Abstract
Objective
To extract drug indications from structured drug labels and represent the information using codes from standard medical terminologies.
Materials and methods
We used MetaMap and other publicly available resources to extract information from the indications section of drug labels. Drugs and indications were encoded by RxNorm and UMLS identifiers respectively. A sample was manually reviewed. We also compared the results with two independent information sources: National Drug File-Reference Terminology and the Semantic Medline project.
Results
A total of 6797 drug labels were processed, resulting in 19 473 unique drug–indication pairs. Manual review of 298 most frequently prescribed drugs by seven physicians showed a recall of 0.95 and precision of 0.77. Inter-rater agreement (Fleiss κ) was 0.713. The precision of the subset of results corroborated by Semantic Medline extractions increased to 0.93.
Discussion
Correlation of a patient's medical problems and drugs in an electronic health record has been used to improve data quality and reduce medication errors. Authoritative drug indication information is available from drug labels, but not in a format readily usable by computer applications. Our study shows that it is feasible to use publicly available natural language processing resources to extract drug indications from drug labels. The same method can be applied to other sections of the drug label—for example, adverse effects, contraindications.
Conclusions
It is feasible to use publicly available natural language processing tools to extract indication information from freely available drug labels. Named entity recognition sources (eg, MetaMap) provide reasonable recall. Combination with other data sources provides higher precision.
Keywords: Drug labeling, Controlled vocabulary, Natural language processing, Knowledge bases, Drug therapy
Background and significance
The effective use of clinical decision support (CDS) in a computerized provider order entry (CPOE) system has been shown to improve the safety and quality of drug prescribing, resulting in significant reduction in medication errors.1–4 For this reason, the Centers for Medicare and Medicaid Services incentive program for the ‘meaningful use’ of electronic health records (EHRs) requires the inclusion of CPOE and CDS elements as the core set of objectives to be achieved by all participants.5 6 To provide timely advice during the drug prescribing process, the CDS function needs access to knowledge about drugs, such as therapeutic class, drug–drug interactions, adverse effects, indications, and contraindications. The availability of standardized drug knowledge bases has been identified as one of the critical elements that can help to realize the benefits of CDS.7
Drug knowledge bases are generally derived from three types of sources.8 Home-grown sources are laborious to create and maintain. Commercial sources can be expensive, and their use often implies ‘locking-in’ to a specific company or vendor. Both home-grown and commercial knowledge bases often use proprietary rather than national or international data standards, and this hinders their interoperability and potential for sharing across institutions or practice settings. Publicly available knowledge bases do not have some of the above drawbacks. However, the existing public resources are limited. The NDF-RT (National Drug File-Reference Terminology) developed by the Veterans Administration offers a classification of drugs by their chemical structure, mechanism of action, physiologic effect, and therapeutic intent.9–12 It also provides some links from drugs to disease entities which can be indications or contraindications. However, NDF-RT has been found to be deficient in some aspects, such as drug class information.13 14
One of the most comprehensive, current and authoritative sources of drug information is already available to the public in the form of drug package inserts (drug labels). The drug labels for most prescription drugs, and many over-the-counter drugs, can be found on the DailyMed website,15 collaboratively created and maintained by the FDA (Food and Drug Administration) and the National Library of Medicine. To date, DailyMed delivers drug label information for almost 40 000 drugs. These electronic drug labels follow the SPL (structured product labeling) standard that enhances their machine readability.8 16 17 However, SPL only provides a structure to separate the drug label into sections (eg, Clinical pharmacology, Indications and Usage, Contraindications, Precautions, and Adverse reactions), and the content of the individual sections is still in free narrative text. To unleash the power locked in these narrative texts, they need to be transformed into data encoded in standard terminologies, which can then be used by the inference engine in a CDS application. We have explored an automated way to do this by a well-established natural language processing (NLP) tool (MetaMap).18 19 Here we report our method and its evaluation.
For this study, we focused on the indications section of the drug label. A comprehensive reference table linking drugs to their indications can be used in various ways to enhance data quality in an EHR. It provides the basis on which a patient's medical conditions can be correlated with their medications for the purpose of mutual validation.20 Carpenter and Gorman studied the discrepancies between a patient's problem list and medications list in diabetic patients and suggested that this could be a good way to improve the accuracy and completeness of both lists.21 Burton et al22 derived an algorithmic way to link medications and diagnoses in an EHR with high overall sensitivity and specificity. Poissant et al23 reported that single-indication drugs could help to generate a more complete interinstitutional patient-specific health problem list. Galanter et al implemented an alert mechanism embedded in their CPOE system that prompted the physician to update the problem list when the indication for a prescribed drug was not present. They reported that 96% of the alerts were valid, and 95% of the problems added as a result of the alert were accurate.24 In a subsequent study, they showed that indication-based prescribing could help to reduce medication errors.25 In addition to evaluating our indication extraction method, we compared our results with drug indication information obtained from two independent data sources—namely, NDF-RT and the Semantic Medline project, which extracts semantic predications from the medical literature using NLP.26–28
Methods
Extraction of drug indications
The extraction of drug indication information from drug labels was done in three steps:
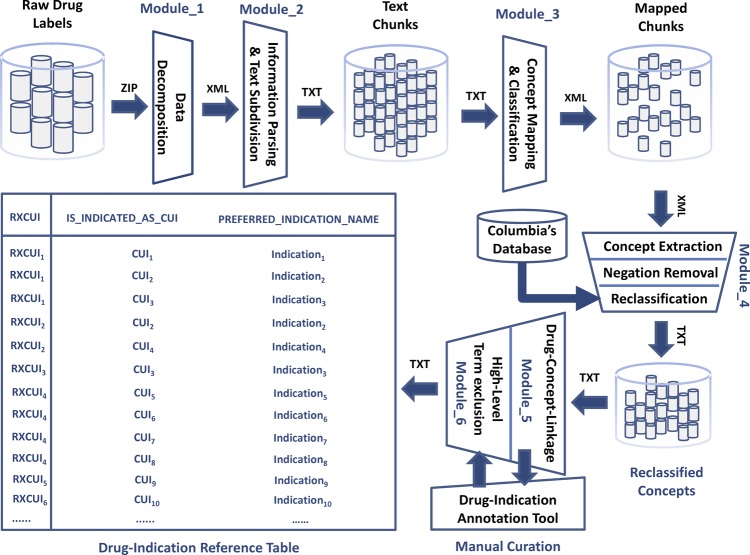
- Preprocessing of drug label information—all available drug labels (in XML format) were downloaded as a zip file from the DailyMed website. The individual files were automatically extracted and passed to a parser (written in Java) which:
- Identified and extracted the ‘Indications’ section labeled by the LOINC code ‘34067-9’ meaning ‘FDA package insert indications and usage section’
- Broke down the section into the smallest text segments identifiable by the XML tags (eg, <section>, <paragraph>, <item>). Each segment was stored as a separate file. The purpose of this step was to facilitate processing by MetaMap, which performed better with short chunks of texts than with long paragraphs.
Identifying medical concepts by MetaMap—making use of the Java application programming interface to the MetaMap's scheduler, the text segment files were sequentially submitted to MetaMap. Each MetaMap output was stored as a separate file for further processing.
- Postprocessing of MetaMap output—this included the following steps:
- All negated concepts were removed—based on its implementation of NegEx, MetaMap flagged negated Unified Medical Language System (UMLS) concepts. Only UMLS concepts that were not marked negated in the MetaMap output were retained.29
- Filtering of high-level concepts—Some high-level UMLS concepts (eg, disease, inflammation, infection) were not considered useful as indications. We identified 34 such concepts, which were removed from the results list.
The above tasks were performed by a suite of Java programs, which we called the Structured Product Labels eXtractor or SPL-X in short (figure 1).
Figure 1.
Structured Product Labels eXtractor system process diagram.
Linking the extracted indications to drug codes
We used RxNorm as our reference drug terminology and the RxNorm concept unique identifier (RxCUI) to encode the drugs.34 In RxNorm, a drug could be represented at different levels of abstraction, depending on whether additional aspects of the drug, such as dose form and strength, were specified (figure 2). For our study, we decided that the semantic clinical drug form (SCDF) was the appropriate level of abstraction for the following reasons. The ingredient level was potentially ambiguous as the indications for the same chemical compound could be different depending on the dose form (eg, steroids as ointments, inhalants or tablets would have different indications). On the other hand, difference in strength alone (distinguished at the SCD, or semantic clinical drug level) seldom affected the indications of a drug.
Figure 2.
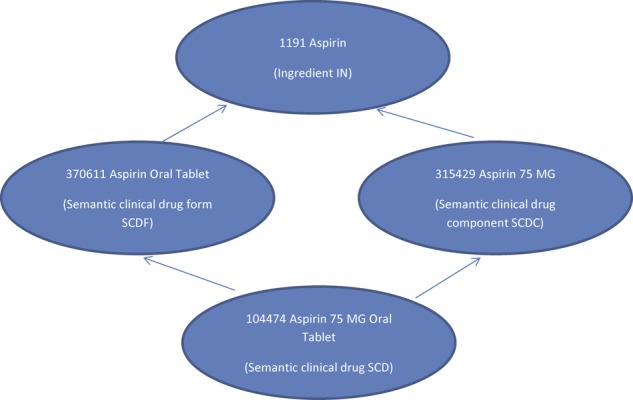
RxNorm drug entities at different levels of abstraction.
Each drug label was identified by a unique identifier called SPL_SET_ID, which was found in the RxNorm data files. By following the relationships between the RxNorm tables and drug entities, a drug label could be linked to a drug at the SCDF level. One label could be used in multiple drugs (eg, the same label could apply to multiple dose forms of the same drug, such as aspirin oral tablet and aspirin oral solution). Conversely, multiple drug labels could be linked to the same drug (eg, different manufacturers of the same generic drug would have different drug labels).
Evaluation of the results
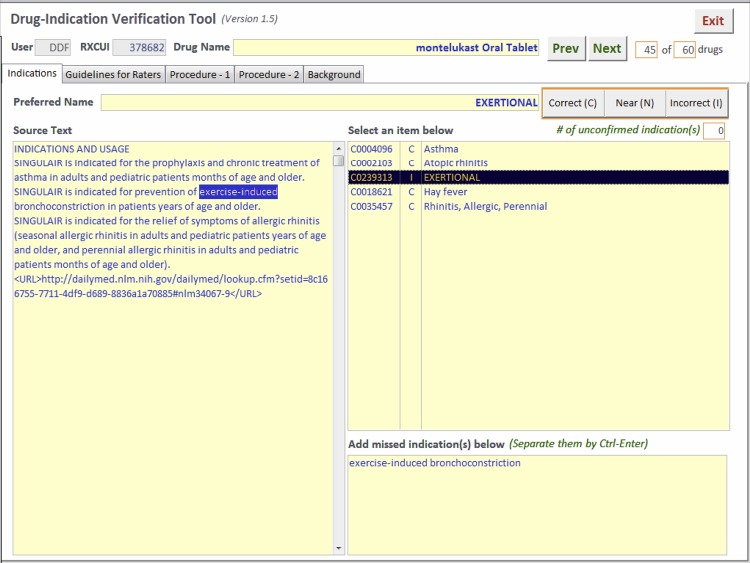
A sample of the drug indications extracted by SPL-X was reviewed manually by physicians. Based on pharmacy prescription filling data used in a separate project, we identified the drugs that were frequently prescribed. The indications for the most frequently prescribed drugs were reviewed, using an evaluation tool built with Microsoft Access (figure 3). The reviewer could see the drug name, all extracted indications, and the indications sections of the drug labels, with the specific words that resulted in the extraction of a particular indication highlighted. Each indication was rated as correct, near or incorrect. Owing to feedback from reviewers that it was often difficult to differentiate between near and correct, in the final analysis the near scores were converted into correct scores (see ‘Results’ below). The reviewers also recorded any indication that was mentioned in the drug label but missed by SPL-X (false negative). Indications that the reviewer knew about but were not mentioned in the drug label were not counted as false negatives. To measure the inter-rater variability among the reviewers, 20 drugs were reviewed by all reviewers, the rest were reviewed by a single reviewer. A failure analysis was done on the false positive and false negative indications for the 20 shared drugs.
Figure 3.
Drug-indication verification tool.
Comparison with other independent data sources
Drug-indication pairs were extracted from the NDF-RT through the UMLS by following the relationships ‘may_treat’ and ‘may_prevent’. The NDF-RT indications were mostly applied to the IN or SCD level, and were propagated to the SCDF level through the RxNorm relationships. Semantic associations between drugs and the diseases they might treat were extracted from Medline citations in the Semantic Medline project. These associations were mostly at the IN level and were similarly propagated to the SCDF level. These two datasets were compared with the output from SPL-X.
Statistical analysis
The performance of SPL-X was measured by the standard precision, recall and F score statistics. Inter-rater agreement was measured by pairwise κ statistics and the Fleiss κ statistic for multiple raters, calculated using the R package for statistical computing.35 36
Results
We downloaded 7063 drug labels from the DailyMed website in March 2010. Among them, 266 labels were non-human drugs and were excluded. SPL-X processed the 6797 labels and submitted the text chunk files to MetaMap. After applying the negation, semantic type, and high-level concept filters, we generated a final table with 19 473 unique drug–indication pairs. In the table, the drugs were identified by their RxNorm RxCUIs (at the SCDF level), and their indications were identified by UMLS CUIs. There were altogether 2104 unique RxCUIs and 2910 unique CUIs. The full table and results of the manual review can be downloaded as an appendix A to this article.
The 300 most frequently prescribed drugs were chosen for manual review by seven physicians. Each physician reviewed 60 drugs, of which 20 were common to all reviewers. Owing to a software glitch, two of the unshared drugs did not display correctly in the tool, so only 298 drugs were finally reviewed. The 20 shared drugs contained a total of 172 indications. Each indication was given a score of ‘C’ (correct), ‘N’ (near), or ‘I’ (incorrect) by each reviewer. After the review, some reviewers commented that it was often difficult to differentiate between ‘N’ and ‘C’; the ‘N’ category was used only sparingly. Among the 1204 scores for the 172 indications of the shared drugs, there were only 108 (9%) ‘N’ scores. For the 62 indications that contained at least one ‘N’, in the majority of cases (55 indications) there were more ‘C’ than ‘I’ for that particular indication among the reviewer scores. In 46 indications with at least one ‘N’, the rest of the scores were all ‘C’. For this reason, we decided to collapse our categories and convert all ‘N’ to ‘C’. We used the results from the 20 shared drugs to assess inter-rater agreement. Pairwise κ values based on the two-category scores ranged from 0.515 to 0.914 (table 1). The overall Fleiss κ for multiple raters was 0.713 for the two-category scores.
Table 1.
Pairwise κ values for the two-category scores (R1–R7, the seven reviewers)
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | |
|---|---|---|---|---|---|---|---|
| R1 | 1 | 0.914 | 0.800 | 0.783 | 0.661 | 0.787 | 0.561 |
| R2 | 1 | 0.772 | 0.752 | 0.629 | 0.787 | 0.527 | |
| R3 | 1 | 0.690 | 0.693 | 0.844 | 0.561 | ||
| R4 | 1 | 0.764 | 0.769 | 0.654 | |||
| R5 | 1 | 0.648 | 0.693 | ||||
| R6 | 1 | 0.515 | |||||
| R7 | 1 |
The results of the manual evaluation are summarized in table 2. The final score for the 20 shared drugs was calculated by simple majority vote for each indication from the seven reviewers. Using the consensus judgments for the 20 shared drugs and individual judgments for the 278 unshared drugs, we evaluated SPL-X extraction results. The overall recall, precision, and F score of SPL-X were 0.95, 0.77, and 0.85, respectively.
Table 2.
Results of manual review of the extracted indications
| Shared drugs (reviewed by all reviewers) | Unshared drugs (reviewed by one reviewer) | Combined | |
|---|---|---|---|
| Number of drugs | 20 | 278 | 298 |
| Number of indications | 172 | 3296 | 3468 |
| Average indications/drug (range) | 8.6 (5–16) | 11.9 (1–130) | 11.6 (1–130) |
| Correct indications | 128 | 2546 | 2674 |
| Incorrect indications | 44 | 750 | 794 |
| Missing indications | 15 | 127 | 142 |
| Recall | 0.90 | 0.95 | 0.95 |
| Precision | 0.74 | 0.77 | 0.77 |
| F score | 0.81 | 0.85 | 0.85 |
Failure analysis of the false-positive cases (incorrect indications) among the shared drugs disclosed four types of errors (table 3). In 25 cases (57%), MetaMap identified the wrong concept. Word sense disambiguation remained a major weakness of MetaMap in the presence of ambiguous terms, such as the example shown in which ‘(bacterial) strain’ was confused with ‘(muscle) strain’ (note that to achieve higher recall we allowed over-matching of the terms and we did not use the MetaMap word-sense disambiguation option).18 NegEx failed to pick up some negated concepts, which could be related to insufficiency of the word-length window.
Table 3.
Failure analysis for false-positive indications
| Type of error | Example | Number of cases (%) | ||
|---|---|---|---|---|
| Drug | Original text (matching string italicized) | Concept identified | ||
| 1. Wrong concept identified | Dexamethasone/tobramycin ophthalmic suspension | ‘Staphylococci, … penicillin-resistant strains’ | Muscle strain | 25 (57) |
| 2. Negated concepts | Dexmethylphenidate extended release capsule | ‘Stimulants are not intended for use in …primary psychiatric disorders, including psychosis’ | Psychotic disorders | 2 (5) |
| 3. Not an indication | Potassium chloride extended release tablet | ‘The use of potassium salts in patients receiving diuretics for uncomplicated essential hypertension is often unnecessary…’ | Uncomplicated hypertension | 14 (32) |
| 4. High level concepts | Cimetidine oral tablet | ‘Treatment is indicated for 12 weeks for healing of lesions…’ | Lesion | 3 (7) |
The remaining two types of false-positive results were not related to NLP itself. The concepts identified by MetaMap correctly represented the meaning in the drug label, but they were not considered correct indications for our use case. Some medical conditions mentioned in the indications section were not indications. For example, hypertension was mentioned in the indication section of the drug label for potassium chloride. This was because some patients with hypertension could be taking diuretic agents, which could lead to hypokalemia, which might then require prescription of potassium chloride. However, hypertension itself was not an indication for this drug. For our study, we compiled a list of very general (or high-level) concepts (such as disease, infection) that were not useful as indications, and excluded them from our results. However, the list was incomplete and some high-level concepts (eg, lesion) were missed, contributing to the incorrect indications.
Among the 15 false-negative results (missing indications) in the shared drugs, nine indications were present in the UMLS (eg, anemia of chronic renal failure patients) but not found by MetaMap. The other six indications were not present in the UMLS (eg, recurrent calcium oxalate calculi).
The NDF-RT data provided indications for a total of 6554 drugs (at the SCDF level), of which 1755 drugs were present in our drug–indication table (83% of the 2104 drugs in our table). For these 1755 drugs, there were 9935 unique drug–indication pairs, of which 1966 pairs were exactly the same as our results (ie, the same drug paired with the same indication). Among the 1966 drug–indication pairs of this concordant subset, 421 had been manually reviewed and 409 were found to be correct, representing a precision of 97% for this subset. Semantic Medline data yielded indications for a total of 6074 drugs (at the SCDF level), among which 1608 drugs were also in our drug–indication table. Among these 1608 drugs, there were 148 116 unique drug–indication pairs, of which 4040 were exact matches of our results. Among the 4040 drug–indication pairs in the concordant subset, 977 were manually reviewed and 905 were found to be correct, representing a precision of 93% (table 4).
Table 4.
Comparison of SPL-X output with drug–indication pairs extracted from NDF-RT and Semantic Medline
| NDF-RT data | Semantic Medline data | |||
|---|---|---|---|---|
| Drugs | Drug–indication pairs | Drugs | Drug–indication pairs | |
| Indications extracted | 6554 | 42507 | 6074 | 696297 |
| Drug also in SPL-X output | 1755 | 9935 | 1608 | 148116 |
| Drug–indication pair same as SPL-X output (concordant subset) | 1108 | 1966 | 1264 | 4040 |
| Manually reviewed | 209 | 421 | 234 | 977 |
| Correct indications | 208 | 409 | 231 | 905 |
| Precision (concordant subset) | 0.97 | 0.93 | ||
NDF-RT, National Drug File Reference Terminology; SPL-X, Structured Product Labels eXtractor.
Discussion
Many studies that linked medical problems to medications used special home-grown drug–indication reference tables. These tables were often hand-crafted by physicians or pharmacists, and the process was labor-intensive and time-consuming. The scalability of the manual approach to thousands of drugs is questionable. NDF-RT fills this gap to some extent by providing semantic relationships between drugs and indications which are encoded in standard terminologies. However, NDF-RT has been found to be incomplete in some areas. Information extracted from the drug labels can be a useful alternative source of drug information. Duke and Friedlin used NLP to extract adverse reactions from drug labels and to map them to MedDRA terms, and they achieved very good results.37 38 But their NLP program was not available to others, nor did they describe their algorithm in sufficient detail to allow it to be replicated elsewhere. In this study, we report our method based on publicly available resources.
One potential use of SPL-X is to supplement or augment NDF-RT. For drugs that are not linked to indications in NDF-RT, SPL-X can provide some useful information. For drugs that are covered by NDF-RT, SPL-X can suggest additional indications that have been missed. When our results were corroborated by Semantic Medline predications, which were also computationally generated (no human review involved), the precision increased from 0.77 to 0.93. By reviewing a small sample of indications discovered by SPL-X, corroborated by Semantic Medline predications but not found in NDF-RT, we found that a lot of them could be useful additions to NDF-RT. Some examples are ‘coronary heart disease’ for nitroglycerin patch, ‘bronchial spasm’ for albuterol inhaler, and ‘atrophy of vagina’ for estradiol patch. Our method is not limited to drug indications and can be applied, with minor modifications, to other sections of the drug label whose primary content is within the realm of findings, symptoms, and diseases, such as contraindications and adverse effects.
In this study, we achieved an overall recall of 0.95 and precision of 0.77. One possible way to improve the performance of our method might be to employ machine learning techniques. For example, when we parsed the labels, we gathered information about where a text chunk came from (eg, a table, list or paragraph) based on the XML tags. It is possible that the text chunks coming from lists or tables, which are normally shorter and more concise, will contain less extraneous information which caused some false-positive results in our study. Theoretically, we could have given more weight to the results derived from lists or tables to improve precision.
Another possible way to tweak weighting is by the MetaMap score, which indicates the level of match between the input text and the target UMLS concept. The fact that a drug can have more than one drug label can also be exploited. Although drug labels provided by different drug companies for the same drug are often verbatim copies, when substantively different labels for the same drug are available they could potentially be used to validate each other. False-positive results due to undiscovered high-level concepts can be avoided by updating our exclusion list. Word-sense disambiguation and negation detection are the main causes of NLP-related errors in our study. They are not specific to our application, but are common, known challenges in NLP.
Further discussion of these problems is beyond the scope of this paper. Improving the recall will be more difficult since the baseline is already quite high. A considerable portion of the missed indications (six out of 15) is not present in the UMLS and will not be recognizable by our method. Some of the corpus-based methods that infer semantic types of the out-of-vocabulary terms could be used to deal with the coverage problem in the future.39
We have also carried out a systematic appraisal of the manual review process, which was not reported by many authors. In order to measure the inter-rater agreement, our evaluation process had a 33% overlap of the reviewed drugs between reviewers. Initially, we thought that a third category of ‘near match’ (in addition to ‘correct’ and ‘incorrect’) would be necessary to capture the marginal cases. It turned out that the extra category was not necessary. In many cases, a ‘near’ score was given because additional information in the drug label was not covered by the UMLS concept. For example, MetaMap identified the concept ‘gastroesophageal reflux disease’ where the drug label said ‘erosive gastroesophageal reflux disease’. For most intents and purposes, the identified concept would be good enough to be the indication. In this case, all reviewers rated ‘correct’ except for one who rated ‘near’. Since the ‘near’ score was used infrequently, and most of them were associated with more ‘correct’ than ‘incorrect’ scores, we removed the extra category and converted all ‘near’ to ‘correct’ scores. The two-category Fleiss κ of 0.713 shows that there is good agreement between the reviewers and supports the reliability of the review process.
Conclusion
In order to make use of the rich information in drug labels for CDS or other purposes, it is necessary to extract and encode the information in standard terminologies. Using a publicly available NLP resource (MetaMap), we built a drug label information extractor, SPL-X. We processed the ‘Indications’ section of over 6000 drug labels and developed a reference table linking drugs to their indications, encoded in RxNorm codes (RxCUI) and UMLS codes (CUI), respectively. Manual review of the results showed that SPL-X has a recall of 0.95 and precision of 0.77. Combination with other independent data sources improves the precision. The same method can be used to extract information from other sections of the drug label, such as contraindications and adverse effects.
Acknowledgments
The authors thank Swapna Abhyankar, Alan Aronson, Olivier Bodenreider, Francois Lang, Dwayne McCully, Christopher Miller, Jim Mork, Tom Rindflesch, Willie Rogers, Laritza Taft, Julia Xu, and Vivienne Zhu for their assistance in this study.
Footnotes
Contributors: KWF and DD-F conceived and designed the study; supervised the conduct of the study and data processing; performed the data analysis. CSJ designed the software for collection, cleansing, and preparation of the data for analysis. KWF drafted the manuscript. All authors contributed substantially to revision of the manuscript.
Funding: This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Kaushal R, Barker KN, Bates DW. How can information technology improve patient safety and reduce medication errors in children's health care? Arch Pediatr Adolesc Med 2001;155:1002–7 [DOI] [PubMed] [Google Scholar]
- 2.Bates DW, Teich JM, Lee J, et al. The impact of computerized physician order entry on medication error prevention. J Am Med Inform Assoc 1999;6:313–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates DW, Pappius E, Kuperman GJ, et al. Using information systems to measure and improve quality. Int J Med Inform 1999;53:115–24 [DOI] [PubMed] [Google Scholar]
- 4.Bates DW, Leape LL, Cullen DJ, et al. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. JAMA 1998;280:1311–16 [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal D, Tavenner M. The "meaningful use" regulation for electronic health records. N Engl J Med 2010;363:501–4 [DOI] [PubMed] [Google Scholar]
- 6.Department of Health and Human Services Health Information Technology: Initial Set of Standards, Implementation Specifications, and Certification Criteria for Electronic Health Record Technology; Final Rule, July, 2010 [DOI] [PubMed]
- 7.Teich JM, Osheroff JA, Pifer EA, et al. Clinical decision support in electronic prescribing: recommendations and an action plan: report of the joint clinical decision support workgroup. J Am Med Inform Assoc 2005;12:365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadow G. Structured product labeling improves detection of drug-intolerance issues. J Am Med Inform Assoc 2009;16:211–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences. Proc AMIA Symp 2002:116–20 [PMC free article] [PubMed] [Google Scholar]
- 10.Brown SH, Elkin PL, Rosenbloom ST, et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Stud Health Technol Inform 2004;107:477–81 [PubMed] [Google Scholar]
- 11.Rosenbloom ST, Awad J, Speroff T, et al. Adequacy of representation of the National Drug File Reference Terminology Physiologic Effects reference hierarchy for commonly prescribed medications. AMIA Annu Symp Proc 2003:569–78 [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak J, Chute CG. Analyzing categorical information in two publicly available drug terminologies: RxNorm and NDF-RT. J Am Med Inform Assoc 2010;17:432–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak J, Chute CG. Further revamping VA's NDF-RT drug terminology for clinical research. J Am Med Inform Assoc 2011;18:347–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phansalkar S, Desai AA, Bell D, et al. High-priority drug-drug interactions for use in electronic health records. J Am Med Inform Assoc 2012;19:735–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DailyMed Website, National Library of Medicine. http://dailymed.nlm.nih.gov/dailymed/about.cfm (accessed 4 Mar 2013).
- 16.Schadow G. Assessing the impact of HL7/FDA Structured Product Label (SPL) content for medication knowledge management. AMIA Annu Symp Proc 2007:646–50 [PMC free article] [PubMed] [Google Scholar]
- 17.Schadow G. HL7 Structured Product Labeling—electronic prescribing information for provider order entry decision support. AMIA Annu Symp Proc 2005:1108. [PMC free article] [PubMed] [Google Scholar]
- 18.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 20.Jao C, Hier D, Galanter W. Using clinical decision support to maintain medication and problem lists: A pilot study to yield higher patient safety. IEEE International Conference on Systems, Man and Cybernetics, 2008:739–43 [Google Scholar]
- 21.Carpenter JD, Gorman PN. Using medication list—problem list mismatches as markers of potential error. Proc AMIA Symp 2002:106–10 [PMC free article] [PubMed] [Google Scholar]
- 22.Burton MM, Simonaitis L, Schadow G. Medication and indication linkage: a practical therapy for the problem list? AMIA Annu Symp Proc 2008:86–90 [PMC free article] [PubMed] [Google Scholar]
- 23.Poissant L, Taylor L, Huang A, et al. Assessing the accuracy of an inter-institutional automated patient-specific health problem list. BMC Med Inform Decis Mak 2010;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galanter WL, Hier DB, Jao C, et al. Computerized physician order entry of medications and clinical decision support can improve problem list documentation compliance. Int J Med Inform 2010;79:332–8 [DOI] [PubMed] [Google Scholar]
- 25.Galanter W, Falck S, Burns M, et al. Indication-based prescribing improves problem list content and medication safety. AMIA Annu Symp Proc 2012:1610 [Google Scholar]
- 26.Kilicoglu H, Rosemblat G, Fiszman M, et al. Constructing a semantic predication gold standard from the biomedical literature. BMC Bioinformatics 2011;12:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rindflesch TC, Fiszman M, Libbus B. Semantic interpretation for the biomedical literature. In: Chen H, Fuller S, Hersh WR, Friedman C, eds. Medical informatics: advances in knowledge management and data mining in biomedicine. New York: Springer-Verlag, 2005:399–422 [Google Scholar]
- 28.Rindflesch TC, Pakhomov SV, Fiszman M, et al. Medical facts to support inferencing in natural language processing. AMIA Annu Symp Proc 2005:634–8 [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 30.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform 2001;84:216–20 [PMC free article] [PubMed] [Google Scholar]
- 31.The UMLS Semantic Groups. http://semanticnetwork.nlm.nih.gov/SemGroups/ (accessed 4 Mar 2013).
- 32.Fan JW, Friedman C. Semantic reclassification of the UMLS concepts. Bioinformatics 2008;24:1971–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan JW, Friedman C. Semantic classification of biomedical concepts using distributional similarity. J Am Med Inform Assoc 2007;14:467–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleiss JL, Levin B, Paik MC. Statistical methods for rates and proportions. New York: John Wiley & Sons, 2003 [Google Scholar]
- 36.The R Project for Statistical Computing. http://www.r-project.org/ (accessed 4 Mar 2013).
- 37.Duke J, Friedlin J, Ryan P. A quantitative analysis of adverse events and "overwarning" in drug labeling. Arch Intern Med 2011;171:944–6 [DOI] [PubMed] [Google Scholar]
- 38.Duke JD, Friedlin J. ADESSA: a real-time decision support service for delivery of semantically coded adverse drug event data. AMIA Annu Symp Proc 2010;2010:177–81 [PMC free article] [PubMed] [Google Scholar]
- 39.Xu R, Supekar K, Morgan A, et al. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. AMIA Annu Symp Proc 2008:820–4 [PMC free article] [PubMed] [Google Scholar]