Abstract
Objective
Medication safety requires that each drug be monitored throughout its market life as early detection of adverse drug reactions (ADRs) can lead to alerts that prevent patient harm. Recently, electronic medical records (EMRs) have emerged as a valuable resource for pharmacovigilance. This study examines the use of retrospective medication orders and inpatient laboratory results documented in the EMR to identify ADRs.
Methods
Using 12 years of EMR data from Vanderbilt University Medical Center (VUMC), we designed a study to correlate abnormal laboratory results with specific drug administrations by comparing the outcomes of a drug-exposed group and a matched unexposed group. We assessed the relative merits of six pharmacovigilance measures used in spontaneous reporting systems (SRSs): proportional reporting ratio (PRR), reporting OR (ROR), Yule's Q (YULE), the χ2 test (CHI), Bayesian confidence propagation neural networks (BCPNN), and a gamma Poisson shrinker (GPS).
Results
We systematically evaluated the methods on two independently constructed reference standard datasets of drug–event pairs. The dataset of Yoon et al contained 470 drug–event pairs (10 drugs and 47 laboratory abnormalities). Using VUMC's EMR, we created another dataset of 378 drug–event pairs (nine drugs and 42 laboratory abnormalities). Evaluation on our reference standard showed that CHI, ROR, PRR, and YULE all had the same F score (62%). When the reference standard of Yoon et al was used, ROR had the best F score of 68%, with 77% precision and 61% recall.
Conclusions
Results suggest that EMR-derived laboratory measurements and medication orders can help to validate previously reported ADRs, and detect new ADRs.
Keywords: Pharmacovigilance, Post-marketing drug surveillance, Adverse drug reactions, Automated laboratory signals, Electronic medical records
Introduction
Pirmohamed et al1 defined an adverse drug reaction (ADR) as ‘any undesirable effect of a drug beyond its anticipated therapeutic effects occurring during clinical use.’ ADRs comprise a major public health problem accounting for up to 5% of hospital admissions,1 28% of emergency department visits,2 and 5% of hospital deaths.3 Most ADRs due to prescribing errors are potentially preventable.4 However, premarketing clinical drug trials may not detect other types of ADRs because such studies are often small, short, and biased by the exclusion of patients with comorbid disease. Thus, premarketing trials do not reflect actual clinical medication use situations for diverse (eg, inpatient) populations. Those ADRs not discovered until the post-marketing phase may result in patient morbidity due to the slow and incomplete spontaneous reporting systems (SRSs) currently used to detect such ADR signals.5 Moreover, the current regulatory environment encourages efficacious drugs to be brought to market as soon as possible.6 Thus real-time clinical post-marketing surveillance is increasingly important, as exemplified by recent market withdrawals of numerous drugs such as Rofecoxib (Vioxx) and Cerivastatin (Baycol).7 8
Many previous post-marketing surveillance analyses were based on adverse event reports voluntarily submitted to the national SRSs via industry sponsored phase IV clinical trials or through prospective clinical registries. The SRSs have served as the core data-generating method for drug safety surveillance since 1960. This system relies solely on healthcare professionals, consumers, and manufacturers to identify and report suspected ADRs. Although the reports have provided valuable information for clinical decision making, the SRS approach has the serious limitation of under-reporting. While phase IV clinical trials and clinical registries can address some of the limitations of patient case mix inherent in phase III trials, they remain restrictive in terms of surveillance outcomes. To supplement previous methods, researchers have recently begun to explore alternative ADR signal discovery approaches such as mining structured and unstructured data in electronic medical record (EMR) systems.9 Institutional EMRs have emerged as a prominent resource for observational research as they contain not only detailed patient information but also copious longitudinal clinical data.
In this study, we evaluated several statistical measures widely applied to spontaneous reporting databases in order to identify ADR signals from retrospective EMR data. We focused on EMR records that included laboratory test results and medication orders. For example, elevated levels of serum creatinine and blood urea nitrogen (BUN) following a medication order (eg, for an aminoglycoside or an ACE inhibitor) may signal ADRs causing renal dysfunction. The use of EMR-derived laboratory signals in drug safety studies may overcome premarketing errors caused by biased reporting. This approach has the potential to support active, real-time surveillance.
Background
Analytical methods applied to post-marketing surveillance include biostatistical and data mining algorithms. In SRSs, straightforward pharmacovigilance methods involve the calculation of frequentist metrics such as proportional reporting ratio (PRR),10 report OR (ROR),11 and Yule's Q.12 These methods were developed to determine whether a drug–ADR pair is a signal or not based on disproportionate reporting. There are in addition more complex algorithms based on Bayesian statistics such as the gamma Poisson shrinker (GPS),13 14 the multi-item gamma Poisson shrinker (MGPS),15 and empirical Bayesian geometric means (EBGMs).16 17 Moreover, Bayesian confidence propagation neural network (BCPNN) analysis was developed based on Bayesian logic where the relation between prior and posterior probability was expressed as the ‘information component’ (IC).18–20 Studies have also investigated James-Stein type shrinkage estimation strategies in a Bayesian logistic regression model to analyze spontaneous adverse event reporting data.21 More recently, Ahmed et al22 23 proposed false discovery rate (FDR) estimation for frequentist methods to address the limitation of arbitrary thresholds. Data mining algorithms have also been developed to mine drug–ADR associations from spontaneous reports,9 which include association rule mining24 and biclustering.25 Tatonetti et al26 proposed an algorithm to identify drug–drug interactions from adverse event reports by analyzing latent signals that indirectly provide evidence for ADRs.
Recently, investigators have explored the use of EMRs for ADR signal detection. Wang et al27 employed natural language processing (NLP) techniques to extract drug–ADR candidate pairs from narrative EMRs and applied the χ2 test to detect ADR signals. Other groups have applied statistical and data mining methods on structured or coded data in EMRs28–36 for ADR signal detection. Jin et al37 proposed a new interestingness measure called residual-leverage for association rule mining to identify ADR signals from healthcare administrative databases. Ji et al38 introduced potential causal association rules to generate potential causal relationships between a drug and ICD-9 coded signs or symptoms in EMRs.
However, there has been little work exploring large EMR databases for ADR signals involving drug–laboratory test interactions. Tegeder et al39 found that laboratory data could identify up to two-thirds of all ADRs; however, only one third of the ADRs which could have been detected using abnormal laboratory signals were recognized by attending physicians. Ramirez et al40 implemented a prospective pharmacovigilance program called the Pharmacovigilance Program from Laboratory Signals at Hospital (PPLSH) based on predefined abnormal laboratory values. PPLSH starts with all abnormal laboratory signals, and then filters out alternative causes (non-drug related causes) with EMR review. After a year of operation at La Paz University Hospital in Madrid, Spain, PPLSH was found to be useful for the detection and evaluation of specific severe ADRs associated with increases in morbidity and duration of hospitalization.40 In a recent publication, Yoon et al41 demonstrated laboratory abnormality to be a valuable source for pharmacovigilance by examining the OR of laboratory abnormalities between a drug-exposed and a matched unexposed group. Schildcrout et al33 analyzed the relationship between insulin infusion rates and blood glucose levels (GLUC) in patients in an intensive care unit. Nevertheless, established analytical methods for laboratory-based ADR signal detection are lacking.
Furthermore, it remains unclear how effective the SRSs-based ADR detection methods are when they are applied to EMR data. In a related publication, Zorych et al42 explored the application of various disproportionality measures for pharmacovigilance by mapping the EMR data to drug-condition two-by-two tables. However, the adverse events in their study are not restricted to laboratory tests and the study design does not require an event-free ‘clean’ period prior to the first condition occurrence.42 In addition, no reference standard was utilized to formally evaluate the disproportionality methods. In this study, we implemented six statistical measures widely used for spontaneous reporting databases, namely PRR, ROR, Yule's Q, the χ2 test, BCPNN, and GPS, and applied them to detect associations between drug intake and laboratory-based ADR signals from the Vanderbilt University Medical Center's (VUMC) inpatient EMRs. These methods were systematically evaluated using two independently developed reference standard datasets.
Methods
Data source
The VUMC has created a comprehensive relational database called the Synthetic Derivative that contains clinical information derived from the VUMC EMRs and other clinical systems, and includes laboratory values, imaging and pathology reports, billing codes, and clinical narratives from all VUMC inpatient and outpatient settings. All Synthetic Derivative records have been de-identified as personal identifiers were stripped and each record was labeled with a research unique identifier. The Synthetic Derivative contains data derived from approximately 1.9 million individuals (growing at a rate of 150 000 new individuals per year), with highly detailed longitudinal data for about 1 million. For this study, we obtained laboratory test results and drug ordering data for all inpatients from 1999 to 2011. At the time of data acquisition in August 2011, the Synthetic Derivative contained hospitalization records for 634 238 patients who stayed at VUMC between 1999 and 2011, of which 303 589 had documented laboratory test records, 225 778 had medication orders, and 187 595 had both. This study was approved by the VUMC institutional review board.
Study design
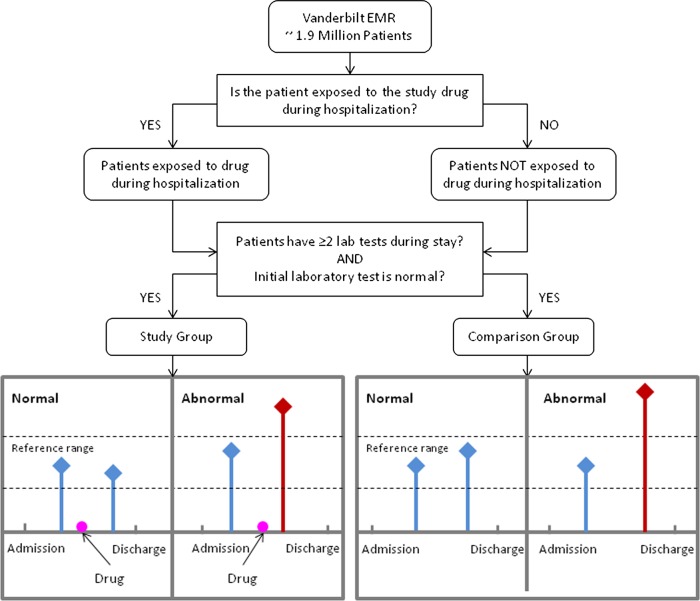
The study aimed to correlate abnormal laboratory results with specific drug administration by comparing the outcomes of patients who were exposed with the outcomes of those unexposed to the study drug where the exposed group consists of patients who had been administered the drug during hospitalization. Patients in the exposed and unexposed groups may have an abnormal laboratory test value or may not have the condition at all. To increase the likelihood that a laboratory test abnormality signified an ADR event, the study only considered a test result abnormal if the test value fell outside of the reference normal range by at least the width of the stated normal reference range. For example, the normal range for the creatinine test in VUMC's EMR is 0.7–1.5. Our study would consider a patient's creatinine measurement elevated if its value is above 2.3 (since 1.5–0.7=0.8, and 0.8+1.5=2.3. An analogous rule applied for lower than normal laboratory test results. The reference normal ranges of each laboratory test used in our study and corresponding abnormal thresholds are provided in online supplementary table S1. On the other hand, a laboratory test was considered normal if its value was within the laboratory-reported normal range.
The study only included data for patients who had at least two results for a specific laboratory test during one admission, with the first laboratory test result being normal (figure 1). The time of admission was set as ‘day zero’ and all drug administration and laboratory result timings were represented as days elapsed since admission until discharge. Each patient in the exposed group was randomly matched to four unexposed patients by age group, gender, race, and major diagnoses (ICD-9 codes grouped according to the Clinical Classifications Software (CCS)43 diagnosis categories by the Agency for Healthcare Research and Quality). The patient ages in the de-identified database were classified as: (1) 18–25; (2) 26–35; (3) 36–45; (4) 46–55; (5) 56–65; (6) 66–75; and (7) >75 years. For two patients to be considered a match, they must be in the same age group and also have the same gender and race. In addition, we attempted to match on diagnoses by searching for patients with the most similar disease codes at admission. For instance, each patient is associated with a vector corresponding to the CCS codes whereby a value of 1 indicates the patient has the disease and a 0 indicates no disease. Patients with the largest overlap of 1's are considered most similar.
Figure 1.
Study design overview. Correlation of abnormal laboratory results with specific drug administration through comparison of the outcomes of patients who were exposed with the outcomes of those who were unexposed to the study drug. Patients must have at least two laboratory measurements during one hospitalization, the first of which must be normal. EMR, electronic medical record. This figure is only reproduced in colour in the online version.
We assumed that a potential drug–laboratory test ADR involved an individual with a normal pre-drug laboratory test result who later, after drug administration, had an abnormal laboratory result. A large number of abnormal laboratory test events in a specific exposed (study) group compared to the unexposed (comparison) group would support an association between the study drug and an ADR. This study examined some of the most commonly performed laboratory tests (based on the institution conducting a threshold number of tests each month). Since laboratory abnormalities have direction (ie, out-of-range high values or out-of-range low values), the study separated the ADRs into ‘increasing’ and ‘decreasing’ signals; therefore, we tabulated two contingency tables for each drug–laboratory test pair (table 1). In this study, a minimum of 25 patients was required per drug–laboratory test pair for signal detection. In the event that measures could not be calculated, the missing combinations were excluded from the analysis.
Table 1.
Example of a 2×2 contingency table for the drug lisinopril and elevated creatinine test
| Laboratory of interest | |||
|---|---|---|---|
| Abnormal (increasing) | Normal | ||
| Drug of interest | Taking | a=45 | b=4851 |
| Not taking | c=109 | d=19475 | |
Signal detection algorithms
In this study, we investigated six signal detection measures: PRR, ROR, Yule's Q (YULE), the χ2 test (CHI), BCPNN, and GPS. For all of these methods, our study calculated an index value for each drug–laboratory test pair from a two-way contingency table such as shown in table 1 (detailed formulas for each method appear in the online supplementary appendix). If the index value exceeded a certain critical value, it was considered to be a signal. The indices and standard critical values used in our study are taken from van Puijenbroek et al12 and Matsushita et al44 and are shown in table 3.
Table 3.
Signal detection method evaluation results on two reference standard datasets using standard 95% CI critical values for each method: SE, SD
| Method | Critical values | VUMC reference dataset (all evidence) | Yoon et al al41 reference dataset | ||||
|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | ||
| CHI | p<0.05 | 0.57 | 0.69 | 0.62 | 0.73 | 0.57 | 0.64 |
| PRR | PRR: 1.96 SE>1 | 0.58 | 0.67 | 0.62 | 0.74 | 0.61 | 0.67 |
| ROR | ROR: 1.96 SE>1 | 0.58 | 0.67 | 0.62 | 0.77 | 0.61 | 0.68 |
| YULE | YULE: 1.96>0 | 0.57 | 0.67 | 0.62 | 0.74 | 0.61 | 0.67 |
| BCPNN | IC: 2 SD>0 | 0.58 | 0.62 | 0.60 | 0.76 | 0.46 | 0.58 |
| GPS | EB05: > 2 | 0.85 | 0.29 | 0.43 | 0.67 | 0.22 | 0.33 |
BCPNN, Bayesian confidence propagation neural networks; CHI, χ2 test; EB05, lower one-sided 95% CI for GPS (please refer to the online supplementary appendix for detailed definition); F, F score; GPS, gamma Poisson shrinker; IC, information component; P, precision; PRR, proportional reporting ratio; R, recall; ROR, reporting OR; SD, standard deviation; SE, standard error; YULE, Yule's Q.
Evaluation
Since there is no gold standard for ADR signal detection in general, systematic evaluation is often difficult and most studies must rely on literature sources or physicians to verify top ranked predictions. Here, our study created an evidence-based reference standard for nine selected drugs based on the strength of evidence for previously known associations, and conducted a systematic evaluation of the six commonly applied pharmacovigilance methods for ‘discovering’ known ADRs. A total of nine drugs were selected from five drug classes based on their common use in clinical practice, and their known ability to produce laboratory-measurable side effects (table 2).
Table 2 .
Nine drugs chosen for the VUMC reference standard dataset
| Drug class | Drug name |
|---|---|
| Aminoglycosides | Gentamicin |
| Sulfamethoxazole/trimethoprim | Sulfamethoxazole |
| ACE inhibitors | Enalapril |
| Lisinopril | |
| Antifungals | Fluconazole |
| Amphotericin | |
| NSAIDs | Aspirin |
| Ibuprofen | |
| Ketorolac tromethamine |
NSAIDs, non-steroidal anti-inflammatory drugs; VUMC, Vanderbilt University Medical Center.
First, project members presented the list of study drugs and laboratory tests to a board-certified internist with a decade of practice experience in an academic setting. The physician manually indicated the existence of any drug–laboratory test associations based on individual and general medical knowledge. The study then supplemented the physician-derived list with evidence found in Epocrates, a widely used drug–adverse event source for medical professionals (available at http://www.epocrates.com) and DoubleCheckMD (available at http://doublecheckmd.com). The study physician classified the strength of evidence for each drug–laboratory test association in the reference standard into two categories: (1) high, if confirmed by Epocrates and/or a physician; or (2) likely, if evidence was found in DoubleCheckMD, the association was related to known effects (eg, abnormal bilirubin levels reported but not specified in the information resource as being conjugated bilirubin or unconjugated bilirubin; all three test results (bilirubin, conjugated bilirubin, and unconjugated bilirubin) then permitted as ADRs), or there was an indirect association (eg, if an abnormal red blood cell count is a direct drug effect, then the packed cell volume could be indirectly related because it is the proportion of blood volume occupied by red blood cells). Associations with ‘high’ strength of evidence are public knowledge, while those in the ‘likely’ category are potential new ADRs. Our final reference set contained 378 drug–event pairs (nine drugs and 42 laboratory abnormalities). There were 55 drug–laboratory test associations in the ‘high’ class and 107 associations in the ‘likely’ class.
Moreover, a reference standard of 470 drug–laboratory event pairs developed by Yoon et al41 was also used in the study for evaluation. Their dataset consists of 10 drugs: ciprofloxacin, clopidogrel, eptoposide, fluorouracil, ketorolac, levofloxacin, methotrexate, ranitidine, rosuvastatin, and valproate.
Evaluation metric
For each method, the analysis compared the detected drug–laboratory test associations with the ‘true’ pairs in both the VUMC reference dataset and the Yoon et al41 dataset. To assess the performance of each method, the study calculated precision (P), recall (R), and F score (F). The study calculated precision as the number of `true' ADRs retrieved by a method divided by the total number of ADRs found by that method (ie, P=TP/(TP+FP)) where TP is the true positive and FP is the false positive. Recall was the number of `true' ADRs returned by a method divided by the total number of true ADRs (ie, R=TP/(TP+FN)) where FN is the false negatives. There is often a trade-off between precision and recall, and it is important to obtain a balance between the two. To address this, we also calculated the F score, which is a weighted average of precision and recall (ie, F=2PR/(P+R)) where the best F score is 1 and worst value is 0.
Results
The six pharmacovigilance methods (CHI, YULE, PRR, ROR, BCPNN, and GPS) were systematically evaluated against the VUMC reference dataset and the reference dataset created by Yoon et al.41 The performance metrics were calculated using all evidence in the reference standard and evaluation results are summarized in table 3.
Evaluation against the VUMC reference dataset showed that CHI, PRR, ROR, and YULE performed similarly with an F score of 62%, while BCPNN had a lower F score of 60%, and GPS had the lowest F score of 43%. However, GPS had a higher precision of 85% but its low F score was due to a low recall of 29%. Evaluation on the reference standard dataset by Yoon et al showed that ROR achieved the best F score of 68% with an overall precision of 77% and recall of 61%.
The study also correlated the physician-designated evidence classes for drug–laboratory test pairs in our reference standard to evaluate the methods’ ability to ‘discover’ new associations. The evaluation results on each category are summarized in table 4. Of note, when an evaluation study labels one category of evidence as a ‘true’ association, the opposing category of evidence is assumed to be ‘false.’ In such cases, precision is biased by the size of the evidence category as it is the fraction of detected signals that are true. On the other hand, recall is the fraction of true ADRs detected; therefore, it should be the measure we focus on here and the higher the recall the better.
Table 4.
Evaluation results using different evidence
| Method | ‘High’ evidence | ‘Likely’ evidence | ||||
|---|---|---|---|---|---|---|
| P | R | F | P | R | F | |
| CHI | 0.37 | 0.65 | 0.47 | 0.42 | 0.72 | 0.53 |
| PRR | 0.37 | 0.59 | 0.45 | 0.45 | 0.72 | 0.55 |
| ROR | 0.37 | 0.59 | 0.45 | 0.45 | 0.72 | 0.55 |
| YULE | 0.36 | 0.59 | 0.44 | 0.44 | 0.72 | 0.55 |
| BCPNN | 0.35 | 0.53 | 0.42 | 0.45 | 0.68 | 0.54 |
| GPS | 0.50 | 0.29 | 0.37 | 0.60 | 0.29 | 0.39 |
BCPNN, Bayesian confidence propagation neural networks; CHI, χ2 test; F, F score; GPS, gamma Poisson shrinker; P, precision; PRR, proportional reporting ratio; R, recall; ROR, reporting OR; YULE, Yule's Q.
Discussion
Our ADR detection analysis using inpatient laboratory test results illustrates the potential utility of this approach for future ADR monitoring. To identify relationships between drugs and laboratory tests, the study compared association measures: CHI, ROR, PRR, YULE, BCPNN, and GPS. Among these methods, CHI, ROR, PRR, and YULE consistently performed better than BCPNN and GPS on two independently constructed reference standard datasets. In addition, most methods had a higher precision when evaluated on the reference dataset by Yoon et al compared with the VUMC dataset. This may imply performance depends on the study drug selected. Moreover, different evidence classes for drug–laboratory test pairs were analyzed and results indicated that our approach can detect laboratory–ADR signals otherwise missed by physicians.
The study design implemented here was different from the Comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm used in Yoon et al.41 Instead of using the laboratory test result immediately after drug initiation, the CLEAR algorithm calculated a representative value from an observation period between the drug initiation time and patient discharge. For further analysis, we implemented the CLEAR algorithm on the VUMC EMR data and observed lower performance compared to our actual study design. For example, when evaluating our algorithm on the VUMC reference set using the χ2 test, we obtained an F score of 62% with 57% precision and 69% recall; however, the CLEAR algorithm achieved an F score of 55% with 48% precision and 63% recall on the same dataset. Moreover, when the ROR method for signal detection was used on the reference dataset of Yoon et al, our algorithm achieved an F score of 68% (77% precision and 61% recall), while the CLEAR algorithm achieved an F score of 44% (61% precision and 34% recall), which is lower than reported in their study. The difference in performance may be due to differences in the study populations. In their study, each patient in the exposed group was matched to four unexposed patients by age (±1 year), gender, admitting department, and major diagnosis based on the blocks of categories in ICD-10. Differences in clinical practice patterns between the USA and Korea may also partially explain the difference in performance.
The term ‘ADR’ is broad and includes various subtypes. Some ADRs can only be detected through changing laboratory test results. Other ADRs may identified when patients describe their symptoms to clinicians, such as depression, muscle stiffness, or sleeplessness, which are typically recorded in narrative notes. To identify such ADR signals from narrative reports in the EMR, Wang et al27 developed an NLP framework and employed the χ2 test. Their study achieved 31% precision and 75% recall (ie, an F score of 44%). Although the data utilized were completely different, the current study results still suggest that application of simple association measures to laboratory data for ADR detection can potentially achieve better performance than with subjective findings mined via NLP.
The goal of our study was to develop data-driven methods to identify existing and new drug–ADR pairs. Therefore, our evaluation focused on detecting drug–ADR pairs rather than single ADR events. However, the laboratory-related ADRs defined in this study may not have been caused by drugs. To assess whether the cases extracted from the EMRs in our study design (figure 1) are truly drug related, we conducted a manual chart review for the drug–laboratory test pair ‘lisinopril versus elevated creatinine,’ which was selected based on the reviewer's clinical expertise. This manual evaluation included review of inpatient creatinine measurements to determine whether acute kidney injury (AKI) occurred after admission (thus ensuring that the creatinine increase criteria were indeed met), then an assessment of whether the patient was on the lisinopril prior to admission, along with whether the medication was given at least once 1 day before the AKI event, and finally a manual chart review of the progress notes and discharge summaries for other factors that are known to cause AKI or could have synergistically caused AKI. Although comprehensive ADR causality assessment scales45 such as the Naranjo instrument46 exist, we used a relatively simple approach based on expert judgment, similar to the Swedish method,47 as we had only one reviewer. The physician reviewer used a Likert scale of 1–5 to rate the likelihood of lisinopril as the cause of the elevated creatinine level. Scores of 1 and 2 indicate that it is unlikely lisinopril is related to the AKI. The rating 3 suggests contributory and multiple mechanisms as a group are likely to have caused the AKI. Finally, scores of 4 and 5 signify causative relationships because no other possible causes were identified in the EMR review. The chart review criteria were developed by the physician reviewer based on his clinical expertise. Detailed definitions of the chart review criteria are given in online supplementary table S2. We then randomly selected 23 of the 45 patients in our dataset who had experienced elevated creatinine after taking lisinopril during their hospital stay for chart review (table 1). As a result, two of the 23 cases were assigned scores of 4–5 (9%), 10 a rating of 3 (44%), and seven scores of 1–2 (30%), with four unknowns (17%). The unknowns were due to insufficient clinical notes. The percentage of solely drug-caused ADR cases (9%) in our study is higher than the 6.35% reported in Ramirez et al,40 possibly because our study design is more restrictive (figure 1). A lot of cases had a score of 3, where the medication contributed to the adverse event but might not be the only contributor. If we also include those cases as drug related, the percentage of true cases would be 53%, which is significantly higher than reported in Ramirez et al.40
Furthermore, for error analysis, our project team consulted another physician to analyze the false positive predictions made by the χ2 test. The physician speculated that many false positive associations were due to confounding of the indication for the drug treatment for a particular disease. For example, elevated creatinine was falsely detected to be associated with fluconazole, but acute renal failure is a common effect of systemic fungemia and fluconazole is often used to treat that condition. Similarly, BUN levels were also found to be elevated after fluconazole initiation. However, this elevation also may be due to systemic fungemia, which is not a direct drug effect. As another example, GLUC was identified to be elevated after fluconazole initiation; however, infection can increase systemic stress, which in turn increases glucose. Although we tried to mitigate the effect of confounders by matching the patient groups, confounders may still exist due to the fuzzy match on patient disease profile at admission. In addition, as only the diagnosis codes entered at admission were considered, any change in patient disease status would have been missed and so a new disease may have developed and acted as a confounder. False positive signals may also be generated because of the widespread use of a particular drug by a specific group of patients. For instance, lisinopril was detected to be associated with GLUC; however, most patients with diabetes are on ACE inhibitors. Thus , more accurate patient populations should be defined in future studies.
The current study had a number of flaws and limitations. First, the reference standards used for ADR detection were neither fully authoritative nor complete. Second, clinicians often prescribe drugs to counteract known anticipated side effects (eg, starting potassium supplements to counteract potential hypokalemia when diuretics are prescribed). Such behaviors can reduce the strength of ADR signals. This may partially explain the lower recall for the ‘high’ evidence class compared to the ‘likely’ class. Additionally, ‘confounding by indication’ may occur. If a disease typically elevates the values of a specific laboratory test, and a medication known to treat the disease is given early in the course of the disease, the value may rise as a result of the underlying disease process, and not because of the medication. Conversely, if the medication is started as the disease is resolving, the observed fall in laboratory test result values may be due to the underlying disease process and not the medication. Moreover, we can only detect ADRs with a short latency period because the study design required ADRs to have occurred during the hospital stay. In addition, the study assessed various pharmacovigilance methods based on the reference standards, not on patient outcomes. Thus, different results may be obtained if other data are used. Lastly, inpatients have acute diseases that predispose them to have abnormal laboratory results due to their weakened condition. Also, they are more likely to take multiple drugs, and some drugs may interact with others to affect laboratory outcomes. Future studies should more fully explore risk adjustment methods to correct for biases and comorbidities.
Conclusion
Identifying ADRs accurately and in a timely manner is a constant challenge, and critical for patient safety. This paper demonstrated that it is feasible to detect known ADR signals as well as identify new ADRs using inpatient laboratory results and time-stamped records of medication orders. Pharmacovigilance methods commonly used on SRSs (CHI, ROR, PRR, YULE, BCPNN, and GPS) were compared as regards to the detection of drug–laboratory test associations on two independently constructed reference standard datasets. Evaluation on the VUMC reference dataset showed that CHI, ROR, PRR, and YULE all had the same F score (62%). When the reference standard dataset by Yoon et al was used, ROR had the best F score of 68% with an overall precision of 77% and recall of 61%. The study suggests that analytical laboratory measurements from clinical practice may in the future contribute greatly to pharmacovigilance.
Acknowledgments
We would like to thank Dr Dukyong Yoon and Dr Rae Woong Park for sharing their reference standard evaluation dataset of drug–event pairs and for helping us through the implementation of their CLEAR algorithm. The dataset used for the analyses described in this study was obtained from Vanderbilt University Medical Center's Synthetic Derivative which is supported by institutional funding and by Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH.
Footnotes
Contributors: ML, MEM, and HX were responsible for the overall design, development, and evaluation of this study. RAM and JCD contributed to the study design. ERMH constructed the reference standard evaluation dataset using VUMC's EMR. ML, JSS, and HX worked on the methods and carried out the experiments. MEM designed and performed the chart review for the drug–ADR pair ‘lisinopril and creatinine.’ ML and HX wrote most of the manuscript, with MEM, RAM, and JCD also contributing to the writing and editing. All authors reviewed the manuscript critically for scientific content, and all authors gave final approval of the manuscript for publication.
Funding: The study was partially supported by NLM-NIH training grant 3T15LM007450-08S1 and grants NLM R01-LM007995 and NCI R01CA141307. Dr Matheny was supported by a Veterans Administration HSR&D Career Development Award (CDA-08-020).
Competing interests: None.
Ethics approval: Vanderbilt University Medical Center Institutional Review Board approved this study.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Pirmohamed M, Breckenridge AM, Kitteringham NR, et al. Adverse drug reactions. BMJ 1998;316:1295–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel P, Zed PJ. Drug-related visits to the emergency department: how big is the problem? Pharmacotherapy 2002;22:915–23 [DOI] [PubMed] [Google Scholar]
- 3.Juntti-Patinen L, Neuvonen PJ. Drug-related deaths in a university central hospital. Eur J Clin Pharmacol 2002;58:479–82 [DOI] [PubMed] [Google Scholar]
- 4.Bates DW, Cullen DJ, Laird N, et al. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE Prevention Study Group JAMA 1995;274:29–34 [PubMed] [Google Scholar]
- 5.Cullen DJ, Sweitzer BJ, Bates DW, et al. Preventable adverse drug events in hospitalized patients: a comparative study of intensive care and general care units. Crit Care Med 1997;25:1289–97 [DOI] [PubMed] [Google Scholar]
- 6.Edwards IR. The accelerating need for pharmacovigilance. J R Coll Physicians Lond 2000;34:48–51 [PMC free article] [PubMed] [Google Scholar]
- 7.Moore TJ, Cohen MR, Furberg CD. Serious adverse drug events reported to the Food and Drug Administration, 1998–2005. Arch Intern Med 2007;167:1752–9 [DOI] [PubMed] [Google Scholar]
- 8.Giacomini KM, Krauss RM, Roden DM, et al. When good drugs go bad. Nature 2007;446:975–7 [DOI] [PubMed] [Google Scholar]
- 9.Harpaz R, Dumouchel W, Shah NH, et al. Novel Data-Mining Methodologies for Adverse Drug Event Discovery and Analysis. Clin Pharmacol Ther 2012;91:1010–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans SJ, Waller PC, Davis S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol Drug Saf 2001;10:483–6 [DOI] [PubMed] [Google Scholar]
- 11.Szarfman A, Machado SG, O'Neill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA's spontaneous reports database. Drug Saf 2002;25:381–92 [DOI] [PubMed] [Google Scholar]
- 12.van Puijenbroek EP, Bate A, Leufkens HG, et al. A comparison of measures of disproportionality for signal detection in spontaneous reporting systems for adverse drug reactions. Pharmacoepidemiol Drug Saf 2002;11:3–10 [DOI] [PubMed] [Google Scholar]
- 13.DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. The American Statistician 1999;53:177–202 [Google Scholar]
- 14.Ahmed I, Haramburu F, Fourrier-Reglat A, et al. Bayesian pharmacovigilance signal detection methods revisited in a multiple comparison setting. Stat Med 2009;28:1774–92 [DOI] [PubMed] [Google Scholar]
- 15.Hauben M. Application of an empiric Bayesian data mining algorithm to reports of pancreatitis associated with atypical antipsychotics. Pharmacotherapy 2004;24:1122–9 [DOI] [PubMed] [Google Scholar]
- 16.DuMouchel W, Smith ET, Beasley R, et al. Association of asthma therapy and Churg-Strauss syndrome: an analysis of postmarketing surveillance data. Clin Ther 2004;26:1092–104 [DOI] [PubMed] [Google Scholar]
- 17.Gould AL. Accounting for multiplicity in the evaluation of “signals” obtained by data mining from spontaneous report adverse event databases. Biom J 2007;49:151–65 [DOI] [PubMed] [Google Scholar]
- 18.Bate A, Lindquist M, Edwards IR, et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur J Clin Pharmacol 1998;54:315–21 [DOI] [PubMed] [Google Scholar]
- 19.Lindquist M, Edwards IR, Bate A, et al. From association to alert—a revised approach to international signal analysis. Pharmacoepidemiol Drug Saf 1999;8(Suppl 1):S15–25 [DOI] [PubMed] [Google Scholar]
- 20.Lindquist M, Stahl M, Bate A, et al. A retrospective evaluation of a data mining approach to aid finding new adverse drug reaction signals in the WHO international database. Drug Saf 2000;23:533–42 [DOI] [PubMed] [Google Scholar]
- 21.An L, Fung KY, Krewski D. Mining pharmacovigilance data using Bayesian logistic regression with James-Stein type shrinkage estimation. J Biopharm Stat 2010;20:998–1012 [DOI] [PubMed] [Google Scholar]
- 22.Ahmed I, Dalmasso C, Haramburu F, et al. False discovery rate estimation for frequentist pharmacovigilance signal detection methods. Biometrics 2010;66:301–9 [DOI] [PubMed] [Google Scholar]
- 23.Ahmed I, Thiessard F, Miremont-Salame G, et al. Pharmacovigilance data mining with methods based on false discovery rates: a comparative simulation study. Clin Pharmacol Ther 2010;88:492–8 [DOI] [PubMed] [Google Scholar]
- 24.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics 2010;11(Suppl 9):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harpaz R, Perez H, Chase HS, et al. Biclustering of adverse drug events in the FDA's spontaneous reporting system. Clin Pharmacol Ther 2010;89:243–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011;90:133–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Hripcsak G, Markatou M, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc 2009;16:328–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown JS, Kulldorff M, Chan KA, et al. Early detection of adverse drug events within population-based health networks: application of sequential testing methods. Pharmacoepidemiol Drug Saf 2007;16:1275–84 [DOI] [PubMed] [Google Scholar]
- 29.Berlowitz DR, Miller DR, Oliveria SA, et al. Differential associations of beta-blockers with hemorrhagic events for chronic heart failure patients on warfarin. Pharmacoepidemiol Drug Saf 2006;15:799–807 [DOI] [PubMed] [Google Scholar]
- 30.Hauben M, Madigan D, Gerrits CM, et al. The role of data mining in pharmacovigilance. Expert Opin Drug Saf 2005;4:929–48 [DOI] [PubMed] [Google Scholar]
- 31.FitzHenry F, Peterson JF, Arrieta M, et al. Medication administration discrepancies persist despite electronic ordering. J Am Med Inform Assoc 2007;14:756–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson JF, Shi Y, Denny JC, et al. Prevalance and clinical significance of discrepancies within three computerized pre-admission medication lists. AMIA Annu Symp 2010;2010:642–6. [PMC free article] [PubMed] [Google Scholar]
- 33.Schildcrout JS, Haneuse S, Peterson JF, et al. Analyses of longitudinal, hospital clinical laboratory data with application to blood glucose concentrations. Stat Med 2011;30:3208–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, Spickard A, 3rd, Miller RA, et al. Identifying UMLS concepts from ECG impressions using KnowledgeMap. AMIA Ann Symp 2005;2005:196–200. [PMC free article] [PubMed] [Google Scholar]
- 35.Denny JC, Peterson JF. Identifying QT prolongation from ECG impressions using natural language processing and negation detection. Stud Health Technol Inform 2007;129:1283–8 [PubMed] [Google Scholar]
- 36.Denny JC, Miller RA, Waitman LR, et al. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform 2009;78(Suppl 1):S34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin HD, Chen J, He HX, et al. Mining unexpected temporal associations: applications in detecting adverse drug reactions. IEEE T Inf Technol B 2008;12:488–500 [DOI] [PubMed] [Google Scholar]
- 38.Ji YQ, Ying H, Dews P, et al. A Potential Causal Association Mining Algorithm for Screening Adverse Drug Reactions in Postmarketing Surveillance. IEEE T Inf Technol B 2011;15:428–37 [DOI] [PubMed] [Google Scholar]
- 39.Tegeder I, Levy M, Muth-Selbach U, et al. Retrospective analysis of the frequency and recognition of adverse drug reactions by means of automatically recorded laboratory signals. Br J Clin Pharmacol 1999;47:557–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez E, Carcas AJ, Borobia AM, et al. A pharmacovigilance program from laboratory signals for the detection and reporting of serious adverse drug reactions in hospitalized patients. Clin Pharmacol Ther 2010;87:74–86 [DOI] [PubMed] [Google Scholar]
- 41.Yoon D, Park MY, Choi NK, et al. Detection of adverse drug reaction signals using an electronic health records satabase: Comparison of the Laboratory Extreme Abnormality Ratio (CLEAR) algorithm. Clin Pharmacol Ther 2012;91: 467–74. [DOI] [PubMed] [Google Scholar]
- 42.Zorych I, Madigan D, Ryan P, et al. Disproportionality methods for pharmacovigilance in longitudinal observational databases. Stat Methods Med Res 2011. doi:10.1177/0962280211403602 [DOI] [PubMed] [Google Scholar]
- 43.Elixhauser A, McCarthy EM. Clinical classifications for health policy research, version 2: hospital inpatient statistics. Healthcare Cost and Utilization Project (HCUP 3) Research Note 1. Rockville, MD: AHCPR Pub., 1996 [Google Scholar]
- 44.Matsushita Y, Kuroda Y, Niwa S, et al. Criteria revision and performance comparison of three methods of signal detection applied to the spontaneous reporting database of a pharmaceutical manufacturer. Drug Saf 2007;30:715–26 [DOI] [PubMed] [Google Scholar]
- 45.Agbabiaka TB, Savovic J, Ernst E. Methods for causality assessment of adverse drug reactions: a systematic review. Drug Saf 2008;31:21–37 [DOI] [PubMed] [Google Scholar]
- 46.Naranjo CA, Cappell H, Sellers EM. Pharmacological control of alcohol consumption: tactics for the identification and testing of new drugs. Addict Behav 1981;6:261–9 [DOI] [PubMed] [Google Scholar]
- 47.Wiholm BE. The Swedish drug-event assessment methods. Special workshop—regulatory. Drug Info J 1984;18:267–9 [DOI] [PubMed] [Google Scholar]