .037 |
| hsa-mir-615-3p | 0.619 | 1.536 | .050 |
| hsa-mir-766 | −3.656 | −12.608 | .023 |
Abbreviation: ddCT, δδCT. The comparison with which the P value applies refers to the 2 gestational ages within the BPA-treated group.
Gestational BPA-induced changes in expression of miRNA
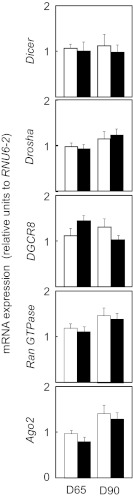
Maternal BPA treatment induced a decrease in fetal ovarian miRNAs with none of the miRNA showing up-regulation. On fetal day 65, this BPA-induced decline was evident for 45 different miRNAs. The range of down-regulation varied from 1:128 of the control to our arbitrary cutoff of greater than 2-fold (ie, half of control) difference (Table 3). Similarly, on fetal day 90, down-regulation was evident for 11 miRNAs. In the gestational day 90 fetal ovaries, the range of down-regulation ranged from 1:275 to 1:2.8 of the control levels (Table 4). Interestingly, only a single miRNA, miR-203, was down-regulated at both gestational day 65 and gestational day 90, and in both instances it was the gene with the greatest differential expression. No differences were found in the expression of miRNA processing endoribonucleases, Drosha and Dicer, or other miRNA biogenesis components including the DGCR8 and Ran GTPase between BPA-treated fetal ovaries and control ovaries (Figure 4). Similarly, expression of Ago2, a critical component on the RNA-induced silencing complex, which mediates the mechanistic effects of miRNA, was not altered in BPA-treated ovaries (Figure 4).
Table 3.
List of miRNAs Differentially Expressed Between C and BPA-Treated Groups at Day 65
| miRNA | ddCT | Fold Change | P Value | Targets | Papers, n |
|---|---|---|---|---|---|
| hsa-mir-203 | −7.005 | −128.465 | .015 | IRS2, INSIG1, ESR1 | 80 |
| hsa-mir-566 | −6.809 | −112.149 | .012 | 2 | |
| hsa-mirplus-d1061 | −6.232 | −75.154 | .006 | ||
| hsa-mir-219-1-3p | −5.942 | −61.473 | .005 | TGFBR2, NR5A2 (SF1), SOX6, INSIG1, ESR1, PPARa, RXRa | 1 |
| hsa-mirplus-c1070 | −5.840 | −57.270 | .046 | ||
| hsa-mir-508-3p | −5.195 | −36.622 | .005 | TGFBR2, NR5A2 (SF1), SOX6, INSIG1, ESR1, PPARa, RXRa 219 family member | 2 |
| hsa-mir-365a-5p | −5.049 | −33.116 | .020 | ||
| hsa-mir-371-3p | −5.024 | −32.529 | .005 | IGF2BP1 | 1 |
| hsa-mir-1911 | −4.775 | −27.373 | .011 | 0 | |
| hsa-mir-551a | −4.628 | −24.734 | .001 | 1 | |
| hsa-mir-380 | −4.623 | −24.635 | .031 | IGF1, KITLG, IGFBP5, PAPPA | 1 |
| hsa-mir-1263 | −4.446 | −21.800 | .001 | 0 | |
| hsa-mir-649 | −4.198 | −18.350 | .017 | KITLG, PAPPA, SOX6, IGF2BP2, PGR | 0 |
| hsa-mir-582-3p | −4.121 | −17.396 | .031 | SOX9, IGFBP4, ACVR2B | 2 |
| hsa-mir-1179 | −4.010 | −16.115 | .025 | 1 | |
| hsa-mir-220b | −3.836 | −14.280 | .016 | No longer considered miR | |
| hsa-mir-92a-2-5p | −3.805 | −13.975 | .038 | ||
| hsa-mir-519e | −3.717 | −13.148 | .050 | SOX4, SOX11 | 4 |
| hsa-mir-297 | −3.124 | −8.716 | .029 | IGFBP5, SOX12 | 5 |
| hsa-mir-518d-5p | −2.870 | −7.309 | .022 | PTGS1 | 5 |
| hsa-mir-449b | −2.862 | −7.269 | .039 | AREG, KITLG, SOX4, ADIPOR1, IGFBP3, INHBB | 7 |
| hsa-mir-1909 | −2.826 | −7.089 | .045 | RARa, IGFBP4, IGF2 | 1 |
| hsa-mir-650 | −2.730 | −6.636 | .027 | 9 | |
| hsa-mir-639 | −2.706 | −6.523 | .032 | 0 | |
| hsa-mir-182-3p | −2.595 | −6.040 | .009 | ||
| hsa-mir-26a-1-3p | −2.542 | −5.825 | .041 | ||
| hsa-mir-605 | −2.501 | −5.662 | .034 | SOX9, ADIPOR2 | 8 |
| hsa-mir-296-3p | −2.392 | −5.250 | .042 | 5 | |
| hsa-mir-571 | −2.345 | −5.079 | .024 | SOX4 | 2 |
| hsa-mir-23b-5p | −2.227 | −4.681 | .011 | ||
| hsa-mir-885-5p | −2.106 | −4.304 | .024 | 4 | |
| hsa-mir-1471 | −2.096 | −4.276 | .040 | 1 | |
| hsa-let-7a-2-3p | −1.867 | −3.649 | .047 | ||
| hsa-mir-431 | −1.805 | −3.494 | .028 | ||
| hsa-mir-558 | −1.696 | −3.240 | .048 | ||
| hsa-mir-548m | −1.694 | −3.235 | .036 | ||
| hsa-mir-518d-3p | −1.672 | −3.187 | .029 | ||
| hsa-mir-342–5p | −1.461 | −2.753 | .041 | ||
| hsa-mir-1201 | −1.457 | −2.746 | .014 | ||
| hsa-mir-193b-5p | −1.313 | −2.484 | .030 | ||
| hsa-mir-200b-5p | −1.290 | −2.445 | .003 | ||
| hsa-mir-505 | −1.199 | −2.296 | .047 | ||
| hsa-mir-134 | −1.121 | −2.175 | .049 | ||
| hsa-mir-500 | −1.053 | −2.074 | .021 | ||
| hsa-mir-940 | −1.023 | −2.032 | .038 |
Abbreviation: ddCT, δδCT. The comparison with which the P value applies is to the control group.
Table 4.
List of miRNAs Differentially Expressed Between C and BPA-Treated Groups at Day 90
| miRNA | ddCT | Fold Change | P Value | Targets | Papers, n |
|---|---|---|---|---|---|
| hsa-mir-203 | −8.103 | −274.945 | .032 | IRS2, INSIG1, ESR1 | 80 |
| hsa-mir-641 | −6.601 | −97.066 | .029 | SOX6, IGFBP4, SOX11 | 0 |
| hsa-mir-135b-3p | −5.091 | −34.086 | .002 | ||
| hsa-mir-100-3p | −4.296 | −19.645 | .047 | ||
| hsa-mir-192-3p | −4.055 | −16.626 | .028 | ||
| hsa-mir-302b | −3.409 | −10.621 | .011 | PPARa, ESR1 | 10 |
| hsa-mir-515-5p | −3.163 | −8.959 | .043 | ESR1, SOX12 | 2 |
| hsa-mir-520g | −3.074 | −8.423 | .016 | TGFBR3 | |
| hsa-mir-524-5p | −2.511 | −5.699 | .048 | SOX8, SOX9, INSIG2, SOX4, LDLR, RARB, SOX11, BMPR1A, GDF10, INSR, BMP6, IGF1R, SOX2, SOX1, IRS1 | 1 |
| hsa-mir-509-3-5p | −2.025 | −4.069 | .049 | IDE (insulin degrading enzyme), IGF1R, AR | 1 |
| hsa-mir-187 | −1.504 | −2.836 | .007 | 8 |
Abbreviation: ddCT, δδCT. The comparison with which the P value applies is to the control group.
Figure 4.
Impact of gestational BPA treatment on the expression of the miRNA processing enzymes Ago2, Dicer, DGCR8, Drosha, and Ran GTPase from fetal day 65 (left panels) and day 90 (right panels) control (C; open bars) and BPA-treated (closed bars) females. Results are expressed as mean ± SEM. Control (n = 4 and 5, respectively, at day 65 and day 90) and prenatal BPA-treated (n = 5, both ages) groups.
Bioinformatic analysis of predicted mRNA targets of miRNA differentially expressed in fetal ovaries exposed to BPA revealed at least 13 miRNA (Tables 3 and 4) targeting kit ligand or members of 3 of the subgroups of the Sry-related high-mobility-group box (SOX) family of genes [SOXC (SOX4, SOX11, and SOX12), SOXD (SOX6), and SOXE (SOX8 and SOX9)]. In addition, 15 differentially expressed miRNAs are potential target genes involved in the regulation of insulin signaling, including IGFs (Igf1 and Igf2) and related binding proteins (Igfbp3, Igfbp4, Igfbp5, Igf2bp1, and Igf2pb2) and the insulin-induced gene (Insig1).
Discussion
This study demonstrates that fetal BPA exposure, at an environmentally relevant exposure level, disrupts the expression of critical steroidogenic enzymes and a number of miRNAs implicated in estrogen-signaling and insulin homeostasis. To our knowledge, this is the first documentation of the impact of developmental exposure to BPA on ovarian miRNA gene expression. The specific mRNA and miRNA changes as they relate to early onset of reproductive and metabolic defects and their potential translational value are discussed below.
Age-specific changes in expression of key ovarian regulators
The age-specific pattern of expression of steroidogenic enzymes, gonadotropin receptors, steroid receptors, and GDF9 mRNA found in this study in fetal ovaries is in agreement with previous studies (31–33) as is the age-dependent increase in expression of these genes (25). Observed changes are consistent with the progression of ovarian development (34) and the concomitant maturation of the steroidogenic enzymatic activity in the ovary (35).
Gestational BPA-induced changes in expression of key ovarian regulators
Changes in expression of steroidogenic enzymes during a critical window of ovarian differentiation may impair its development and lead to reproductive disruptions later in life. Our finding that BPA increases Cyp19 is consistent with estrogenic impact, as predicted from our earlier studies comparing the impact of prenatal T with prenatal T plus androgen antagonist. This earlier study found that cotreatment of T with antiandrogen flutamide did not prevent the increase in Cyp19 expression caused by gestational T treatment (25), supportive of mediation via estrogenic actions of the aromatizable androgen T. Consistent with this, Cyp19 mRNA was also overexpressed in ovaries of CD-1 mice treated with 25 mg/kg·day or 50 mg/kg·day BPA from gestational day 1 through postnatal day 20 (36). In vitro studies also show that BPA treatment increases aromatase activity in placental and kidney cells (37) and embryonic brain cells (38), suggestive of the potential for systemic BPA effects. In contrast, another study with Wistar rats found that in utero exposure to 0.1 or 1 mg/L of BPA in drinking water failed to increase circulating concentrations of estradiol at 12 weeks (39); expression of ovarian Cyp19 was not evaluated in this study. Such generalized effects may stem from the ubiquity of the signaling pathways that BPA may activate to trigger Cyp19 overexpression. In support of this concept, Cyp19 overexpression after BPA exposure was found to be correlated with the up-regulation of cyclooxygenase-2, via the activation of cAMP response element-binding protein, protein kinase A, protein kinase B, and MAPK signaling pathways in rat testicular Leydig cells (40).
Findings from our study also provide the first in vivo evidence that BPA can induce changes in the expression of 5α-reductase. The transient increase in 5α-reductase at fetal day 65 in BPA-treated fetuses is similar to findings seen at the same developmental time point in ovaries from fetuses treated with excess T (25). These effects are consistent with BPA being an estrogen mimic because the cotreatment of T with antiandrogen flutamide was found not to prevent this transient up-regulation of 5α-reductase (25). To what extent the perturbations in 5α-reductase as well as Cyp19 contribute to the postnatal reproductive defects seen in prenatal BPA-treated sheep (29) remains to be determined.
Other than the changes with Cyp19 and 5α-reductase discussed above, gestational BPA treatment had no effect on the expression of other steroidogenic enzymes or steroid receptors evaluated. Although a similar lack of effect was found on postnatal day 50 of prenatal BPA-treated mice (36), evidence exists in rodents supportive of BPA having effects on the expression of other steroidogenic enzymes (36, 41). In vitro studies addressing the effects of BPA on granulosa cells or antral follicles have reported a disruption in the estradiol biosynthesis pathway in the ovary (42–44). For instance, in vitro studies in granulosa cells have found that BPA treatment does have an impact on the expression of steroidogenic acute regulatory protein and Cyp11a1 (43, 44). Differences between studies likely relate to the species being studied, timing of BPA treatment relative to ovarian development, age at which measures were made, and/or internal dose of BPA achieved.
Relative to the gonadotrophic control of the ovary, maternal exposure to BPA had no effect on the mRNA expression of gonadotropin receptors at either fetal age studied. These findings parallel observations of fetal sheep exposed to excess of native steroid T (25). No changes in the mRNA expression of ovarian gonadotropin receptors (LH receptor and FSH receptor) were also reported after the prenatal treatment of the mice with BPA (36).
Age-specific changes in miRNA expression
microRNAs have been identified as key regulators of posttranscriptional gene regulation in tissue development (26). In mice, the knockout of Dicer, a ribonuclease that modulates miRNA maturation, results in dysfunctional folliculogenesis (45) and decreased ovulation rate (46, 47), accounting for the relevance of miRNA in ovarian function. A few studies have identified differential expression of miRNA in the developing gonads of the sheep (48), chicken (49), and mouse (50). The only study involving fetal ovine ovaries found that both miR-22 and miR-let-7 are differentially expressed during early fetal ovarian differentiation and bioinformatics analysis (TargetScan) implicated these miRNAs in ESR1 and Cyp19 regulation, respectively (48). The study by Torley et al (48) found increased expression of several let-7 family miRNAs (eg, let-7a, let-7c, let-7d, let-7e, and let-7g) between days 42 and 75 of gestation. Our study, in contrast, found increased let-7b only between days 65 and 90. Because of the marked differences in timing of fetal ovarian collection between these studies and the dynamic nature of the developing gonad, not much additional information can be gained by comparing the studies. The time points examined in our study correspond to time after meiotic resumption (day 55) and after primordial follicle differentiation (day 75). In contrast, Torley et al (48) examined day 42 and day 75 ovaries, which correspond to the period before meiotic resumption (day 55) and prior to primordial follicle differentiation (day 75). Although each study provides a logical rationale for selection of the time points examined, our study has the added benefit of having a treatment regimen (maternal BPA exposure) to determine whether fetal ovarian gene (miRNA) expression can be differentially modulated over time in response to an environmental disruptor.
Two miRNAs, miR-217 and -608, with the greatest increase in expression between day 65 and day 90 control fetal ovaries, might be involved in early ovarian differentiation, specifically in the formation of the first primordial follicles. Earlier studies have shown that primordial follicle assembly begins at fetal day 75 and is completed by fetal day 100 (34). Previous studies have implicated miR-217 in induction of a premature senescence-like phenotype in endothelial cells (51) and miR-608 in cell cycle arrest, DNA repair, and cell survival (52). Therefore, the up-regulation of both miRNAs observed in this study suggests a potential role for these miRNAs in the active remodeling of the ovarian tissue during midgestation (34).
Age-specific changes in the expression of miRNA in BPA-treated ovaries were different from control ovaries supportive of potential impact of BPA in regulating miRNA machinery during early ovarian development. Given the key role of miRNA in regulating mRNA stability and translation, the alteration of miRNA expression during this critical window of development may permanently alter subsequent function. Although we cannot definitively demonstrate the roles of miRNAs in the present study with respect to function, these studies provide the first look into the potential pathophysiology mediated by miRNA induced by BPA exposure in unborn female offspring.
Gestational BPA impact on miRNA expression
This is the first in vivo report of which we are aware that examines miRNA expression in any tissue after BPA exposure. Studies with cell lines point to the potential for endocrine disruptors to alter miRNA expression (53, 54). Impact of BPA on miRNA expression has been reported with placental cell lines (55) and breast cancer cells (56). The finding that BPA, an estrogen mimic, alters miRNA expression during ovarian development provides support for hormonal regulation of miRNA expression. In support of this contention, native steroids (T and estradiol) have been found to be potential regulators of miRNA expression in the developing ovary in fetal sheep (25) and chicken (57).
Among the miRNAs that were found to be down-regulated in prenatally BPA-treated ovaries, miR-137 was reported to suppress production of progesterone, androgens, and estrogens (58), and miR-200 has been reported to be up-regulated in ovarian cancer (59). miR-765 was found to be down-regulated in premature ovarian failure patients (60). In a recent study, a predicted functional analysis pointed to Cyp19 as a gene target for miR-let-7 (let-7a, let-7c, let-7d, let-7e, and let-7g). In the current study, let7a-2-3p was found to be down-regulated in BPA-treated ovaries at day 65. This finding together with the up-regulation of Cyp19 mRNA expression at day 65, opens up the possibility that let-7 may regulate Cyp19 mRNA expression during early ovarian development in this species. Further, in-depth studies would be needed to show causality and whether let7a-2-3p directly or indirectly targets the Cyp19 gene.
Because functional profiling of miRNAs is at its infancy, the establishment of direct links between miRNA changes and mRNA targets is difficult. The prediction of mRNA target based on bioinformatic analysis suggests a potential role for differentially expressed miRNA in regulating the expression of SOX family genes, which are also expressed in the ovary (61–65). Importantly, SOX family genes play a key role in sex determination and embryonic development (66). As such, the down-regulation of miR-582-3p, miR-605, and miR-524-5p by BPA may reflect a potential compromise in the germinal line. Many of the differentially expressed miRNAs in BPA-treated ovaries appear to be involved in the regulation of insulin signaling, which is consistent with previous studies showing disruption of insulin homeostasis by prenatal BPA treatment (22). ESR1 is a putative target for miR-203, the only miRNA down-regulated at both fetal day 65 and day 90. However, we found no changes at the mRNA level in ESR1 or insulin-related genes. Because miRNAs may regulate translation independent of effects on mRNA stability, the effects of a particular miRNA cannot be derived solely by mRNA expression analysis. Indeed, very few miRNAs appear to correlate with their predicted targets (67).
The finding that gestational BPA exposure has a predominant effect to reduce the expression of most miRNAs within fetal ovaries suggests that BPA may be impacting the processing machinery of miRNAs. Hormonal regulation of miRNA biogenesis enzymes by steroids, including estradiol (68) has been demonstrated previously. The lack of changes in the processing enzymes studied (Drosha, Dicer, and DGCR8) indicates that BPA may impact miRNA processing machinery at a different level, such as nuclear export of miRNAs (Exportin) or in the complex that helps the mature miRNA to exert its function independent of Ago2 (RNA induced silencing complex) (69). Alternatively, BPA could be affecting the miRNA biosynthetic pathway by modulating the protein expression or the protein activity of 1 or multiple components. Another possibility is that these particular miRNA genes may have estrogen response elements within their promoters, potential target for BPA.
Translational relevance
The dose used in the current study (0.5 mg/kg·day BPA) produced 2.62 ± 0.52 ng/mL of free BPA in umbilical arterial samples from female fetuses at day 90 of gestation. Internal dose exposure reported in this study is in line with reports in human cord blood (10, 70, 71) and within the range reported in maternal urine (reviewed in Reference 9). It is important to recognize that considerable controversy exists regarding internal BPA exposure levels in humans due to the potential for contamination of sample from various sources (72). Similarly, usefulness of animal studies using injections as a route of administration has been questioned due to the belief that in humans, exposure is predominantly via diet (73). This premise is also highly controversial because evidence exists to indicate humans are ubiquitously exposed to BPA via air, water, thermal paper, etc (72). Nonetheless, recent studies have found that internal levels of BPA achieved via diet and sc injections are similar (74). Considering that internal exposure levels are what is important in the context of programming, the comparative studies of Prins et al (74) recognize the relevance of animal studies involving sc administration.
Although a large body of work has been published in the past few years regarding the potential for BPA to interfere with physiological functions, the mechanisms by which BPA elicits its actions remains uncertain (75). The ubiquitous exposure of humans to BPA and the potential for BPA to act at very low doses (76) emphasize the need for careful mechanistic investigation of organ systems, targets of BPA insult. Considering that miRNAs can target several genes, the finding from this study that BPA, at an environmentally relevant dose, down-regulates several ovarian miRNAs during early ovarian differentiation raises concerns relative to impact of BPA at a system level. To what extent such perturbations in ovarian miRNAs may underlie the compromised reproductive function of the prenatally BPA-treated sheep (29) remains to be determined.
Acknowledgments
We are grateful to Mr Douglas Doop for his expert animal care, facility management, and help with generation of the experimental fetuses; Ms Alyse DeHaan, Ms Carol Herkimer, Ms Alexandra Spencer, and Ms Genevieve Ray for assistance with prenatal BPA treatment and/or the collection and processing of fetal ovaries. The authors thank Dr Kurunthachalam Kannan (Wadsworth Center, New York State Department of Health, and Department of Environmental Health Sciences, School of Public Health, State University of New York at Albany) for the BPA measurements.
This work was supported by Grant R01 ES 016541 (to V.P.) and the Hall Family Foundation (to L.K.C.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Akt
- protein kinase B
- AR
- androgen receptor
- Ago2
- Argonaute 2
- BPA
- Bisphenol A
- C
- control
- CT
- cycle threshold
- Cyp19
- aromatase
- DGCR8
- DiGeorge syndrome critical region gene 8
- EDC
- endocrine disrupting chemical
- ESR
- estrogen receptor
- FDX
- ferredoxin
- GDF9
- growth differentiation factor 9
- HSD
- hydroxysteroid dehydrogenase
- IGFRI
- IGF-I receptor
- Insig1
- insulin-induced gene
- IRS
- insulin receptor substrate
- miRNA
- microRNA
- PPAR
- peroxisome proliferator-activated receptor
- Ran GTPase
- rat sarcoma-related nuclear protein
- RNU6
- small RNA U6
- SOX
- Sry-related high-mobility-group box.
References
- 1. Hartgens F, Kuipers H. Effects of androgenic-anabolic steroids in athletes. Sports Med. 2004;34:513–554 [DOI] [PubMed] [Google Scholar]
- 2. Waller DK, Gallaway MS, Taylor LG, et al. Use of oral contraceptives in pregnancy and major structural birth defects in offspring. Epidemiology. 2010;21:232–239 [DOI] [PubMed] [Google Scholar]
- 3. Hotchkiss AK, Rider CV, Blystone CR, et al. Fifteen years after “Wingspread”—environmental endocrine disrupters and human and wildlife health: where we are today and where we need to go. Toxicol Sci. 2008;105:235–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heindel JJ, vom Saal FS. Role of nutrition and environmental endocrine disrupting chemicals during the perinatal period on the aetiology of obesity. Mol Cell Endocrinol. 2009;304:90–96 [DOI] [PubMed] [Google Scholar]
- 5. vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environ Health Perspect. 2005;113:926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuch HM, Ballschmiter K. Determination of endocrine-disrupting phenolic compounds and estrogens in surface and drinking water by HRGC-(NCI)-MS in the picogram per liter range. Environ Sci Technol. 2001;35:3201–3206 [DOI] [PubMed] [Google Scholar]
- 7. Rudel RA, Brody JG, Spengler JD, et al. Identification of selected hormonally active agents and animal mammary carcinogenesis in commercial and residential air and dust samples. J Air Waste Manage Assoc. 2001;51:499–513 [DOI] [PubMed] [Google Scholar]
- 8. Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA). Reprod Toxicol. 2007;24:139–177 [DOI] [PubMed] [Google Scholar]
- 9. Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJ, Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118:1055–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Padmanabhan V, Siefert K, Ransom S, et al. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol. 2008;28:258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikezuki Y, Tsutsumi O, Takai Y, Kamei Y, Taketani Y. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum Reprod. 2002;17:2839–2841 [DOI] [PubMed] [Google Scholar]
- 12. Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110:A703–A707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edlow AG, Chen M, Smith NA, Lu C, McElrath TF. Fetal bisphenol A exposure: concentration of conjugated and unconjugated bisphenol A in amniotic fluid in the second and third trimesters. Reprod Toxicol. 2012;34:1–7 [DOI] [PubMed] [Google Scholar]
- 14. Sun Y, Irie M, Kishikawa N, Wada M, Kuroda N, Nakashima K. Determination of bisphenol A in human breast milk by HPLC with column-switching and fluorescence detection. Biomed Chromatogr. 2004;18:501–507 [DOI] [PubMed] [Google Scholar]
- 15. Calafat AM, Weuve J, Ye X, et al. Exposure to bisphenol A and other phenols in neonatal intensive care unit premature infants. Environ Health Perspect. 2009;117:639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quesada I, Fuentes E, Viso-Leon MC, Soria B, Ripoll C, Nadal A. Low doses of the endocrine disruptor bisphenol-A and the native hormone 17β-estradiol rapidly activate transcription factor CREB. FASEB J. 2002;16:1671–1673 [DOI] [PubMed] [Google Scholar]
- 17. Zsarnovszky A, Le HH, Wang HS, Belcher SM. Ontogeny of rapid estrogen-mediated extracellular signal-regulated kinase signaling in the rat cerebellar cortex: potent nongenomic agonist and endocrine disrupting activity of the xenoestrogen bisphenol A. Endocrinology. 2005;146:5388–5396 [DOI] [PubMed] [Google Scholar]
- 18. Sohoni P, Sumpter JP. Several environmental oestrogens are also anti-androgens. J Endocrinol. 1998;158:327–339 [DOI] [PubMed] [Google Scholar]
- 19. Kitamura S, Suzuki T, Sanoh S, et al. Comparative study of the endocrine-disrupting activity of bisphenol A and 19 related compounds. Toxicol Sci. 2005;84:249–259 [DOI] [PubMed] [Google Scholar]
- 20. Moriyama K, Tagami T, Akamizu T, et al. Thyroid hormone action is disrupted by bisphenol A as an antagonist, J Clin Endocrinol Metab. 2002;87:5185–5190 [DOI] [PubMed] [Google Scholar]
- 21. Prasanth GK, Divya LM, Sadasivan C. Bisphenol-A can bind to human glucocorticoid receptor as an agonist: an in silico study. J Appl Toxicol. 2010;30:769–774 [DOI] [PubMed] [Google Scholar]
- 22. Alonso-Magdalena P, Vieira E, Soriano S, et al. Bisphenol A exposure during pregnancy disrupts glucose homeostasis in mothers and adult male offspring. Environ Health Perspect. 2010;118:1243–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Padmanabhan V, Sarma HN, Savabieasfahani M, Steckler TL, Veiga-Lopez A. Developmental reprogramming of reproductive and metabolic dysfunction in sheep: native steroids vs. environmental steroid receptor modulators. Int J Androl. 2010;33:394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Padmanabhan V, Veiga-Lopez A. Developmental origin of reproductive and metabolic dysfunctions: androgenic versus estrogenic reprogramming. Semin Reprod Med. 2011;29:173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luense LJ, Veiga-Lopez A, Padmanabhan V, Christenson LK. Developmental programming: gestational testosterone treatment alters fetal ovarian gene expression. Endocrinology. 2011;152:4974–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114 [DOI] [PubMed] [Google Scholar]
- 27. Ortega HH, Salvetti NR, Padmanabhan V. Developmental programming: prenatal androgen excess disrupts ovarian steroid receptor balance. Reproduction. 2009;137:865–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ortega HH, Rey F, Velazquez MM, Padmanabhan V. Developmental programming: effect of prenatal steroid excess on intraovarian components of insulin signaling pathway and related proteins in sheep. Biol Reprod. 2010;82:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Savabieasfahani M, Kannan K, Astapova O, Evans NP, Padmanabhan V. Developmental programming: differential effects of prenatal exposure to bisphenol-A or methoxychlor on reproductive function. Endocrinology. 2006;147:5956–5966 [DOI] [PubMed] [Google Scholar]
- 30. Padmanabhan V, Halubai S, Veiga-Lopez A, Liao C, Domino S, Kannan K. 2011 Maternal-cord blood levels of bisphenol-A (BPA) in human and following BPA administration in pregnant sheep. Paper presented at: Society of Toxicology, 50th Annual Meeting, Washington, DC, March 6–10, 2011; Abstract 2186:310 [Google Scholar]
- 31. Mandon-Pepin B, Oustry-Vaiman A, Vigier B, Piumi F, Cribiu E, Cotinot C. Expression profiles and chromosomal localization of genes controlling meiosis and follicular development in the sheep ovary. Biol Reprod. 2003;68:985–995 [DOI] [PubMed] [Google Scholar]
- 32. Juengel JL, Bodensteiner KJ, Heath DA, et al. Physiology of GDF9 and BMP15 signalling molecules. Anim Reprod Sci. 2004;82–83:447–460 [DOI] [PubMed] [Google Scholar]
- 33. Hogg K, McNeilly AS, Duncan WC. Prenatal androgen exposure leads to alterations in gene and protein expression in the ovine fetal ovary. Endocrinology. 2011;152:2048–2059 [DOI] [PubMed] [Google Scholar]
- 34. Sawyer HR, Smith P, Heath DA, Juengel JL, Wakefield SJ, McNatty KP. Formation of ovarian follicles during fetal development in sheep. Biol Reprod. 2002;66:1134–1150 [DOI] [PubMed] [Google Scholar]
- 35. Payen E, Pailhoux E, Abou Merhi R, et al. Characterization of ovine SRY transcript and developmental expression of genes involved in sexual differentiation. Int J Dev Biol. 1996;40:567–575 [PubMed] [Google Scholar]
- 36. Xi W, Lee CK, Yeung WS, et al. Effect of perinatal and postnatal bisphenol A exposure to the regulatory circuits at the hypothalamus-pituitary-gonadal axis of CD-1 mice. Reprod Toxicol. 2011;31:409–417 [DOI] [PubMed] [Google Scholar]
- 37. Nativelle-Serpentini C, Richard S, Séralini GE, Sourdaine P. Aromatase activity modulation by lindane and bisphenol-A in human placental JEG-3 and transfected kidney E293 cells. Toxicol In Vitro. 2003;17:413–422 [DOI] [PubMed] [Google Scholar]
- 38. Chung E, Genco MC, Megrelis L, Ruderman JV. Effects of bisphenol A and triclocarban on brain-specific expression of aromatase in early zebrafish embryos. Proc Natl Acad Sci USA. 2011;108:17732–17737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kubo K, Arai O, Omura M, Watanabe R, Ogata R, Aou S. Low dose effects of bisphenol A on sexual differentiation of the brain and behavior in rats. Neurosci Res. 2003;45:345–356 [DOI] [PubMed] [Google Scholar]
- 40. Kim JY, Han EH, Kim HG, et al. Bisphenol A-induced aromatase activation is mediated by cyclooxygenase-2 up-regulation in rat testicular Leydig cells. Toxicol Lett. 2010;193:200–208 [DOI] [PubMed] [Google Scholar]
- 41. Ye L, Zhao B, Hu G, Chu Y, Ge RS. Inhibition of human and rat testicular steroidogenic enzyme activities by bisphenol A. Toxicol Lett. 2011;207:137–142 [DOI] [PubMed] [Google Scholar]
- 42. Grasselli F, Baratta L, Baioni L, et al. Bisphenol A disrupts granulosa cell function. Domest Anim Endocrinol. 2010;39:34–39 [DOI] [PubMed] [Google Scholar]
- 43. Peretz J, Gupta RK, Singh J, Hernandez-Ochoa I, Flaws JA. Bisphenol A impairs follicle growth, inhibits steroidogenesis, and downregulates rate-limiting enzymes in the estradiol biosynthesis pathway. Toxicol Sci. 2011;119:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwintkiewicz J, Nishi Y, Yanase T, Giudice LC. Peroxisome proliferator-activated receptor-γ mediates bisphenol A inhibition of FSH-stimulated IGF-1, aromatase, and estradiol in human granulosa cells. Environ Health Perspect. 2010;118:400–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lei L, Jin S, Gonzalez G, Behringer RR, Woodruff TK. The regulatory role of Dicer in folliculogenesis in mice. Mol Cel Endocr. 2010;315:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagaraja AK, Andreu-Vieyra C, Franco HL, et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torley KJ, da Silveira JC, Smith P, Anthony RV, Veeramachaneni DN, Winger QA, Bouma GJ. Expression of miRNAs in ovine fetal gonads: potential role in gonadal differentiation. Reprod Biol Endocrinol. 2011;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bannister SC, Smith CA, Roeszler KN, Doran TJ, Sinclair AH, Tizard ML. Manipulation of estrogen synthesis alters MIR202* expression in embryonic chicken gonads. Biol Reprod. 2011;85:22–30 [DOI] [PubMed] [Google Scholar]
- 50. Mishima T, Takizawa T, Luo SS, et al. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822 [DOI] [PubMed] [Google Scholar]
- 51. Menghini R, Casagrande V, Cardellini M, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–1532 [DOI] [PubMed] [Google Scholar]
- 52. Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in MicroRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biochem. 2008;105:824–834 [DOI] [PubMed] [Google Scholar]
- 53. Hsu PY, Deatherage DE, Rodriguez BA, et al. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutat Res. 2011;714:105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Avissar-Whiting M, Veiga KR, Uhl KM, et al. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Tox. 2010;29:401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tilghman SL, Bratton MR, Segar HC, et al. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One. 2012;7:e32754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bannister SC, Tizard ML, Doran TJ, Sinclair AH, Smith CA. Sexually dimorphic microRNA expression during chicken embryonic gonadal development. Biol Reprod. 2009;81:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sirotkin AV, Ovcharenko D, Grossmann R, Laukova M, Mlyncek M. Identification of microRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219:415–420 [DOI] [PubMed] [Google Scholar]
- 59. Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707 [DOI] [PubMed] [Google Scholar]
- 60. Zhou Y, Zhu Y, Zhang S, Wang H, Wang S, Yang X. MicroRNA expression profiles in premature ovarian failure patients and its potential regulate functions. Chin J Birth Health Hered. 2011;19:20–22 [Google Scholar]
- 61. Hunt SM, Clarke CL. Expression and hormonal regulation of the Sox4 gene in mouse female reproductive tissues. Biol Reprod. 1999;61:476–481 [DOI] [PubMed] [Google Scholar]
- 62. Komatsu N, Hiraoka Y, Shiozawa M, Ogawa M, Aiso S. Cloning and expression of Xenopus laevis xSox12 cDNA. Biochim Biophys Acta. 1996;1305:117–119 [DOI] [PubMed] [Google Scholar]
- 63. Narahara M, Yamada A, Hamada-Kanazawa M, Kawai Y, Miyake M. cDNA cloning of the Sry-related gene Sox6 from rat with tissue-specific expression. Biol Pharm Bull. 2002;25:705–709 [DOI] [PubMed] [Google Scholar]
- 64. Chaboissier MC, Kobayashi A, Vidal VI,. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901 [DOI] [PubMed] [Google Scholar]
- 65. Zhang L, Lin D, Zhang Y, Ma G, Zhang W. A homologue of Sox11 predominantly expressed in the ovary of the orange-spotted grouper Epinephelus coioides. Comp Biochem Physiol B Biochem Mol Biol. 2008;149:345–353 [DOI] [PubMed] [Google Scholar]
- 66. Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007;236:2356–2366 [DOI] [PubMed] [Google Scholar]
- 67. Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nothnick WB, Healy C, Hong X. Steroidal regulation of uterine miRNAs is associated with modulation of the miRNA biogenesis components Exportin-5 and Dicer1. Endocrine. 2010;37:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Davis-Dusenbery BN, Hata A. Mechanisms of control of microRNA biogenesis. J Biochem. 2010;148:381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chou WC, Chen JL, Lin CF, Chen YC, Shih FC, Chuang CY. Biomonitoring of bisphenol A concentrations in maternal and umbilical cord blood in regard to birth outcomes and adipokine expression: a birth cohort study in Taiwan. Environ Health. 2011;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kosarac I, Kubwabo C, Lalonde K, Foster W. A novel method for the quantitative determination of free and conjugated bisphenol A in human maternal and umbilical cord blood serum using a two-step solid phase extraction and gas chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;898:90–94 [DOI] [PubMed] [Google Scholar]
- 72. Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev. 2009;30:75–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shelby M. National toxicology program—CERHR monograph on the potential human reproductive and developmental effects of bisphenol A. NIH publication no 2008;08-5994. Bethesda, Maryland: National Institutes of Health [Google Scholar]
- 74. Prins GS, Ye SH, Birch L, Ho SM, Kannan K. Serum bisphenol A pharmacokinetics and prostate neoplastic responses following oral and subcutaneous exposures in neonatal Sprague-Dawley rats. Reprod Toxicol. 2011;31:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rubin BS. Bisphenol A: an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol. 2011;127:27–34 [DOI] [PubMed] [Google Scholar]
- 76. Vandenberg LN, Colborn T, Hayes TB, et al. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev. 2012;33:378–455 [DOI] [PMC free article] [PubMed] [Google Scholar]