Abstract
The clinical element model (CEM) is an information model designed for representing clinical information in electronic health records (EHR) systems across organizations. The current representation of CEMs does not support formal semantic definitions and therefore it is not possible to perform reasoning and consistency checking on derived models. This paper introduces our efforts to represent the CEM specification using the Web Ontology Language (OWL). The CEM-OWL representation connects the CEM content with the Semantic Web environment, which provides authoring, reasoning, and querying tools. This work may also facilitate the harmonization of the CEMs with domain knowledge represented in terminology models as well as other clinical information models such as the openEHR archetype model. We have created the CEM-OWL meta ontology based on the CEM specification. A convertor has been implemented in Java to automatically translate detailed CEMs from XML to OWL. A panel evaluation has been conducted, and the results show that the OWL modeling can faithfully represent the CEM specification and represent patient data.
Keywords: Ontologies, Semantic Web, OWL, Clinical Element Model, Secondary Use of EHR
Introduction
Background
Healthcare system interoperability is one of the most important goals for meaningful use of the electronic health records (EHR). It is essential to facilitate IT support for workflow management, decision support systems, and evidence-based healthcare, as well as secondary use of EHR across healthcare organizations. The clinical element model (CEM)1 2 was designed to provide a consistent architecture for representing clinical information in EHR systems. The CEM has been adopted in the Strategic Health IT Advanced Research Project, secondary use of EHR (SHARPn)3 as the common unified information model for unambiguous data representation, interpretation, and exchange within and across heterogeneous sources and applications.
The current CEM was represented either in the Clinical Element Modeling Language (CEML), an XML-based language for expressing the structure and constraints of CEM models;4 or in the Constraint Definition Language (CDL),1 which defines a superset of CEML functionality, with extensions including additions of new constraints to the modeling language, formalization of the model specialization capabilities, and a concrete reference object model. Although CDL brings more formalization to the CEM definition, neither CDL nor CEML currently supports inferences or rules, which are critical to add computerized support and reasoning to the CEM models and populated patient data. In addition, there is no open-source tooling available yet to support CDL authoring or querying, and there is a lack of tooling for automatic consistency checking or reasoning for the CEM.
Proposed solution and impact
One way to integrate CEMs with the above functionalities is to translate CEM definitions to an ontology language. In this project, we chose to use the Web Ontology Language (OWL),5 which we believe serves best for our purpose for the following reasons. OWL is built on formalisms that use description logic (DL)6 to allow reasoning and inference. The rule interchange format (RIF)7 can be used to add rules to OWL and can be used to infer new knowledge from an OWL based ontology and reason about OWL individuals. Moreover, the Semantic Web community has developed open source tools for editing, storing, and reasoning over information represented in OWL. The vast ongoing supports and investments on OWL and its related tools make it very valuable to align clinical information model languages such as CEML, CDL, and Archetype Definition Language (ADL)8 to OWL, so that the OWL-related tools can be directly applied to these models.
The OWL representation also provides an environment to seamlessly integrate information models and terminology models used in the clinical context. The CEM is an information model that represents the logical structure of data within an EHR. Information models usually connect to terminologies (eg, SNOMED CT9 for clinical terms, RxNorm10 for medications), which define the domain knowledge of concepts used in the information model. Unfortunately, information models and terminology models are usually developed by different groups and are expressed in very different syntaxes, which complicates the efforts to interface the information expressed in the models.11 OWL and Semantic Web technologies have already been applied in representing and validating many terminology models.12–16 If the CEM can be represented in OWL, the information model (CEM) and terminology models can be represented in the same language and representation environment, thereby facilitating the integration of information and terminology models.
In addition, an OWL representation of the CEM can potentially facilitate the harmonization and transformation between the CEM specification and other clinical data modeling languages. Previous efforts focused on representing general clinical models or EHR standards such as HL7 v3, openEHR, or ISO 13 606 in the Semantic Web environment.12–15 OWL and the Semantic Web can provide an ideal platform for transforming these models that were originally represented in different syntaxes.
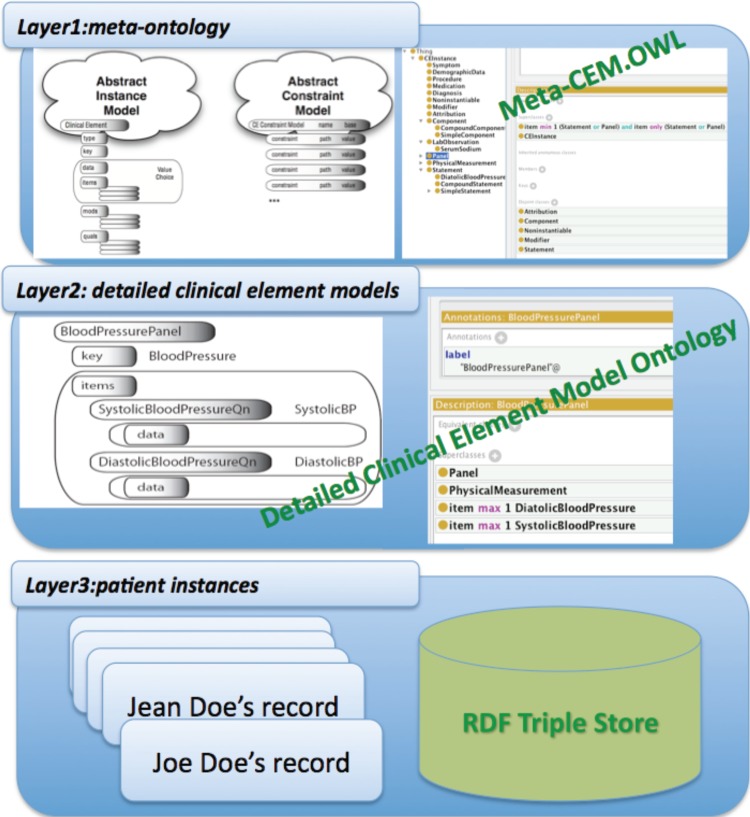
As illustrated in figure 1, we envision a three-layer clinical data representation model in OWL: (1) a meta-level ontology that defines the abstract meta-representation of the CEM, where the basic structures, the properties and their relationships, and the constraints are defined; (2) OWL ontologies for representing each individual detailed CEM; and (3) patient data represented in the Resource Description Framework (RDF)17 with respect to the ontologies on layer 2, which provides the basis of interoperability between applications that exchange individual patient data in a form that machines can understand.18 After representing the CEM as well as patient data in OWL/RDF, we can utilize Semantic Web tools to check for semantic consistency on both the ontology and the instance levels, and to reason over these data for extracting new knowledge.
Figure 1.
System overview.
Our previous work has focused on representing basic CEM components and their relationships in the meta-level ontology.18 The research introduced by the current paper extends this previous work and completes the Semantic Web representation of the whole three-layer model in figure 1. We have implemented a convertor that can automatically convert detailed CEM models to the OWL format. The convertor has been evaluated using the CEM SHARPn release, which is designed specially for the EHR secondary use purpose. The meta-ontology as well as converted detailed CEM ontologies are available on the SHARPn website (http://informatics.mayo.edu/sharp/index.php/CEM_OWL_Project).
Meta-CEM ontology description
CEM basic category classes
Based on the CEM specification, each clinical element may be classified into a basic structural category. These basic categories can be considered as the building blocks for constructing detailed CEMs. Our preliminary work has focused on OWL representation of these CEM basic categories.18 An OWL class has been defined for each category. We also used OWL DL axioms to formally define the semantics, constraints, and relations for these categories. Table 1 briefly introduces these basic categories.
Table 1.
Basic clinical element model (CEM) categories
| Category | Definition | Example |
|---|---|---|
| Statement | A complete assertion about a particular aspect, a characteristic or condition of a patient | DiastolicBloodPressureMeas: a single statement model, where it captures the value of the DBP measurement |
| Panel | A common grouping of clinical observations. It is a collection of other statements or panels | BloodPressurePanel can contain items such as DiastolicBloodPressure and SystolicBloodPressure |
| Component | Is similar to a statement in CEM, except that it cannot be persisted alone in the EHR, but must be persisted as an internal part of another model with type such as Statement or Panel | |
| Attribution | Declares the who, where, why, and when information regarding an action | The model BloodPressurePanel has an attribution called ‘Observed’, which describes when, who, where, and why the blood pressure measurement was obtained |
| Qualifiers | Is used to add useful information to an instance but does not change the meaning of the data or items of the instance | BodyPosition of HeartRateMeas is a qualifier. In this case, whether the patient was sitting or standing during the measurement does not affect the meaning of the measurement itself |
| Modifier | Alters the meaning of the instance | Subject of model HeartRateMeas is a modifier. In this model, the subject changes the semantic interpretation of the measurement |
DBP, diastolic blood pressure; EHR, electronic health records.
In addition to the basic category classes, we also declared a set of properties for defining relationships in CEMs. OWL object properties item, qual, mod, and att are used to represent the relationships between a CEM element and its associated statements/panels, components, qualifiers, modifiers, or attributions respectively. For example, the OWL class BloodPressurePanel has two sub-statements associated with the item property: SystolicBloodPressureMeas and DiastolicBloodPressureMeas. In addition, the OWL object property data is used to define any values for a CEM statement or component. For example, we can declare the actual value of a patient's systolic blood pressure measurement using the data property. The detailed description of how we defined these basic categories and relations has been discussed previously.18
Category classes associations
The basic category classes can be used to represent individual clinical elements. Sometimes there is also a need to represent associations among the defined clinical elements. For example, the information of a particular patient could be stored in one statement instance while the information of the patient's family members could be stored in other statement instances. In this case, we must define the relation between two patient statement instances and annotate the relation as Parent–Child. In this section, we discuss how to define different types of associations among categories using the Association specification in CEM.
An association is used to represent a collection of related statements, components, qualifiers, modifiers, and/or attributions. An association is bound with a vocabulary concept that defines its semantic meaning. Associations may be specialized as panels, collections, sequences, lists, semantic links, and annotations.
A collection or a panel is an association that references a set of associations and/or statements. The primary difference between the two is that a panel represents a very strong relationship between its embedded elements, whereas a collection represents a weak relationship. For example, we could use a blood pressure panel association to represent the systolic and diastolic blood pressure (DBP) observations taken together. In contrast, a collection may be arbitrarily defined and reference associations and/or statements that share limited context. In the OWL representation, we created new OWL classes for collection and panel, where the semantic definition for both classes is that they can only associate either associations or statements through the item property.
We can also define a sequence of a set of clinical elements. We adopted the OWL sequence extension19 to express a sequence in OWL vocabulary, which is supported by DL reasoners.i The OWL sequence extension introduces the OWLList class to define a member in a sequence. The hasNext property points to the next member in the sequence and the content of the member is associated using the hasContents property. Figure 2 shows an example of how to use the OWL sequence extension to represent a CEM sequence about the use of a catheter in a patient. In that example, a CEM statement describes the insertion of the catheter, statements describe site maintenance, and a statement describes the removal of the catheter.
Figure 2.
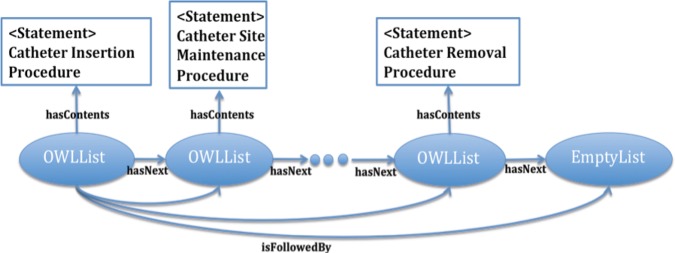
Example of representing sequence using Web Ontology Language list.
In CEM, a list is an association that contains a set of references of some other independent statements or associations. We define a new class called List in CEM-OWL. Since the order of referenced elements in the list is not significantly important, we propose the use of owl:unionOf to represent the association of a list. The List class itself is the union of the referenced statements and/or associations.
A semantic link is an association that represents a strong semantic relationship between two or more statements/associations. Statements and/or associations referenced in a semantic link are assumed to serve some role in the semantic link. For example, a semantic link can be used to represent the relationship between a diagnosis statement and a medication administration statement indicating that the medication was administrated to treat the diagnosis condition, or the relationship between the subjects of two patient statements is Parent–Child. Figure 3A shows an example of how a semantic link could be represented in OWL. In the example, the class Treatment is a semantic link that references two statements Diagnosis and Med_admin and the definition includes ‘a treatment must treat some diagnosis with some medication’.
Figure 3.
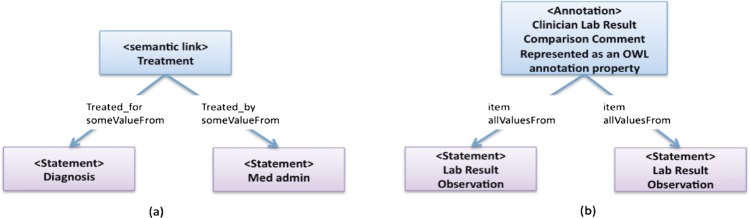
Examples of Semantic Link (A) and Annotation (B).
In CEM, an annotation is an association that adds independent textual annotation information to the referenced statements and/or associations. For example, a clinician can comment on observation such as laboratory results. Figure 3B shows an example of how we represent a CEM annotation in OWL. The class Clinician_Lab_Result_Comparison_Comment is a subclass of Annotation that references two statement classes, which must be lab result observations. A textual annotation value can be appended as an OWL Annotation Property to the Annotation class to store any comments to this annotation.
Detailed model representation
The CEM-OWL meta-ontology described above can be used to represent each detailed CEM in OWL. The representation of a detailed CEM has constraints on the values that can be legally stored in a particular element. A constraint can (1) limit an element to contain instances of a specific reference class (eg, qualifier, modifier, attribution), (2) limit the data contained in the element to that described by a specific model (eg, a body position model), (3) define the cardinalities, and (4) restrict the domain from which data values in the element are derived (eg, a value set).
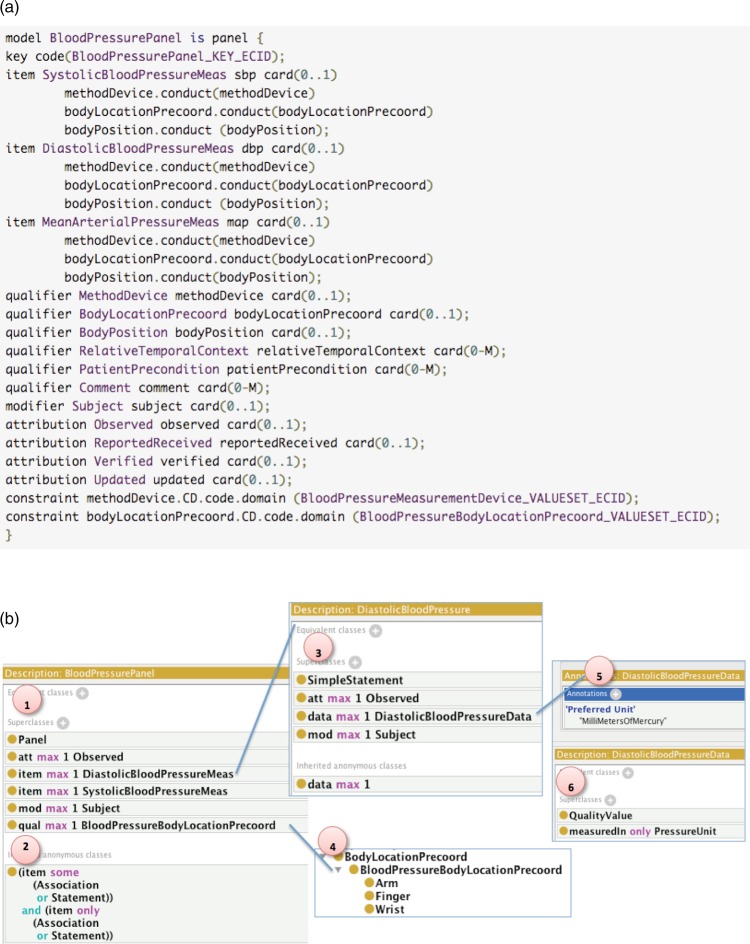
To limit an element to contain instances of a specific reference class, any detailed CEM that belongs to a category should be declared as a subclass of the corresponding category class (figure 4, reference point 1). It therefore inherits all the semantic definitions of its parent classes. Figure 4 shows an example of how we represent the BloodPressurePanel model. Figure 4A shows the BloodPressurePanel model in the CDL format and figure 4B shows its OWL representation (partial).ii Since CDL defines that BloodPressurePanel is a Panel, we define the class BloodPressurePanel to be a subclass of the Panel class so that it inherits the semantic definition of the Panel class (see figure 4, reference point 2).
Figure 4.
Example representation of a detailed clinical element model in Constraint Definition Language (CDL) and Web Ontology Language: (A) the CDL representation of the blood pressure panel model; (B) the Web Ontology Language representation of the blood pressure panel model (partial).
To limit the data contained in the element to that described by a specific model, we can link the referenced elements in each detailed CEM using the properties item, qual, mod, and att. Figure 4A lists a set of items, qualifiers, attributions, and one modifier. As figure 4B, reference point 1 shows, we can define that the BloodPressurePanel class has items DiastolicBloodPressureMeas and SystolicBloodPressureMeas, attribution (via att) Observed, modifier (via mod) Subject, qualifier (via qual) BloodPressureBodyLocationPrecoord, etc.
CDL defines cardinality constraints of each embedded CEM. OWL provides three constructs for cardinality constraints: owl:cardinality, owl:minCardinality, and owl:maxCardinality. These constraints can be used to faithfully represent the cardinality constraints defined by CDL. As figure 4B reference point 1 shows, we can define that each blood pressure panel can only have maximally one item referencing DiastolicBloodPressure using owl:maxCardinality.
In CEM, a value set is a category of like concepts. A value set constraints the permissible values of a particular value domain. For example, figure 4A shows two such constraints for Blood Pressure Measurement Device and Blood Pressure Body Location. In CEM-OWL, we define a value set following the guideline proposed by the W3C.20 The W3C guideline proposes two options to represent a value set: representing values in a value set as individuals or as subclasses. Here we follow the second approach,iii where each value set is presented as an OWL class and the features of the class represent a space that is partitioned by the values (ie, permissible values) in the value set. Each permissible value in the value set is represented as individual OWL classes, and defined as subclasses of the OWL class for the value set itself. These individual classes are defined as disjoint to each other, and the value set is defined as an exhaustive union of its permissible value classes. In figure 4B, reference point 4, for example, the class BloodPressureBodyLocationPrecoord represents a value set with three permissible values: Arm, Finger, and Wrist, each of which is a subclasses of the BloodPressureBodyLocationPrecoord class and is disjoint from the other two. In addition, the class BloodPressureBodyLocationPrecoord is defined as an exhaustive union of the three subclasses. This way we can restrict the values for blood pressure body location to only these three choices.
We also propose how to represent units of measure defined in CEMs. In order to ensure semantic interoperability, we adopted the Measurement Unit Ontology (MUO)21 and the Unified Code for Units of Measure (UCUM).22 For each physical quality data value we define a new class that is a subclass of the QualityValueiv class in MUO. We can then use the MUO property measuredIn to define the allowed units of this quantity value. In the example in figure 4B, reference point 3, we defined that each DiastolicBloodPressure can have at most one DiastolicBloodPressureData, which is defined as a subclass of MUO:QualityValue (figure 4B, reference point 6). A DiastolicBloodPressureData can only be measured in terms of a PressureUnit (figure 4B, reference point 6), which is a value set that contains all of the pressure units that are defined in UCUM. We can also use the MUO property Preferred Unit to define the preferred unit (MilliMetersOfMercury) for this quantity value (figure 4B, reference point 5).
Use case illustration
After the semantic representation of the CEMs as well as the populated CEM patient data, Semantic Web technologies can be used to perform consistency checking, automatic classification, and automatic inference by linking to existing domain ontologies. In this section, we use a few use cases to illustrate the above semantic functionalities.
Consistency checking
We can perform consistency checking on both the model level and the instance level. On the model level, since the meta-ontology semantically defines the basic category class, any detailed CEM must follow the restriction defined. Figure 5A shows an example. As the bottom part of figure 5A shows, a panel is defined to associate with either associations or statements as its item. If a CEM modeler decided to create a new Panel that has at least one component as its item (rather than an Association or a Statement), a semantic reasoner will return an error due to this inconsistency. This illustrates how the OWL representation of the CEM meta-model can be used to ensure the correct implementation of detailed CEMs.
Figure 5.

Examples with inconsistencies: (A) model inconsistency; (B) value set inconsistency.
On the instance level, we can check if a particular instance has the correct number of linked components as defined by the cardinality constraints, or has a value within the correct data ranges and with appropriate units of measurement, or has appropriate values as defined by the specific value sets. Figure 5B shows an example of value inconsistency. The blood pressure body location has been defined as arm, finger, or wrist, as the upper part of figure 5B shows. The lower part of figure 5B shows sample data with the blood pressure body location as leg, which is not an instance of arm, finger, or wrist. In this case, a reasoner will return an error due to this inconsistency.
Automatic classification
We can semantically define a class using OWL equivalent classes and axioms. Then a semantic reasoner can automatically determine whether an individual belongs to the class. Figure 6 shows an example. We can define that normal DBP data must be within a particular range as the upper part of figure 6 shows. The lower part of figure 6 shows two sample DBP data values: one is 120 and the other one is 65. As we can see the system automatically classified the second one as a NormalDBPData, but not the first one. Similarly, we can define other classes using OWL DL.
Figure 6.
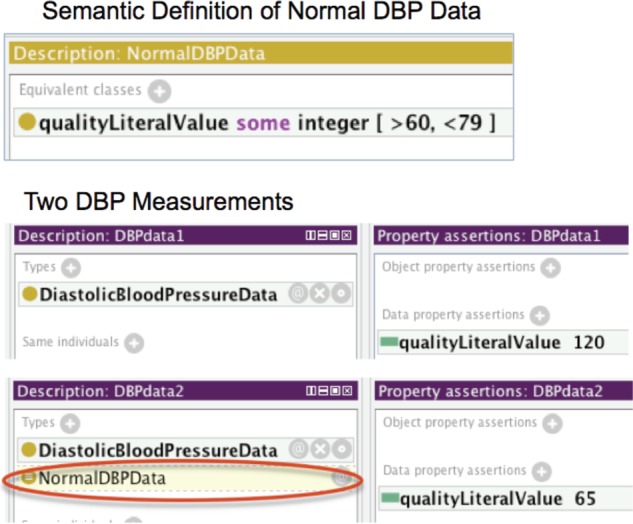
An example of automatic classification.
Semantic reasoning
The CEM-OWL representation can also provide a more natural connection to existing ontologies and semantic technologies. EHR data representations, such as CEMs, need to reference domain ontologies such as SNOMED CT, RxNorm, and LOINC, for the semantic meaning of each element. OWL can serve as a common platform to link information models such as CEM to the domain ontologies. In addition, the populated patient data with respect to the CEM-OWL makes it easier for us to leverage existing Semantic Web based tools to further semantic reasoning.
Here we use one example in figure 7 to illustrate how CEM-OWL, domain ontologies, and semantic reasoners need to be used together to infer information for a phenotyping algorithm. The upper part of figure 7 shows one of the conditions a patient must qualify to be classified as a resistant hypertension patient. The lower part of figure 7 shows a few sample sentences from a patient's clinical notes. We want to semantically represent the patient's medication history and blood pressure measurement history in a structured way using CEM. More specifically we can use the SecondaryUseNotedDrug class to represent medication information and the BloodPressurePanel class to represent the values of systolic blood pressure measurements and DBP measurements, as well as the patient location (inpatient or outpatient). As we discussed above, each detailed CEM has a key that links to its corresponding concept in a domain ontology. Labetalol 100 mg, for example, can be linked to SNOMED CT concept SCT318445000. Based on SNOMED-CT, the system can infer that ‘labetalol 100 mg’ (SCT318445000) is an antihypertensive drug (SCT1182007). In addition, we can leverage existing Semantic Web tooling to perform reasoning after representing the patient data with respect to the CEM-OWL. For example, we can use the Clinical Narrative Temporal Relation Ontology (CNTRO)23 and its associated temporal-relation reasoning framework to handle time aspects in this query. As this simple example illustrates, retrieving answers to clinically important queries usually involves comprehensive inference processes.
Figure 7.

Conditions and sample patient data for resistant hypertension.
Implementation results and evaluation
CEM-OWL evaluation
We have created the CEM-OWL meta-ontology based on the CEM specification.1 A convertor has been implemented in Java to automatically translate all the detailed CEMs from XML to OWL, which was used to covert all the 183 detailed CEMs for SHARPn to OWL.
We followed the ontology evaluation criteria introduced by Brank et al24 to evaluate the CEM-OWL modeling. The ontology evaluation criteria cover several levels: syntactic, lexical, hierarchical, semantic relations, and context and application. The OWL ontology has been validated using HermiT reasoner V.0.8.1 embedded in Protégé 4.1 (http://protege.stanford.edu/) for syntactic and consistency checking. For other evaluation criteria, a review panel with two CEM experts (TAO and TC) and one OWL expert (GJ) conducted manual evaluations followed by extensive discussions. The members of the review panel are independent of the development of the CEM-OWL model. Each reviewer used a scoring system from 1 to 5 (strongly disagree, disagree, unable to assess, agree, strongly agree) to evaluate the ontological representation for each criterion. After evaluating each criterion, the reviewers compared and discussed their scores until agreement was reached on a single score. We considered a score of 4 or 5 to indicate that the OWL representation was satisfactory and could faithfully represent the original intended meaning of the CDL specification.
Table 2 shows the evaluation results. The review panel assigned a score 5 (strongly agree) to five out of the seven evaluation criteria and a score 4 (agree) to the remaining two evaluation criteria. For the two criteria with a score 4, justifications are provided in the comments column.
Table 2.
Clinical element model (CEM)-Web Ontology Language (OWL) representation evaluation result
| Criterion | Score | Comments |
|---|---|---|
| Definition of classes (lexical) | 5 | The reviewers agreed that the lexical definition of each class is clear |
| Hierarchical structure (sub-classes hierarchy) | 4 | The reviewers agreed that the sub-class hierarchy faithfully represents the CEM specifications. There is a concern about defining the basic category classes (eg, SimpleStatement and ComponentStatement) and the specialized classes (eg, Patient, Activity, etc) as sibling classes under the Statement class since they represent different types of classifications of CEMs. An alternative solution could be to create two parent classes under the Statement class to separate the two groups of classes |
| Definition of classes (semantic) | 5 | The reviewers agreed that the semantic definition of each class is correct |
| Semantic relations (object property definitions) | 5 | The reviewers agreed that the domain and range defined for each property are correct |
| Context and application (validation of the usage of the CEM-OWL model in a detailed CEM) | ||
| Class usage | 4 | The reviewers agreed that defining each detailed CEM (eg, BloodPressurePanel) as a new class and a subclass of the category it belongs to (eg, Panel) is appropriate and semantically correct. There was a discussion about the option on defining each detailed CEM as an individual of the category class (eg, BloodPressurePanel rdf:type Panel). The reviewers agreed that the current way is more appropriately aligned to the three-layer approach used for this representation |
| Relation usage | 5 | The reviewers agreed that the relations defined in the meta-ontology are sufficient to represent the connection between components in the detailed CEM |
| Cardinality constraints | 5 | The reviewers agreed that the cardinality constraints are defined correctly and the OWL cardinality constraints are sufficient to represent CEM cardinality constraints |
From the evaluation results, we can conclude that the lexical and semantic definitions of the classes, the hierarchical structure, as well as the semantic relation definitions in the meta-ontology are well defined and can faithfully represent the CEM specification. For the context and application criterion, we evaluated the usage of the classes, relations, and cardinality constraints on a detailed CEM (BloodPressurePanel). The review panel agreed that those components were sufficient to faithfully represent the semantic meaning of the detailed CEM model and the detailed CEM model was represented correctly using these components in OWL. Please note that we did not include the usage of value set definition and unit of measurement definition in the evaluation because neither is included in the CDL specification and therefore they are not included as part of the meta-model. This will be included in a future project on data type definition.
OWL convertor evaluation
The evaluation of the automatic detailed CEM convertor contains two steps. The first step is a manual comparison between the automatic generated OWL files and the corresponding original XML file. Three experts (RRF, QZ, and FC, who are independent of the CEM-OWL model development as well as the CEM-OWL converter implementation) evaluated different aspects of the converted OWL files including name spaces, class and property naming conventions, domains and ranges, class hierarchies, cardinality constraints, and relation definitions. All experts have agreed that the converted files can faithfully represent the original contents.
The second step is to test the usage of these converted OWL files. In order to do this, we have represented the medication and demographic information of 400 patients from the SHARPn normalization pipeline in RDF using the CEM-OWL ontologies. The patient data have been successfully represented in RDF. We can therefore conclude that the CEM-OWL ontologies we designed are capable of representing real patient data.
Discussion
Clinical information models harmonization
One of the important implications from our present work is that using OWL semantics to represent a CDL logical model would potentially facilitate accurate transformation of the CEMs specified in CDL into the format of ADL,8 which is a formal language for expressing archetypes. The HL7 Template Architecture standard discusses the semantic mappings between ADL and OWL. Some preliminary investigations have been undertaken to convert the ADL-based model into OWL. For instance, Lezcano et al15 developed an approach to translate definitions expressed in the openEHR ADL to a formal representation expressed using OWL, targeted at facilitating semantic interoperability for the computerized support of alerts, workflow management, and evidence-based healthcare across heterogeneous EHR systems. Martinez-Costa et al represented OpenEHR archetypes using OWL for the purpose of consistency checking. Notably, a recent international collaboration—the Clinical Information Modeling Initiative (CIMI)—committed to use ADL for their Clinical Information Model specifications.25 OWL is also a CIMI recommended output for its final integrated clinical information model. In order to reuse CDL-based CEMs in the CIMI community, there will be a need to convert the CEMs in CDL into the format of ADL. Since both CDL and ADL have been represented using OWL, OWL can serve as a common language to map CDL and ADL together. We are currently investigating the ADL and CDL transformation based on their OWL rendering.26
Semantic definition limitations
In the OWL representation, we tried to capture the semantic definitions of each CEM category. Some of the definitions, however, are difficult to represent in the OWL DL environment. For example, whether a clinical element should be defined as a collection or a panel depends on whether or not the associated elements are strongly semantically related. Unless the ‘strongly semantically related’ elements are defined in a domain ontology or information model, it is difficult for a computer system to automatically make the distinction. In addition, the CDL specification contains a restriction that specifies that the statements and/or associations within a panel cannot be modified independently. This kind of rule is better handled by downstream auditing tooling than by the semantic definition in OWL.
Property definitions
There are multiple ways to define relationships between two detailed clinical elements. One option is to use the properties defined in the meta-ontology (eg, item, qual, mod, att) to declare the relationships between two detailed clinical elements. For example, we can declare
BloodPressurePanel
item max 1
DiastolicBloodPressure
for the relation between the BloodPressurePanel and the DiastolicBloodPressure.
Another option is to define new properties for each pair of relations. For example, we can define a new property diastolicBloodPressureMeas to be a sub-property of the item property and declare the above relation as:
BloodPressurePanel diastolicBloodPressureMeas max 1 DiastolicBloodPressure
The latter approach will introduce a lot more properties, which could potentially lead to interoperability issues. But it will be more straightforward to human users to understand the data representation. The first approach is usually preferred when the data are majorly used to automatic computation whereas the second approach is preferred when the data need to be processed manually.
Conclusion and future work
In this paper, we introduce our efforts to represent the CEM in the OWL. We believe that the OWL representation of the CEM can be beneficial for the following reasons: (1) it lends formal semantic definitions to the CEM specifications; (2) it connects the CEM with various Semantic Web technologies to add authoring, reasoning, and querying capacities to the contents of the CEM; (3) it provides a unified platform to connect the CEM as an information model with different terminologies; and (4) it enables the harmonization of CEM models with clinical models that are represented in other languages such as ADL.
Several directions remain to be pursued. We will work on representing the HL7-based data types used by the CEM using both OWL 1.1 and the new OWL 2.0 data range definition features. Moreover, we plan to investigate the alignment between ADL and CDL to facilitate automatic transformation from detailed CEM models to ADL representation. Our ultimate goal is to represent patient instances in RDF and develop applications on top of this data to leverage the reasoning and storage mechanisms of Semantic Web technologies.
Acknowledgments
We thank Ms. Teresa Conway, RN, MS, and Mr Feichen Shen for their help on evaluating the CEM-OWL representation.
Funding: This work was made possible by funding from the Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinator for Health Information Technology. The contents of the manuscript are solely the responsibility of the authors.
Contributors: CT designed the CEM-OWL model, led the study design and analysis, and drafted the manuscript. GJ, TAO, RRF, and QZ contributed to the evaluation of the model. RRF, GJ, and JP helped draft the manuscript. DS helped with coding the CEM-OWL convertor. SMH and CGC provided institutional support and manuscript editing.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
RDF provides classes such as rdf:list and rdf:seq to represent a collection of items. Although the rdf:seq class is the RDF ‘Sequence’ container, by convention it is used to indicate to human readers that the order of contained items is intended to be significant. There are no logical semantics defined to a DL classifier in a rdf:seq. In addition, RDF vocabulary such as rdf:list cannot be used in OWL-DL18.
Due to space limitation, here we only show partial OWL representation of the BloodPressurePanel model for the purpose of illustrating how we represent detailed CEMs by adding different constraints.
The choice between one and the other is based on each individual case, and is not within the scope of this paper.
This is how MUO named this class to represent the value of an individual quality, for instance the weight of an individual object.
References
- 1.James A. Qualibria Constraint Definition Language (CDL), language guide 2010. Oct. 22, 2010
- 2.GE/Intermountain Clinical Element Model Search. (cited 2012). http://www.clinicalelement.com/.(accessed 22 Nov 2012).
- 3.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: The SHARPn project. J Biomed Inform 2012;45:763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steiner DJ, Coyle JF, Rocha BH, et al. Medical data abstractionism: fitting an EMR to radically evolving medical information systems. Stud Health Technol Inform 2004;107(Pt 1):550–4 [PubMed] [Google Scholar]
- 5.OWL Web Ontology Language Reference. (cited 2012). http://www.w3.org/TR/owl-ref/ (accessed 22 Nov 2012)
- 6.Horrocks I, Patel-Schneider PF, McGuinness DL, et al. OWL: a description logic based ontology language for the semantic web. The description logic handbook: theory, implementation, and applications. 2nd edn Cambridge University Press, 2007 [Google Scholar]
- 7.Rule Interchange Format (RIF) http://www.w3.org/standards/techs/rif (accessed 22 Nov 2012)
- 8.HL7 Template and Archetype Architecture. (02/23/2012). http://www.hl7.org/library/committees/template/HL7_Atlanta_10_20_04.doc (accessed 22 Nov 2012)
- 9.Donnelly K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform 2006;121:279–90 [PubMed] [Google Scholar]
- 10.https://www.nlm.nih.gov/research/umls/rxnorm/ (accessed 22 Nov 2012) [Google Scholar]
- 11.Rector AL. The interface between information, terminology, and inference models. Stud Health Technol Inform 2001;84(Pt 1):246–50 [PubMed] [Google Scholar]
- 12.Santos MR, Bax MP, Kalra D. Building a logical EHR architecture based on ISO 13606 standard and semantic web technologies. Stud Health Technol Inform 2010;160(Pt 1):161–5 [PubMed] [Google Scholar]
- 13.Martinez-Costa C, Menarguez-Tortosa M, Fernandez-Breis JT, et al. A model-driven approach for representing clinical archetypes for Semantic Web environments. J Biomed Inform 2009;42:150–64 [DOI] [PubMed] [Google Scholar]
- 14.Tao C, Jiang G, Wei W, et al. Towards semantic-web based representation and harmonization of standard meta-data models for clinical studies. AMIA Summits Transl Sci Proc 2011;2011:59–63 [PMC free article] [PubMed] [Google Scholar]
- 15.Lezcano L, Sicilia MA, Rodriguez-Solano C. Integrating reasoning and clinical archetypes using OWL ontologies and SWRL rules. J Biomed Inform 2011;44:343–53 [DOI] [PubMed] [Google Scholar]
- 16.Heymans S, McKennirey M, Phillips J. Semantic validation of the use of SNOMED CT in HL7 clinical documents. J Biomed Semantics 2011;2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resource description framework (rdf) (02/23/2012). http://www.w3.org/RDF/ (accessed 22 Nov 2012)
- 18.Tao C, Parker CG, Oniki TA, et al. An OWL meta-ontology for representing the clinical element model. American Medical Informatics Assocication Annual Symposium; 2011 [PMC free article] [PubMed] [Google Scholar]
- 19.Drummond N, Rector A, Stevens R, et al., eds Putting OWL in order: patterns for sequences in OWL. OWL experiences and directions (OWLEd 2006). Athens, GA, 2006 [Google Scholar]
- 20.Representing Specified Values in OWL: “value partitions” and “value sets”. http://www.w3.org/TR/swbp-specified-values/ (accessed 22 Nov 2012)
- 21.Measurement Units Ontology. http://forge.morfeo-project.org/wiki_en/index.php/Units_of_measurement_ontology (accessed 22 Nov 2012)
- 22.The Unified Code for Units of Measure. (cited 2012). http://www.unitsofmeasure.org/ (accessed 22 Nov 2012)
- 23.Tao C, Wei WQ, Solbrig HR, et al. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives. AMIA Annu Symp Proc 2010;2010:787–91 [PMC free article] [PubMed] [Google Scholar]
- 24.Brank J, Grobelnik M, Mladenić D. A survey of ontology evaluation techniques. Proc of 8th Int multi-conf Information Society 2005:. 166–9 [Google Scholar]
- 25.The CIMI wiki: http://informatics.mayo.edu/CIMI/index.php/Main_Page (accessed 22 Nov 2012)
- 26.Legaz-García MC, Tao C, Menárguez-Tortosa M, et al. An approach for the mapping of CEM and ppenEHR archetypes. AMIA Clinical Research Informatics Summit; 2013, submitted [Google Scholar]