Abstract
Neonatal intraventricular injection of adeno-associated virus has been shown to transduce neurons widely throughout the brain, but its full potential for experimental neuroscience has not been adequately explored. We report a detailed analysis of the method’s versatility with an emphasis on experimental applications where tools for genetic manipulation are currently lacking. Viral injection into the neonatal mouse brain is fast, easy, and accesses regions of the brain including cerebellum and brain stem that have been difficult to target with other techniques such as electroporation. We show that viral transduction produces an inherently mosaic expression pattern that can be exploited by varying the titer to transduce isolated neurons or densely-packed populations. We demonstrate that expression of virally-encoded proteins is active much sooner than previously believed, allowing genetic perturbation during critical periods of neuronal plasticity, but is also long-lasting and stable, allowing chronic studies of aging. We harness these features to visualize and manipulate neurons in the hindbrain that have been recalcitrant to approaches commonly applied in the cortex. We show that viral labeling aids the analysis of postnatal dendritic maturation in cerebellar Purkinje neurons by allowing individual cells to be readily distinguished, and then demonstrate that the same sparse labeling allows live in vivo imaging of mature Purkinje neurons at resolution sufficient for complete analytical reconstruction. Given the rising availability of viral constructs, packaging services, and genetically modified animals, these techniques should facilitate a wide range of experiments into brain development, function, and degeneration.
Keywords: Adeno-associated virus, AAV, neonatal viral transgenesis, 2-photon in vivo imaging, mouse
Introduction
The ability to create mosaic animal models in which selected cell populations are both genetically altered and permanently labeled has yielded new insight into cell-autonomous and non-autonomous actions of many normal and disease-associated proteins (Davy & Soriano, 2005; Holtmaat et al., 2009; Holtmaat & Svoboda, 2009; Kanning et al., 2010; Park & Bowers, 2010; Warr et al., 2011). In parallel, the introduction of transgenic mice with sparse mosaic expression of fluorescent proteins (Feng et al., 2000) has afforded unprecedented views of neuronal morphology in vivo that have revised our understanding of structural plasticity in the brain following environmental stimulation and pathophysiological insult. Flexible yet precise control of mosaicism is needed in both of these settings, but serious challenges limit the use of current techniques. Modified genetic elements and fluorescent tags can be easily introduced by in utero or neonatal electroporation, but the range of transfection is limited by the direction of the electric field and the diffusion of DNA (De Vry et al., 2010). Viral transduction overcomes these spatial limits, but requires equally invasive intrauterine access to the embryo (Hashimoto & Mikoshiba, 2003; 2004; Shen et al., 2004; Stott & Kirik, 2006; Rahim et al., 2009; Rahim et al., 2011). Germline genetic manipulation avoids the complications of surgery, but often yields unreliable mosaicism. Random integration-site effects result in variable density, cellular specificity, and regional distribution of transgene expression that made the Thy1-XFP series so useful for imaging, but required screening many lines to identify a few with patterns appropriate for study (Feng et al., 2000). Better control of mosaicism can be obtained using sparsely expressing Cre lines to direct lox-mediated recombination (Guo et al., 2002; Chakravarthy et al., 2008; Rotolo et al., 2008; Young et al., 2008), or more recently, recombination-mediated mosaic analysis with double markers (MADM) (Zong et al., 2005) and mosaic mutant analysis with spatial and temporal control of recombination (MASTR) (Lao et al., 2012). However, both of these approaches require the convergence of multiple independently assorting alleles in a small fraction of the offspring, are limited by the specificity of existing Cre lines, and in the case of MADM, may also necessitate construction of a modified locus for each gene to be studied (Zong et al., 2005; Espinosa et al., 2009; Hippenmeyer et al., 2010). An ideal approach would be easy to use, produce early-onset, long-lasting expression, and permit widespread genetic manipulation throughout the brain.
Here we describe a simple technique to achieve both titratable genetic mosaicism and sparse fluorescent labeling by neonatal intraventricular injection of genetically engineered AAV. The technique was initially developed by John Wolfe and colleagues to create brain-specific transgenic mice that avoided problems associated with developmental expression of ectopic proteins (Passini & Wolfe, 2001; Passini et al., 2003). Unlike germline transgenesis, random transduction by AAV produces a mosaic pattern of expression. At one extreme, injections can be tailored for sparse expression suited to study of cell-intrinsic mechanisms, and at the other provide dense expression designed for cell-extrinsic studies. Dual-transduction of the same or non-overlapping populations can be attained by co-injection of multiple viruses encoding distinct genetic elements. Most importantly, neonatal AAV transduction targets neuronal populations throughout the brain, providing an easy way to manipulate regions that have been intractable by past methods.
Materials and Methods
Viral construction and packaging
Constructs
Four different inserts were cloned into the pAAV expression plasmid for these experiments (Table 1). The first of these construct encoded the enhanced yellow fluorescent protein (YFP) and the tetracycline transactivator (tTA) separated by the Thosea asigna virus TaV 2A sequence, (GGCAGTGGAGAGGGCAGAGGAAGTCTGCTAACATGCGGTGACGTCGAGGAGAATCCTGGCCCA) (Trichas et al., 2008). The second construct was similar, but substituted tdTomato in place of YFP. The third construct encoded three copies of YFP each separated by a 2A sequence. All three of these constructs also contained the cytomegalovirus enhancer/chicken β-actin promoter (CBA), the woodchuck hepatitis post-transcriptional regulatory element (WPRE), and the bovine growth hormone polyadenylation signal (BGH). The final construct contained the elongation factor 1α (EF1α) promoter, WPRE, and human growth hormone polyadenylation signal (hGH), and encoded the mammalian codon-improved Cre recomibinase (iCre) and tdTomato separated by the Porcine teschovirus-1 PTV-1 2A sequence.
Table 1.
List of viral constructs, serotypes, titers, and mouse lines used.
| Expression Cassette | Serotype | Viral particles/ hemisphere |
Mouse strain | Figure |
|---|---|---|---|---|
| CBA-tdTomato-2A-tTA(2S) | AAV8 | 4.0×109 | ICR | Figure 4 |
| 1.0×107 - 1.0×1010 | C57BL/6J | Figure 6 | ||
| 2.0×109 | C57BL/6J | Figure 7 | ||
| 8.0×108 | C57BL/6J | Figure 8 | ||
| 5.0×109 | TetO-LacZ/GFP reporter mice | Not shown | ||
| CBA-YFP-2A-tTA(2S) | AAV8 | 2.0×1010 | C57BL/6J for all | Figure 2 |
| 2.0×109 | Figure 5 | |||
| 4.0×107 - 2.0×1010 | Figure 6 | |||
| CBA-YFP-2A-tTA(2S) | AAV1 | 2.0×109 - 2.0×1010 | C57BL/6J for all | Figure 6 |
| 2.0×1010 | Figure 7 | |||
| 2.0×109 | Figure 8 | |||
| CBA-YFP-2A-YFP-2A-YFP | AAV8 | 4.0×108 | R26R Cre-reporter mice | Figure 9 |
| 1.0×108 | ICR | Figure 10 | ||
| 3.8×109 | ICR | Figure 11 | ||
| 1.0×108 | ICR | Figure 11 and Videos 1, 2 | ||
| CBA-YFP-2A-YFP-2A-YFP | AAV1 | 1.3×1010 | C57BL/6J | Figure 4 |
| 1.3×1010 | ICR | Figure 11 | ||
| EF1α-iCre-2A-tdTomato | AAV8 | 1.2×109 | Ai3 Cre-reporter mice | Figures 3 and 4 |
| 2.0×109 | R26R Cre-reporter mice | Figure 9 | ||
| EF1α-iCre-2A-tdTomato | AAV6 | 1.2×1010 | ICR | Figure 4 |
Viral packaging
AAV1 was prepared as described in Kim et al. (Kim et al., 2008). Briefly, rAAV1 was generated by polyethyleneimine (PEI) transfection of pAAV shuttle vector, cis-plasmid pH21 (AAV1 helper plasmid), and pFΔ6 into a HEK293T cell line. At 48 hr after transfection, cells were harvested and lysed in the presence of 0.5% sodium deoxycholate and 50 U/ml benzonase (Sigma) by repeated rounds of freeze/thaws at −80° C and −20° C. The virus was isolated using a discontinuous iodixanol gradient and then affinity purified on a HiTrap HQ column (GE Healthcare). Samples were eluted from the column and buffer exchanged to PBS using an Amicon Ultra 100 Centrifugation device (Millipore). The genomic titer of each virus was determined by quantitative PCR using an ABI 7900 machine (Applied Biosystems). The viral DNA samples were prepared by treating the virus with DNaseI (Invitrogen), heat-inactivating the enzyme, then digesting the protein coat with proteinase K (Invitrogen), followed by a second heat-inactivation. Samples were compared against a standard curve of supercoiled plasmid diluted between 104 and 107 copies per ml.
AAV8 was generated by calcium-phosphate co-transfection of pAAV shuttle vector, cis-plasmid p5E18 (AAV8 helper plasmid), and pAdΔF6 into HEK293T cells. At 48 hr after transfection, cells were collected and resuspended in 50 mM Tris pH 8.0, 5 mM MgCl2, 0.15 M NaCl. Cells were incubated with DNase I (1 mg/ml) and RNase A (0.1 mg/ml) for 30 min at room temperature and then lysed in the presence of 0.5% sodium deoxycholate for 10 min at 37° C. The virus was purified using a discontinuous iodixanol gradient. The band corresponding to AAV was collected, dialyzed and concentrated in DPBS using an Amicon Ultra 15 Centrifugation filter (Millipore). The genomic titer of each virus was determined by quantitative PCR using a Stratagene Mx3005P machine (Agilent Technologies).
AAV6 was generated by same protocol described above for AAV8 generation. AAV6 was generated by co-transfection of pAAV shuttle vector and pDP-6 (containing AAV6 rep and cap genes and serving as an adenoviral helper plasmid) into HEK293T cells. The recombinant AAV6 then was purified as for AAV8.
Experimental animals and AAV injection
Experimental animals
Four strains of mice were used for these studies. Unless otherwise stated, experiments were done using wild type C57BL/6J (Jackson Laboratories) or ICR (Harlan Laboratories) animals mated in house to generate timed pregnancies. Experiments to test Cre expression used Ai3 ROSA26 CAG-lox-stop-lox-eYFP (Jackson Laboratories stock #7903 (Madisen et al., 2010)) or the R26R lacZ reporter line (Jackson Laboratories stock #3474 (Soriano, 1999)). Experiments to test tTA expression used the tetO-nls-GFP-lacZ reporter line (Mayford et al., 1996). Transgenic offspring for these experiments were generated by mating Ai3, R26R or tetO-nls-GFP-lacZ males with ICR females. Viral injections described below were performed blind to genotype, and transgenic status determined by tail biopsy either at the time of weaning or at harvest. All procedures were reviewed and approved by the Baylor College of Medicine Institutional Animal Care and Use Committee in accordance with the guidelines of the US National Institutes of Health.
Intracranial injections into neonatal mice
Within 6 hr after birth, neonates were collected from the cage and prepared for injection by cryoanesthesia. Following cessation of movement, a solution of rAAV diluted in sterile PBS containing 0.05% trypan blue was injected bilaterally into the ventricles using a 10 µl Hamilton syringe (Hamilton, 7653-01) with 32 gauge needle (Hamilton, 7803-04, RN 6PK PT4). The injection site was located two-fifths of the distance along a line defined between each eye and the lambda intersection of the skull (Fig. 1). The needle was held perpendicular to the skull surface during insertion to a depth of approximately 3 mm. Once the needle was in place, 2 µl of viral solution was manually injected into each lateral ventricle (1 µl for experiments comparing P0 and adult injection). After both injections were complete, pups were placed on a warming pad until they regained normal color and resumed movement. All injected animals were then transferred to an ICR foster mother for care. The ICR fosters had delivered within 4 days prior to the day on which pups were injected. Depending on the number of injected pups needing care, most or all of the pups born to the ICR foster were removed to ensure success of the injected animals. For delayed injection experiments, P1 (24–30 hr old), P2 (48–54 hr old) or P3 neonates (72–78 hr old) were injected as above.
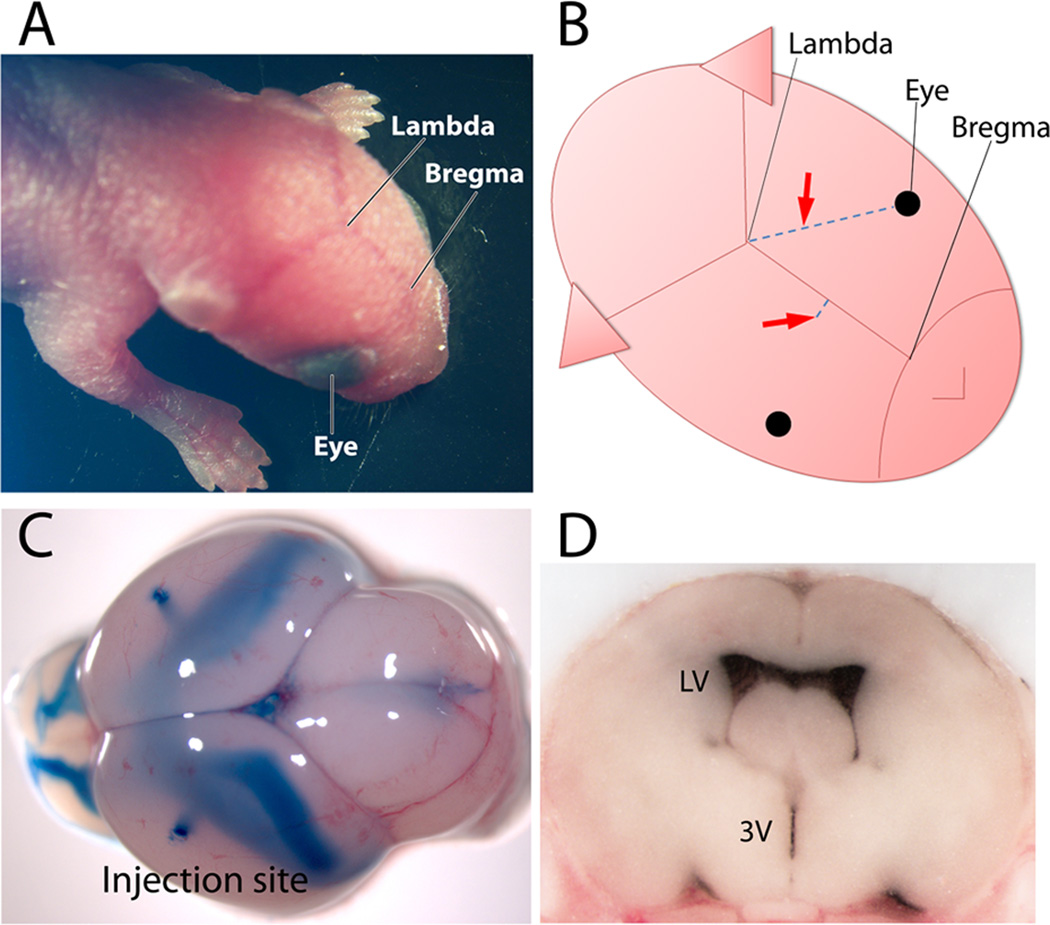
Figure 1. Intracranial injection of adeno-associated virus into the cerebral lateral ventricles of neonatal mice.
The three landmarks used to target the lateral ventricle for injection are easily visible through the translucent skin of P0 C57 neonatal pups (A). Diagrammatic view of mouse head shows the two different target sites (arrows) that can be used for viral injection relative to the eyes and the suture intersections of lambda and bregma. One injection site is located approximately two-fifth of the distance between lambda and each eye; the other site is located approximately 1 mm lateral to the sagittal suture, halfway between lambda and bregma. Both coordinates reliably target the lateral ventricles (B). Neonatal whole brain (C) and coronal cross-section of frozen brain (D) harvested immediately after bilateral dye injection to illustrate the extent and localization of the fill. The two injection sites can be seen at the brain surface from dye that escaped along the needle path, while the filled ventricles are visible through the tissue and can be seen along the rostral-caudal extent of contiguous chambers. LV-lateral ventricle: 3V-third ventricle.
Stereotaxic injections into adult mice
Adult mice (2–4 months) were anesthetized with 1.5 % isoflurane, placed in a stereotaxic apparatus, and prepared for viral injection by a midline scalp incision followed by the opening of a small burr hole in the skull over the desired injection site at 1.5 mm caudal to the bregma, 0.5 mm lateral to the midline, and 1.3 mm deep to the dura mater. 1 µl of AAV diluted in PBS containing 0.05% trypan blue was injected through a pulled glass micropipette at a constant rate of 80 nl/min using a syringe pump (UMP3 with Micro-4 controller, World Precision Instruments). The micropipette was held in place for 5 min after injection and then withdrawn slowly to minimize backflow. The skin was resealed with Vetbond and sutures. Animals were given buprenorphine for analgesia prior to awakening, and were maintained on carprofen-containing chow (Rimadyl chewies, BioServ) for 3 days post-surgery.
Preparation of mouse brains
Mice were sacrificed by carbon dioxide inhalation at 3–4 weeks or 12 months post-injection. Brains were fixed overnight at 4° C in fresh 4% paraformaldehyde and then transferred to 30% sucrose for cryoprotection. Brains were frozen on dry ice and then sectioned at 45 µm using a freezing-sliding microtome. Sections were stored in antifreeze buffer [50 mM NaPO4, pH7.4 containing 25% glycerol and 30% ethylene glycol (v/v)] at −20°C until use.
Immunohistochemistry
Brain sections of mice injected with AAV-YFP at P0 or P3 were immunostained with NeuN, CaMKIIα, S100β, Iba1, GAD67, calbindin D-28k or β-galactosidase antibodies to determine the phenotype of YFP-positive cells. Sections were washed with TBS to remove antifreeze medium, followed by permeabilization and blocking with 3% goat serum plus 0.1% Triton-X 100 in TBS for 1 hr at room temperature. Sections were then incubated with mouse anti-S100β (1:500; Sigma, S2632), rabbit anti-Iba1 (1µg/ml; Wako Chemicals, 019-19741), mouse anti-NeuN (1:1000; Millipore, MAB377), anti-CaMKII (1:1000, Chemicon, MAB3119), rabbit anti-GAD67 (1:1000; Chemicon, AB5992), mouse anti-calbindin D-28k (1:5000, Swant, McAB 300) or chicken anti-β-galactosidase antibody (1:5000, Abcam, ab9361) in blocking solution at 4° C overnight. The following day the sections were washed with TBS and incubated for 2 hr at room temperature with Alexa Fluor 594-conjugated goat anti-rabbit (1:500; Invitrogen, A11012), Alexa Fluor 568-conjugated donkey anti-mouse (1:500; Invitrogen, A10037), or Alexa Fluor 647-conjugated goat anti-chicken secondary antibody (1:500, Invitrogen, A21449) for fluorescent detection.
In vivo structural imaging after P0 virus injections
4- and 8-week-old P0-injected mice were anesthetized with isoflurane, and a 3 mm cranial window was placed as described in previous studies (Holtmaat et al., 2009). For imaging of cortical pyramidal neurons, the window was centered at 2 mm lateral and 2 mm posterior to the bregma. For imaging of cerebellar Purkinje cells, the window was centered 1.5 mm lateral and 2 mm posterior to the lambda. Following 2–5 days’ recovery, in vivo images were acquired using a Prairie View two-photon microscope. A Coherent Ti:Sapphire laser tuned to 890 nm was focused into tissue using a 20× (0.95 NA) or 60× (1.0 NA) Olympus lens at 10–40 mW (0.8 µs dwell time) and emitted photons were collected using a high sensitivity (>8500 A/lumen) Hamamatsu photomultiplier tube. Exposed brain was initially sampled at low resolution (512 × 512 pixels, 620 µm × 620 µm FOV, 5 µm slices) to identify YFP-labeled cells. Labeled cells were then imaged at high resolution (1024 × 1024 pixels, 155 × 155 µm FOV, or 2048 × 2048 pixels, 310 µm × 310 µm FOV), with 0.5 to 1 µm optical slices. Individual dendrites were re-imaged 7 and 14 days later. Images were processed for denoising using a novel polynomial interpolation method (Torskey and Smirnakis, in preparation). Dendrites and dendritic spines were quantified and reconstructed in three dimensions using Neurolucida software (MBF Biosciences).
Results
Targeting viral injections to the lateral ventricles of neonatal mice
A key step in successful P0 intraventricular injection is to precisely target the lateral ventricles with minimal damage to the brain. We have explored a number of different injections, leading us to attain two independent coordinates for intraventricular virus infusion in neonatal mouse. Targeting of the lateral ventricles was accomplished by inserting the injection needle freehand perpendicular to the skull surface and penetrating 3 mm deep. One of the sites was located 2/5 of the distance along an imaginary line between lambda and the eye; the other was located 1 mm lateral to the sagittal suture midway between lambda and bregma (Fig. 1A and B). To develop accuracy with the technique, a dye solution can be injected in place of virus and the brain harvested immediately to examine localization and spread. Following correctly targeted injections, dye will be visible throughout the continuous ventricular chambers spanning the brain from the olfactory bulb to the cerebellum. Cross-section of the brain at the level of the rostral striatum should reveal dye restricted to the ventricles, but within these chambers fills the entire space. Within a few practice sessions, the lateral ventricles can be reliably targeted by free-hand injection (Fig. 1D).
The other requirement for successful use of intracranial injection is good survival with minimal injury. We used small-bore injection needles designed to balance the potential for tissue damage against the possibility of needle clogging: we opted for 32 gauge needles with small neonates such as B6 and 30 gauge with larger strains such as ICR. After injection, ICR pups were returned to their mothers, while B6 pups were fostered to ICR females because we found B6 mothers less likely to accept and nurse the pups after being removed from the nest and handled. These approaches yielded survival rates above 95% with consistent transduction patterns and no evidence of tissue damage.
Neonatal intraventricular injection of AAV8 results in long-lasting, widespread neuronal transduction
We examined the distribution of viral transduction using native fluorescence from recombinant adeno-associated virus serotype 8 (AAV8) encoding eYFP or tdTomato injected into the cerebral lateral ventricles 3–4 weeks before harvest. P0 injection with AAV8 resulted in extensive transduction throughout the brain, with dense labeling apparent in the olfactory bulb, striatum, cerebral cortex, hippocampal formation and cerebellum (Fig. 2A–H). Intraventricular injection of AAV8 at 1010 particles/ventricle transduced 88±3% of NeuN-positive pyramidal neurons in cerebral cortex, 93.3 ± 0.7% of NeuN-positive pyramidal neurons in the CA1 region of the hippocampus and 87 ±2% of calbindin D-28k-positive Purkinje cells in the cerebellum (mean ± s.e.m, n=20–28, 3–5sections/brain from 5–6 animals).
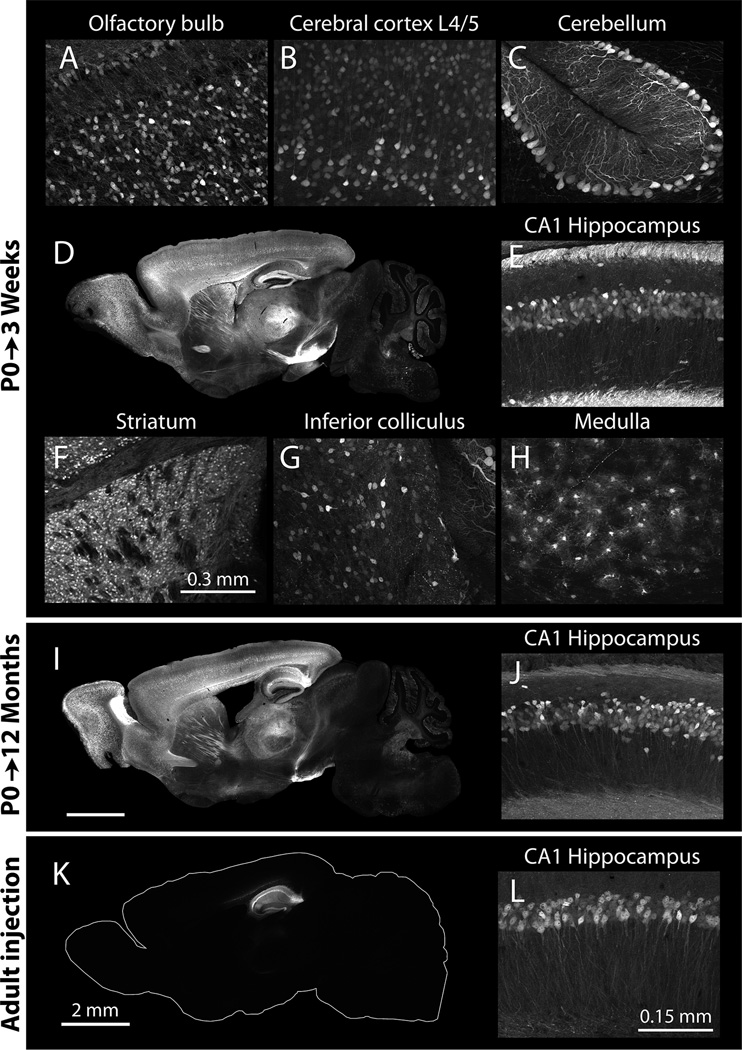
Figure 2. Neonatal intraventricular AAV injection produces widespread and long-lasting transgene expression in the brain.
Images were taken from mice harvested 3 weeks (A–H, K, L) or 10 months (I, J) after intraventricular injection at birth (A–J) or stereotaxic injection as a young adult (K, L). All images were taken from mice injected with 2 µl of AAV8 to deliver 2.0×1010 particles/hemisphere. Transduction is visualized by native YFP fluorescence. Whole-brain sagittal sections and high-magnification views of transduced regions show widespread transduction with dense neuronal labeling (A–H). Transgene expression is maintained for at least 12 months after P0 injection, as shown in this whole brain sagittal section and high magnification view of the hippocampal CA1 region. In contrast to the widespread expression attained at P0, viral injection into the adult brain results in a significantly more restricted spread, with transduction localized within several mm of the injection site (K, L). Scale bar in L applies to panels E, G, H, and J; scale bar in K applies to D and I. Exposure times for whole brain images have been determined based on cortex and hippocampus which fluoresce most brightly, while exposure times for higher magnification panels have been adjusted to match the intensity of labeling in each brain region.
Labeling was also detected in the superior and inferior colliculus, pons, and medulla, with more transduction along superficial structures as expected from viral diffusion in the cerebrospinal fluid through the fourth ventricle and the subarachnoid space (Fig. 2G, H). Fluorescence was occasionally detected in the thalamus, but the density of labeled cells was lower than in other regions of the same brain. To compare the efficiency of neonatal injection with adult injection, adult mice were stereotaxically injected with the same titer and volume of virus. This resulted in a much more limited pattern of viral transduction immediately adjacent to the injection site (n=3, Fig. 2I, J vs. 2K, L).
To examine whether viral transgene expression could be maintained long-term, mice were injected bilaterally at birth and harvested 3 weeks or 12 months later to examine the extent and intensity of the fluorescence label (n=5 for each group). Even 12 months after injection, fluorescence remained at qualitatively similar levels and was located in the same structures as at 3 weeks post-injection. In contrast to the localized injury often seen following adult intracranial injections, we observed no sign of overt malformations or injury to the brain in animals that had undergone neonatal intraventricular injection. Gross neuroanatomy was normal in the mice we studied (>100 P0 injections to date), and virally-labeled neurons displayed normal morphology in all brain regions examined. Immunostaining for astrocytic and microglial markers looked similar to uninjected wild-type animals (data not shown). The injected pups appeared to mature normally and were indistinguishable from uninjected litters at weaning. Although we have not conducted rigorous behavioral assessments, the neonatally injected mice do not display obvious behavioral abnormalities.
Rapid onset of viral expression following intraventricular AAV injection
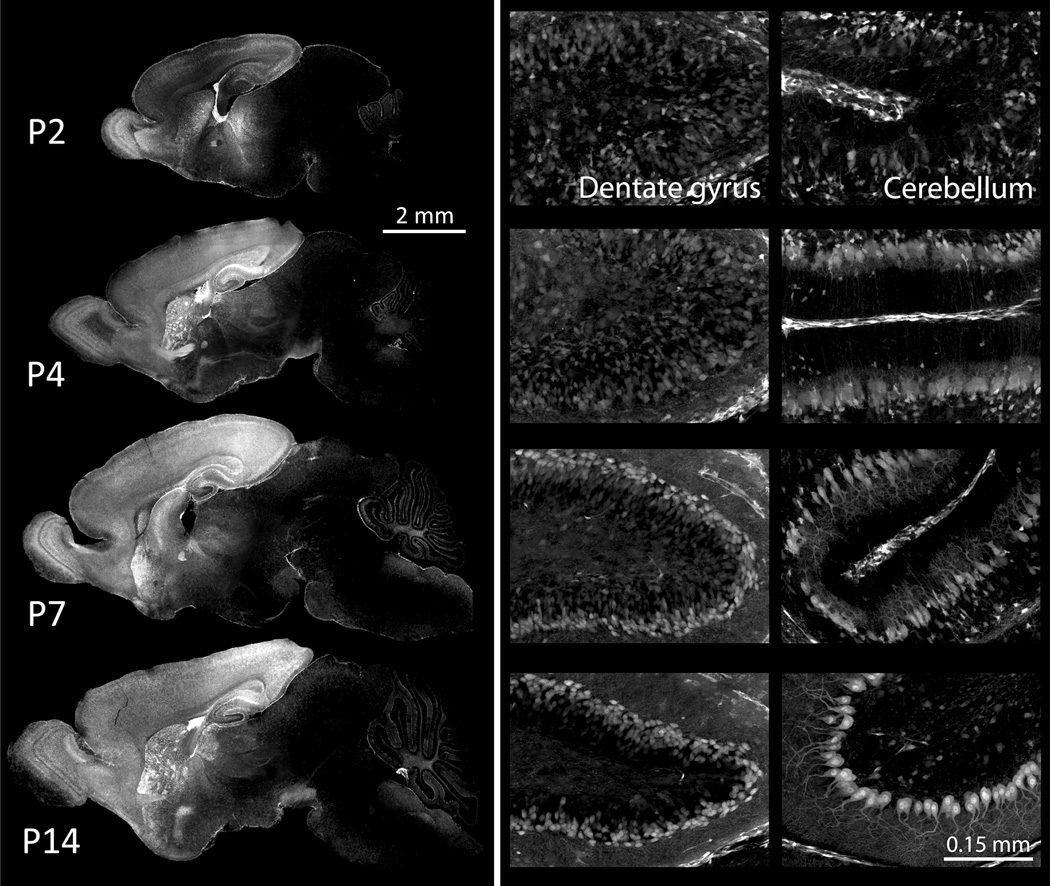
One potential disadvantage of transduction with AAV is its delayed onset of transgene expression, which may limit studies on postnatal development in juvenile mice after P0 injection (Sarra et al., 2002; McCarty et al., 2003; Natkunarajah et al., 2008). We wanted to determine in our own hands when transgene expression began and when it peaked, harvesting brains at P2, P4, P7 and P14 after intraventricular P0 injection with AAV8. We selected a virus encoding both tdTomato and Cre-recombinase under control of the elongation factor 1-αEF-1αpromoter, and injected into Cre-reporter Ai3 mice in which YFP expression is restricted by a loxP-flanked stop cassette (n=2–5) (Madisen et al., 2010). Widespread expression of both native tdTomato fluorescence and Cre-dependent YFP was already apparent by P2 (Fig. 3). The intensity and spread of YFP expression increased over the following week, reaching levels by P7 that were almost identical to the adult brain (Fig. 3). Fluorescent labeling allowed us to observe postnatal neuronal migration and structural maturation throughout the brain. Particularly striking were the formation of the hippocampal dentate gyrus (middle column of Fig. 3) and dendritic outgrowth of cerebellar Purkinje cells (right column of Fig. 3). The unexpected speed of functional transgene expression following intraventricular AAV injection offers a powerful new tool for studying early postnatal brain development.
Figure 3. Rapid onset of a fluorescent Cre reporter following P0 injection of AAV8.
Representative images show complete sagittal sections (left column) and high-magnification views of hippocampal dentate gyrus (middle column) and cerebellum (right column) following P0 injection of AAV8-EF1α-iCre into the Cre reporter line Ai3 (Madisen et al., 2010). Animals were harvested at P2, P4, P7, and P14 to examine the onset and spread of transduction using native YFP fluorescence as an indicator of virally-expressed Cre activity. All images show YFP fluorescence derived from the Cre-activated transgene; no immunostaining was needed to visualize the label. Widespread expression is apparent within days after injection, allowing observation of neuronal migration (dentate gyrus) and neurite maturation (cerebellum) during early postnatal development. Fluorescence increases across ages as more neurons inherit the active reporter, to normalize the image, longer exposures were used for P2 and P4 than P7 and P14. Exposure times for whole brain images were determined based on the cortex and hippocampus, which fluoresce most brightly, while exposure times for the magnified panels were adjusted to show patterns within each region rather than the intensity of expression between them.
Serotype and promoter bias the regional pattern of viral expression
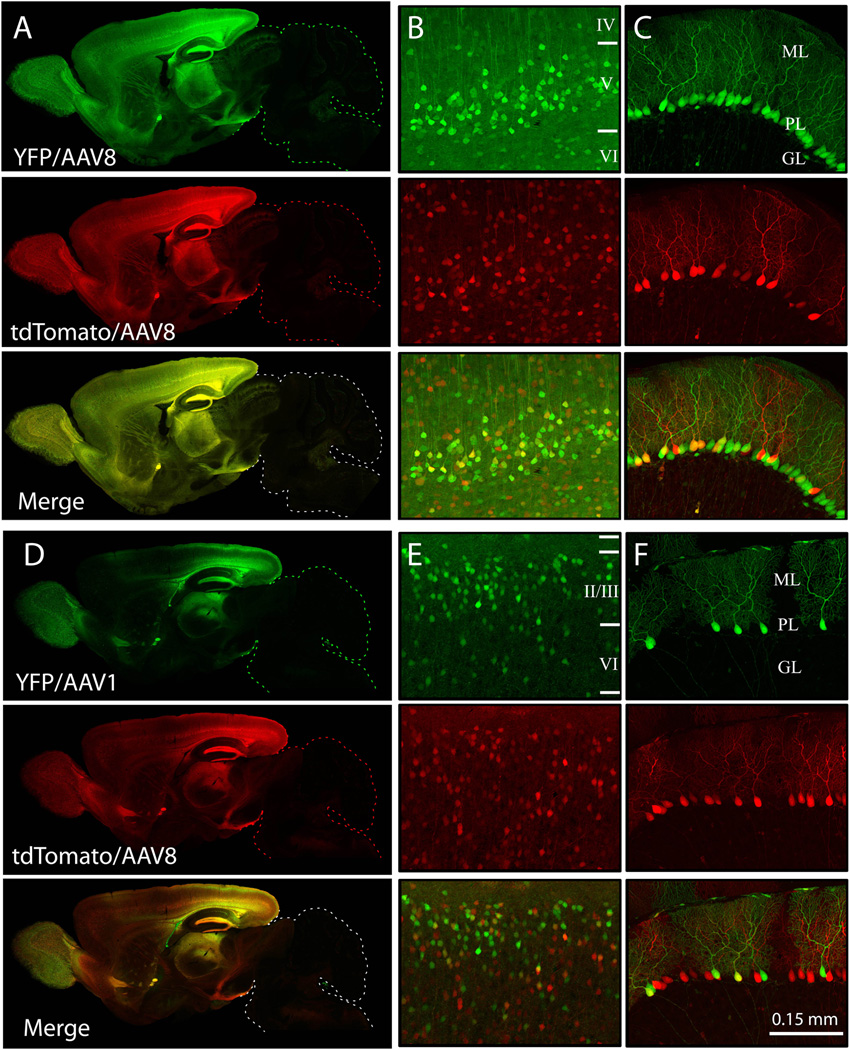
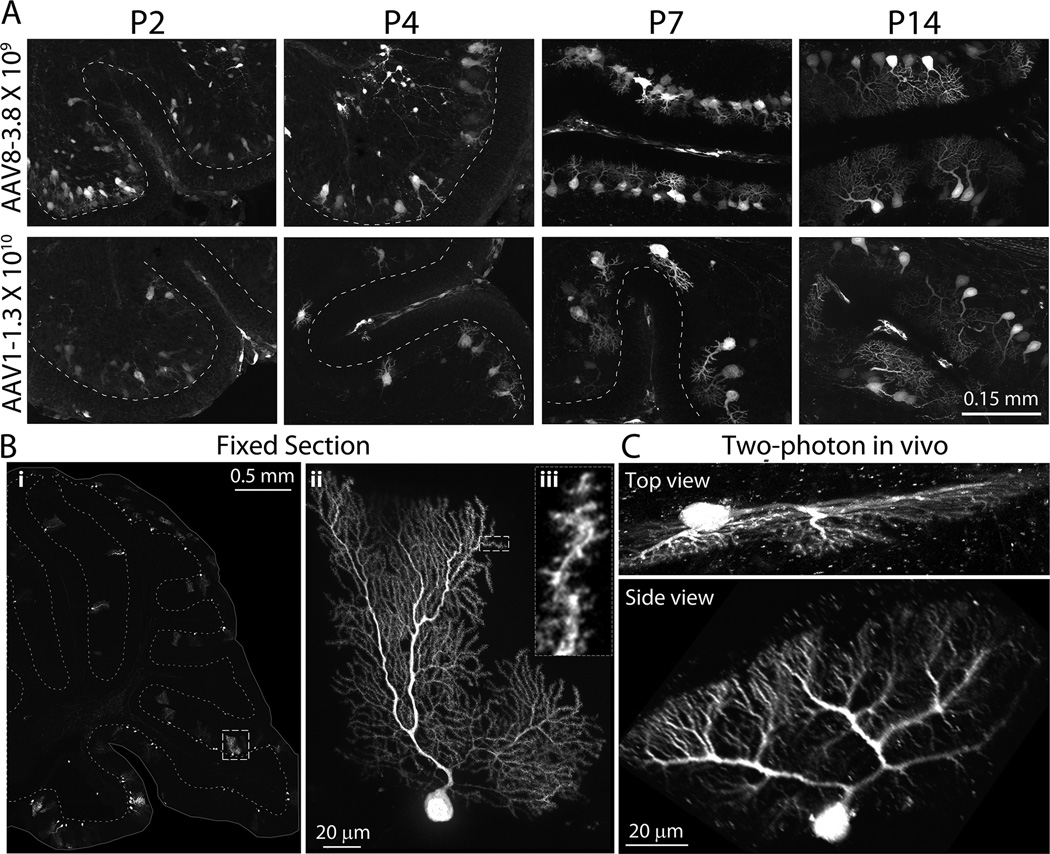
AAV serotype influences tissue tropism, cellular specificity, and transduction efficiency (Passini et al., 2003; Broekman et al., 2006; Wu et al., 2006; Cearley et al., 2008). We set out to determine whether innate serotype properties could be used to bias which neurons or cell types are manipulated by AAV transgenesis, comparing the transduction patterns of AAV8 with AAV1 and AAV6. Preliminary experiments were done to determine what titer of each virus yielded similar intensity expression, ending with ICR pups receiving 1.3×1010 particles/ventricle of AAV1, 1.2×1010 particles/ventricle of AAV6, or 1.3 to 4.0×109 particles/ventricle of AAV8. All vectors were controlled by the CBA promoter and encoded either three-copies of YFP connected by 2A self-cleavage sequence (triple YFP) (AAV1 and AAV8) or tdTomato (AAV6 and AAV8) as a readout for expression. Transduction patterns were analyzed at P2, P4, P7, P14 and P21 (n=4–7 per time point for each serotype). As expected, each serotype produced different expression patterns with varying levels of intensity across different brain regions. AAV1 and AAV6 were both most strongly expressed in the ventricular ependymal cell layer, suggesting that they do not penetrate the parenchyma as well as AAV8 (Fig. 4). Within the neocortex AAV1 expressed most strongly in superficial layers, which contrasted sharply the even distribution of transduced neurons observed with AAV8. AAV1 produced dense transduction within the olfactory bulb and caudal neocortex, but was notably excluded from the rostral neocortex. AAV6 transduction was more sparse than either AAV1 or AAV8, but more evenly distributed throughout the forebrain than AAV1. AAV6 stood out for its relative ability to infect ventral lobules of the cerebellum VIII, IX, and X, where fluorescence within Purkinje cells matched that of pyramidal neurons in the neocortex. Like AAV8, expression of both AAV6 and AAV1 was apparent at the earliest time point examined (P2), although compared to AAV8, both AAV1 and AAV6 reached maximal expression levels later than AAV8 and produced less intense fluorescence overall.
Figure 4. Both serotype and promoter influence the onset and pattern of transgene expression.
Representative images show transduction patterns in whole brain sagittal sections and high-magnification views of cerebral cortex and hippocampus after intraventricular injection of AAV1, AAV6, or AAV8 at P0. Mice were injected with 2 µl of AAV1-CBA-triple YFP to deliver 1.3 ×1010 particles/hemisphere (first column), AAV6-CBA-tdTomato to deliver 1.2 ×1010 (second column), AAV8-CBA-tdTomato to deliver 3.0×109 (third column), or AAV8-EF1α-tdTomato to deliver 1.2×109 (fourth column), and harvested at P2, P4, P7 and P14 for comparison of transduction efficiency and expression onset. High-magnification images were taken from somatosensory cortex for AAV1, primary motor cortex for AAV6 and AAV8. The hippocampal CA1 region is shown for all serotypes; dotted lines indicate the borders of the pyramidal cell layer. Immunostaining for NeuN in P14 tissue confirmed that all three serotypes primarily transduce neurons. Viral expression is visualized by native fluorescence in all panels (YFP for AAV1, tdTomato for AAV6, AAV8-CBA, and AAV8-EF1α) and shown as red in the color panels (AAV1-YFP has been pseudo-colored for this illustration). Immunostaining for NeuN is shown in green (middle row), and the merged channels below. Yellow arrows provide a fiduciary point in each image highlighting one example cell expressing both virally-delivered fluorescent protein and NeuN. Exposure times were determined based on fluorescence intensity within the hippocampus and cortex, and were longer for AAV1 and AAV6 than for AAV8. CBA: chicken β-actin promoter, EF1α: elongation factor 1α promoter.
We also evaluated how promoter choice affected the onset and pattern of transgene expression by comparing two AAV8 virions encoding tdTomato, one using the chick β actin promoter (CBA) and the other, elongation factor 1α (EF1α). Both promoters are known for broad expression, and the overall transduction patterns they produced were similar. AAV8-EF1α expression saturated slightly earlier than AAV8-CBA, while the latter continued to increase in intensity and extent throughout the first 2 weeks after injection (Fig. 4). The most notable differences between the two promoters were subtle biases in the pattern of transduction. AAV8-CBA produced slightly more consistent transduction of the neocortex compared to the caudal bias of AAV8-EF1α, while AAV8-EF1α was superior in transducing cerebellar Purkinje neurons.
Although each combination of serotype and promoter resulted in different transduction patterns throughout the brain, they all shared a bias towards neuronal expression. Cells expressing virally-delivered YFP or tdTomato could often be identified as neurons based on their morphology and location, and this was further confirmed by immunostaining for the pan-neuronal marker NeuN (9–10 sections/brain from 2 animals for each serotype, Fig. 4).
Timing of intraventricular AAV8 injection alters tropism
When intraventricular injection of AAV1 is delayed past P0, the virus does not transfect brain parenchyma efficiently (Chakrabarty et al., 2010). To determine if the timing of injection similarly affected the pattern of AAV8 transduction, 2.0×109 particles/ventricle of AAV8-YFP was injected into the lateral ventricles of littermate mice at P0, P1, P2 or P3. Mice were then sacrificed after 4 weeks and analyzed for transgene expression (n=5 for each condition). Surprisingly, delayed AAV8 injections resulted in substantial transduction throughout the whole brain, although the efficiency decreased at later ages. The transduction attained by P1 injection was nearly identical to that seen after P0 injection. Delayed injection of AAV8 resulted in a diminished spread of virus, particularly in brain structures farthest from the lateral ventricles such as the superficial layers of cerebral cortex, olfactory bulbs, and cerebellum (Figure 5A, C, E). Interestingly, delayed injection of AAV8 transduced a large number of non-neuronal cells, which rarely occurred following P0 injection (Figure 5B, D, F).
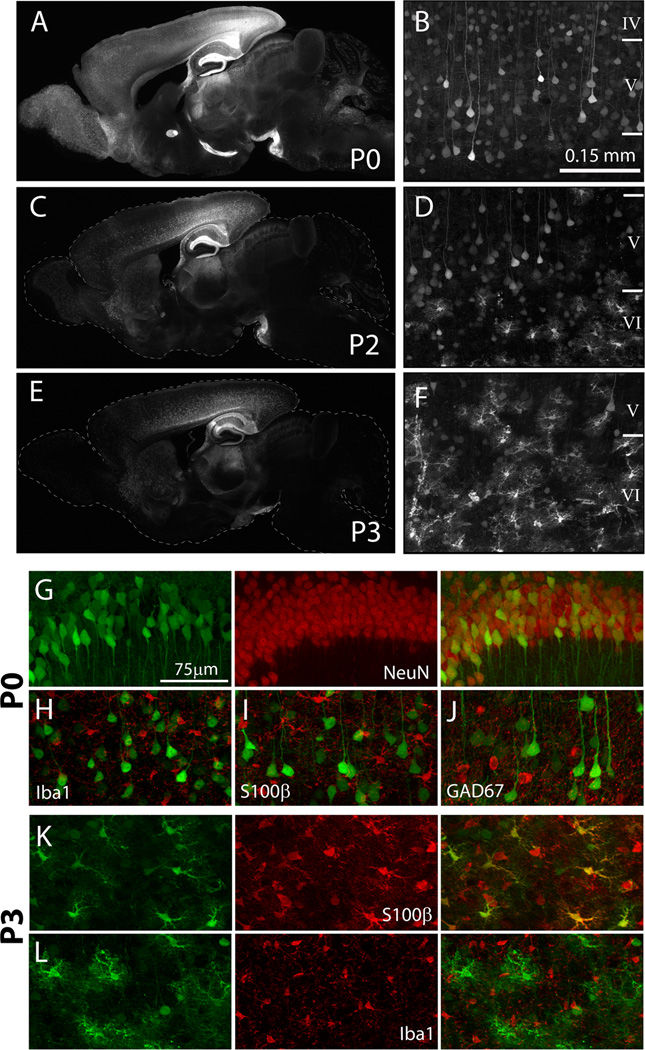
Figure 5. Timing of AAV8 injection alters the pattern and cell-specificity of transduction.
Representative images of whole-brain sagittal sections from mice harvested 3 weeks after P0, P2, or P3 injection show diminished spread of virus with age, particularly in distant structures such as the olfactory bulb and cerebellum (A, C, E). Higher magnification images of coronal sections shows the transduction of non-pyramidal cells in the deep layers of the cortex following delayed AAV8 injection (compare B with D and F). Immunostaining with cell-type-specific markers for neurons (NeuN), astrocytes (S100β), microglia (Iba1), and GABAergic interneurons (GAD67) was used to identify the transduction preference of AAV8 from P0-injected mice (G–J) and P3-injected mice (K, L). Green = YFP native fluorescence (first column); Red = immunomarker (second column); overlay of the two channels is shown in third column of G and H, I, J. Representative images are from hippocampal CA1 (G) or deep layers of cerebral cortex (H–L). Immunofluorescence staining for NeuN (G) shows that AAV8 primarily transduced NeuN-positive neurons in P0 injected mice. In contrast, P3 injection resulted in viral transduction of S100β-positive astrocytes (K). All images were taken from mice bilaterally injected
The extent of non-neuron transduction increased with the age at injection. Labeled non-neuronal cells were detected in most brain structures with the exception of the olfactory bulb. Immunofluorescence staining for the pan-neuronal marker NeuN confirmed that the majority of cells transduced by AAV8 at P0 was neurons (n=3, Figure 5G). Within several areas, including the piriform cortex, amygdala, pons, medulla, and stratum oriens of the hippocampus, a few S100β-positive astrocytes were found expressing the viral label, but these were a small fraction of the transduced cell population (<1%). Within the hippocampal formation and cerebral cortex, most of the transduced cells were morphologically identified as excitatory pyramidal neurons and expressed CaMKIIα. Except within the thalamus, very few labeled cells co-stained for the inhibitory neuronal marker GAD67. Immunofluorescence staining in sections from mice injected at P3 demonstrated that the majority of cells transduced by AAV8 at this age were S100β-positive astrocytes (n=3 Figure 5K). These data indicate that the timing of intraventricular AAV8 injection can strongly influence both the overall transduction efficiency as well as cell-type specificity. This unique property of AAV8 expands the potential repertoire for AAV targeting based on infection time, and provides a novel approach to astrocyte-specific transgene delivery.
Extent of brain mosaicism can be controlled by adjusting the viral titer
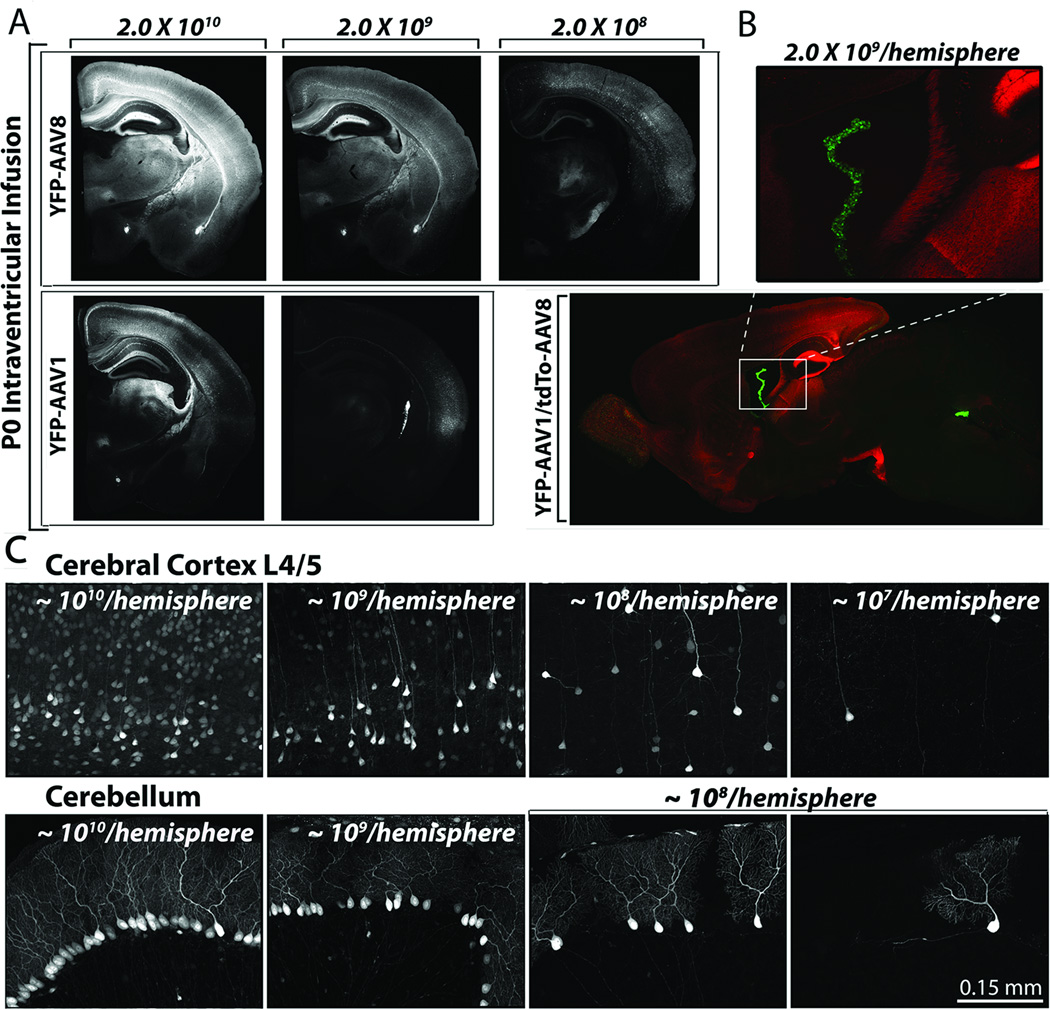
One advantage of viral-mediated gene transfer is that viral titer can be easily adjusted to alter transduction efficiency. We tested whether viral dilution could be reliably harnessed to generate controllable transgene mosaicism, and at what dilutions different serotypes were effective. We prepared serial dilutions of AAV8-YFP and AAV1-YFP from ~1010 to ~108 particles/µl; 2 µl of each dilution was bilaterally injected into the lateral ventricles of P0 pups (n=5–8 for each condition). Dilution of both AAV8 and AAV1 reduced transduction efficiency throughout the brain, with far less fluorescent protein expression at 109 than at 1010 particles/µl (Fig. 6). Dilution of AAV8 resulted in progressively fewer neurons being transduced at 109 particles/µl than at 1010 particles/µl, and fewer still at 108 particles/µl than at 109 particles/µl. However the spread of AAV8 infection was essentially identical among the different dilutions. In contrast, the spread of transduction with AAV1 declined sharply at the first 10-fold dilution to 109 particles/µl (Fig. 6A). To directly compare the transduction efficiency of AAV8 to AAV1, we co-injected the two serotypes at the same titer (109 particles/µl, n=4 per condition). As when injected alone at these titers, AAV8 transduced neurons throughout the brain, while transduction by AAV1 was largely restricted to the choroid plexus (Fig. 6B). The strong transduction of ventricular epithelia suggests that high-affinity binding of AAV1 to these cells left little virus free to enter the rest of the brain.
Figure 6. AAV8 offers a broad dynamic range for mosaicism.
Serial dilution of AAV8 or AAV1 yielded decreasing transduction densities. Representative images show transduction patterns in coronal sections taken 3 weeks after P0 injection of serially diluted AAV8 or AAV1 (A). When injected at the same concentration, AAV8 produced much more efficient transduction compared to AAV1. Co-injection of 2.0×109 particles AAV8-tdTomato and 2.0×109 particles AAV1-YFP per hemisphere provided a direct comparison of their respective infection efficiencies. Sagittal section from a co-injected mouse shows that AAV8 spread throughout the parenchyma, while AAV1 at this titer was largely restricted to the choroid plexus (B), consistent with its distribution when injected alone at this concentration (A, bottom right panel). The inset in B shows a higher magnification view of labeled cells in the third ventricle. Higher magnification images of cortex (upper row) and cerebellum (lower row) show the cellular pattern of transgene mosaicism achieved by serial dilution of AAV8 (C). Sagittal sections were collected 3 weeks after P0 injection.
Next, we optimized the viral titers needed to attain reliable high- and low-density expression with each serotype based on serial dilution of each preparation. High-density neuronal transduction was consistently achieved by intra-ventricular injection of 4.0×109 to 2.0 ×1010 particles of AAV8 per hemisphere or 4.0×1010 particles/hemisphere of AAV1. Injection of virus at these concentrations left only a small population of wild-type cells surrounded by a field of transduced neighbors, ideal for studying cell-extrinsic effects of a virally-delivered transgene. A complementary transduction pattern was attained by low-titer injections using 4.0×107 particles/hemisphere AAV8 or 2.0×109 particles/hemisphere AAV1. At these concentrations, P0 injection consistently yielded sparse transduction in which only a few isolated neurons were transduced, ideal for studying cell-intrinsic effects of a virally-delivered transgene. Thus, the titer of both serotypes could be easily adjusted to control transgene mosaicism in the brain, but over a greater range for AAV8 than AAV1.
Co-injection of two viruses generates dual mosaic patterns of transgene expression
In some experimental settings, it would be helpful to express different transgenes in neighboring cells. We tested whether this could be achieved by co-injecting a mixture of two viruses encoding different fluorescent proteins. We examined the expression attained by combining two viruses of the same serotype (AAV8-YFP with AAV8-tdTomato), as well as viruses of different serotypes (AAV1-YFP with AAV8-tdTomato). All three vectors (AAV8-YFP, AAV1-YFP, and AAV8-tdTomato) use the same promoter and ITRs. Injection of either identical or different serotypes resulted in widespread transduction of both injected vectors. Co-injection of viruses with the same serotype resulted in more cells that were transduced by both viruses (n= 8, Fig. 7A–C), while co-injection of different serotypes yielded more cells that were transduced by one or the other virus (n=4, Fig. 7D–F). AAV1 and AAV8 preferentially targeted different layers of the cortex, resulting in greater transduction of neurons in the deep layers with AAV8 and neurons in superficial layers with AAV1. The pattern of expression for each virus was similar regardless of whether it was used alone or in combination suggesting that different virions sharing the same capsid proteins, promoters, and ITRs act independently in vivo.
Figure 7. Multiple transgenes can be delivered simultaneously by AAV co-injection.
Representative images show the transduction pattern found in whole brain (A, D), cerebral cortex (B, E), and cerebellum (C, F) 3 weeks after co-injection with two different AAVs. Subtle differences in the pattern of transgene expression were found following co-injection of two viruses of the same serotype (AAV8-YFP and AAV8-tdTomato; A, B, and C) or of different serotypes (AAV1-YFP and AAV8-tdTomato; D, E and F). High-magnification views of pyramidal neurons in cortical layers 4–6 (B, E) and Purkinje neurons in the cerebellum (C, F) reveal that cells can be transduced by one or both viruses, although a greater proportion of neurons express both fluorescent labels after co-injection of AAV8 than after a combination of AAV8 and AAV1. AAV8-YFP and AAV1-YFP were delivered at 2.0×1010 particles/hemisphere; AAV8-tdTomato was delivered at 2.0×109 particles/hemisphere. YFP fluorescence is shown in green, tdTomato in red. Cortical layers II/III-VI are marked; ML: molecular layer, PL: Purkinje cell layer, GL: granule cell layer
We next tested whether the density of transduction could be independently controlled when two viruses were co-injected as it could for one virus alone. Co-injection of two viruses of the same serotype each at low titer (4.0×108 particles/hemisphere of each AAV8-YFP and AAV8-tdTomato) resulted in sparse expression of both viruses, and as a result, fewer dually-transduced cells compared with titers ≥2.0×109 (n=4, Fig. 8A–D). Co-injection of two viruses of different serotypes and titers (2.0×109 particles of AAV1-YFP and 8.0×108 particles AAV8-tdTomato per hemisphere) titer also yielded a largely non-overlapping pattern of viral expression, with the higher titer virus displaying correspondingly more dense transduction than the lower titer (n=6, Fig. 8E–H). Thus, both serotype and titer can be adjusted as needed to generate varying transduction patterns for each viral transgene.
Figure 8. Serotype and titer can be independently adjusted to control the pattern of mosaicism obtained by co-injection of multiple viruses.
Representative images show the transduction pattern found in whole brain (A, E), CA1 hippocampus (B), cerebral cortex layer 4, 5 (C, F), cerebral cortex layer 2, 3 (G) and cerebellum (C, F) 3 weeks after co-injection of two different AAVs at reduced titer. High-magnification views of neurons transduced following co-injection of the same serotype (4.0×108 particles of AAV8-YFP and 4.0×108 particles AAV8-tdTomato per hemisphere) in CA1 hippocampus (B), cerebral cortex (C) and cerebellar lobe (D) reveals a greater number of singly-transduced cells than observed when both viruses are injected at high titers. High-magnification views of neurons transduced by co-injection of two different AAV serotypes (2.0×109 particles of AAV1-YFP and 8.0×108 particles AAV8-tdTomato per hemisphere) in cerebral cortex layer 4, 5 (F) and layer 2, 3 (G) and cerebellar lobe (H) demonstrates that both serotype and titer can be independently adjusted to control the density of cells expressing each virally-encoded protein. Cortical layers II/III-VI are marked; SO: stratum oriens, SP: stratum pyramidale, SR: stratum radiatum, GL: granule cell layer, PL: Purkinje cell layer, ML: molecular layer
Viral co-expression of Cre recombinase and fluorescent protein provides a vital label of genetically modified neurons
One potential application for mosaic viral transgenesis is the generation of mice in which neighboring neurons differ only in their expression of a particular gene of interest. This approach has been used extensively to conditionally delete selected genes in a region- or cell-type-specific manner, but has rarely allowed identical cells within the same structure to be manipulated independently.
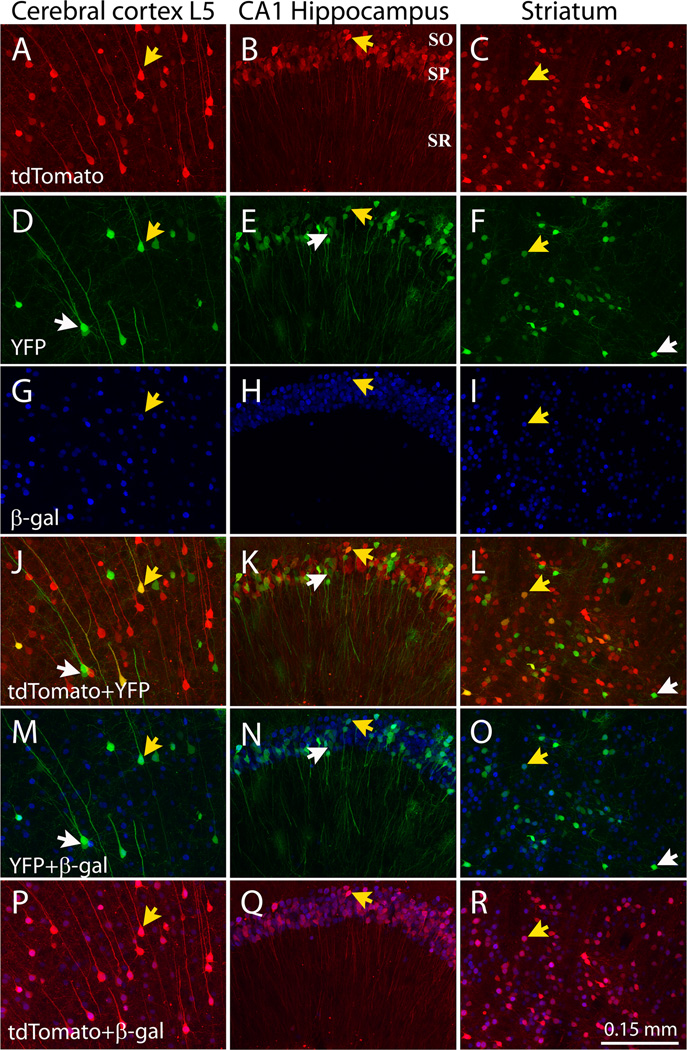
We used a virus co-expressing Cre recombinase and the red fluorescent protein tdTomato using Thosea asigna virus (TaV) 2A self-cleavage sequences (AAV8/iCre-2A-tdTomato) to test the accuracy with which a virally-expressed fluorescent protein labeled cells that underwent Cre-mediated recombination. The AAV8-iCre-2a-tdTomato virus was co-injected at varying titers with AAV8-YFP into P0 pups from the R26R lacZ reporter line (Soriano, 1999). Importantly, we found that tdTomato (Fig. 9A–C) and β-gal (Fig. 9G–I) were reliably co-localized (Fig. 9P–R), indicating that the viral fluorescent protein served as an accurate indicator of Cre-induced genetic modification within most brain regions, including the hippocampus and striatum (96±0.4 % of cells in CA1hippocampus and 94±0.6% of cells in striatum co-expressed tdTomato and β-gal; n=22–25 sections, 4–5 sections/brain from 5 animals). Co-localization was less dependable within the neocortex, where in layers 2/3 and 4, β-gal was often detected in cells with very low red fluorescence (89.4±0.1.0% of cells in cortex co-expressed tdTomato and β-gal, n=29 section, 6 sections/brain from 5 animals).
Figure 9. Viral transduction can be used to genetically modify and fluorescently label neurons.
Representative images show the secondary motor cortex layer 5 (left column), hippocampus (middle column), and striatum (right column) of R26R Cre-reporter mice 3 wk after co-injection at P0 with AAV8-iCre-2A-tdtomato (2.0×109 particles per hemisphere) and AAV8-triple-YFP (4.0×108 particles per hemisphere). Sections were immunostained for β-galactosidase (β-gal) to determine if tdTomato fluorescence accurately labels genetically manipulated cells. The top 3 rows show native fluorescence for tdTomato (red, A–C) and YFP (green, D–F), along with β-galactosidase immunostaining (blue, G–I). The lower 3 rows show overlap between tdTomato + YFP (J–L), YFP + β-gal (M–O), and tdTomato + β-gal (P–R). Reliable co-localization between tdTomato and β-gal is seen in the hippocampus (P) and cerebellum (Q), while reliable overlap in the cortex requires a lower titer of virus (R). White arrows indicate representative cells expressing YFP alone; yellow arrows indicate cells that were transduced by both viruses and which express YFP, tdTomato, Cre, and β-gal.
While the majority of controllable genetic models are based on the Cre-loxP system, transgenic mice that utilize the tTA-rtTA system for inducible, reversible expression of transgenes are becoming increasingly available. To test viral delivery of tTA, we designed a virus encoding tdTomato-2A-tTA (AAV8-tdTomato-2A-tTA) and injected it into GFP tet reporter mice (tetO-nls-GFP-LacZ) (Mayford et al., 1996). We observed reliable co-expression of viral tdTomato and transgenic GFP in most brain areas in these mice (Fig. 10). Neonatal injection of 5.0×109 particles of AAV8-tdTomato-2A-tTA into GFP tet reporter mice resulted in highly reliable co-expression of tdTomato and GFP in individual neurons, with very low mismatch in hippocampus and cerebellum and slightly higher mismatch in cortex (98±0.5% of pyramidal neurons in CA1, 95±1.0% of Purkinje cells in cerebellum, and 83±2.1% of neurons in cerebral cortex co-expressed tdTomato and GFP; n=12–19 sections, 3–4 sections/brain from 4–5 animals).
Figure 10. Viral co-expression of tTA and tdTomato induces expression of tet-responsive genes and fluorescently labels transgenic cells.
TetO-LacZ/GFP reporter mice were injected at P0 with AAV8-tdTomato-2A-tTA (5.0×109 particles per hemisphere) to determine if virally-expressed transactivator was capable of activating transgene expression from the tetO promoter and whether the transgenic cells could be reliably identified by the viral fluorescent label. Representative images from cerebral cortex (A), hippocampal CA1 (B), CA3 (C), and cerebellum (D) showing the viral reporter tdTomato in red (top row) and the transgenic reporter GFP in green (middle row), along with their co-localization (bottom row).
These data provide proof-of-principle that neonatal injection of viruses co-expressing a transgene and fluorescent marker can be employed to genetically manipulate a subset of cells in brain tissue and accurately identify these cells for further study.
Neonatal injection of a novel AAV8-triple-YFP vector produces sparse bright labeling suitable for high-resolution chronic in vivo imaging of neuronal microstructure
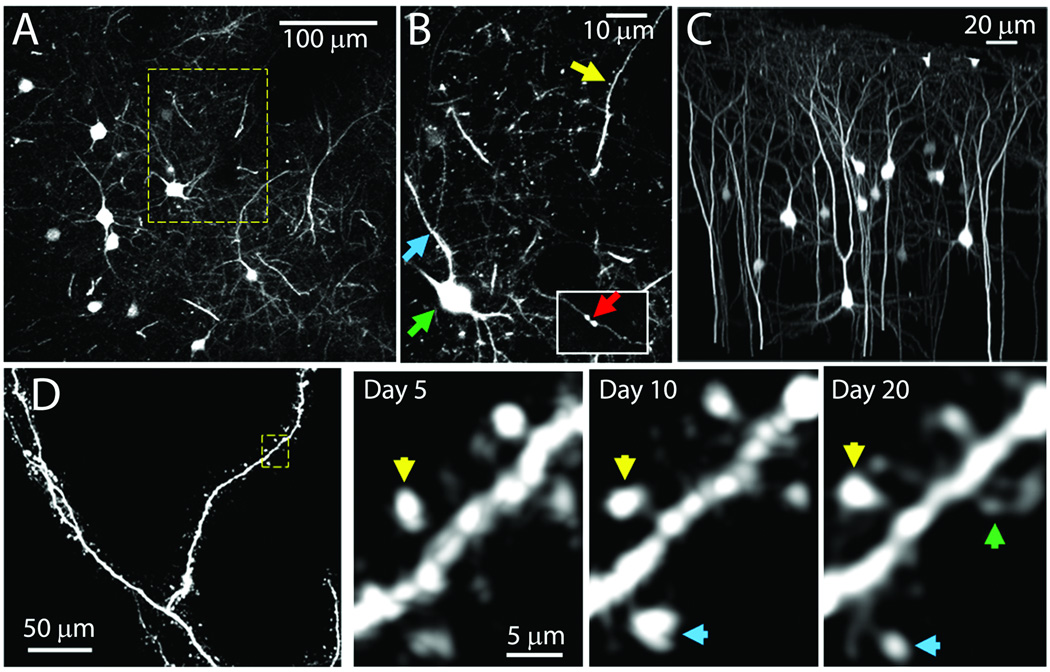
We recognized that the distributed transduction pattern of our low titer injections resembled the sparse labeling seen in the Thy1-GFP transgenic M and S mouse lines that have been widely used for imaging dendritic processes in vivo (Holtmaat et al., 2009; Holtmaat & Svoboda, 2009). We wondered if viral transgenesis might allow live imaging of neurons in the intact brain as had these two Thy1 transgenic lines. However, the low viral titers needed to replicate the sparse labeling of the Thy1-GFP mice also resulted in low levels of transgene expression that were below the threshold needed for visualization by two-photon in vivo microscopy. To increase the intensity of fluorescent labeling, we designed an AAV viral vector containing three copies of the YFP coding sequence connected by 2a sequences. In vivo imaging 4 weeks after P0 injection demonstrated that all major anatomical features of cortical pyramidal neurons could be readily resolved in AAV8-triple-YFP infected cells (Fig. 11). Cell bodies, apical and basal dendrites, axons, and even individual spines were visible in our preparations (Fig. 11A, B, and C). In many cases, apical dendrites could be traced all the way to their origin in cortical layer 5 (500–600 µm depth). An important advantage of this labeling technique compared to the Thy1-GFP mice is the relatively large number of labeled pyramidal cells in L2/3. Labeled L2/3 pyramids could be imaged in their entirety (Fig. 11D), allowing in vivo comparisons of apical (the primary recipients of feedback inputs) and basal (the primary recipients of feed-forward inputs) dendritic arbors, which has not yet been possible in the Thy1-GFP lines (Holtmaat et al., 2009). These data, along with the finding that fluorescence endures for more than 12 months in injected mice, indicate that P0 injection with AAV-triple-YFP provides an efficient method for labeling the processes of cortical pyramidal neurons for chronic in vivo two-photon imaging.
Figure 11. P0 injection of AAV8-triple-YFP produces fluorescent neuronal labeling suitable for in vivo two-photon imaging.
All images were taken from a 4-week old mouse injected at P0 with AAV8-triple-YFP (1.0×108 particles/hemisphere). (A) Z-projection of imaging volume from 70 to 85 µm cortical depth (seven 2-µm-thick optical slices). (B) Higher-magnification image of boxed section shown in panel A demonstrating labeled cell processes (three 1-µm-thick optical slices). Neuronal cell bodies (green arrow), apical dendrites (yellow arrow), basal dendrites (blue arrow), and axonal boutons (inset - red arrow) are all visible. (C) Three-dimensional projection of a 250 × 250 × 300 µm volume demonstrating the diversity of pyramidal neuron morphologies resolvable in P0-injected mice, with background fluorescence digitally masked in Imaris Bitplane. (D) Chronic time-lapse imaging of AAV8-triple-YFP-labeled apical dendrites, imaged at 5, 10, and 20 days post-surgery. Left panel: Z-projection of a chronically-imaged dendrite (0.3 µm-per-pixel in X–Y plane, 40 1-µm-thick optical slices). Right panel: Magnified view of a single optical section from the region boxed in panel D, imaged over subsequent days. New spines (green arrows), stable spines (yellow arrows), and changes in spine morphology (blue) are visible. Nonspecific background fluorescence was removed in C and D.
Sparse fluorescent labeling of Purkinje neurons offers a window into cerebellar development and plasticity
In addition to transducing cortical layers that are not labeled in the Thy1-XFP transgenic lines, neonatal viral injection also reaches areas of the brain that aren’t visible in the Thy1 mice. Specifically, as shown in Figures 2, 3, 4, and 5, viral transgenesis strongly labels cerebellar Purkinje neurons in both juvenile and adult. Moreover, viral expression begins within days after injection, at a time when Purkinje neurons are just beginning to form their mature dendritic arbors. Compared with cortical neurons, few tools exist to sparsely label or genetically manipulate Purkinje neurons. The natural tropism of several AAV serotypes for these cells might offer an easy way to overcome this limitation. We injected AAV8-triple-YFP (109 particles/hemisphere) or AAV1-YFP (1010 particles/hemisphere) at P0 and harvested pups 2, 4, 7, and 14 days later (Fig. 12). While arborization is still immature, individual cells can be easily identified at these dilutions. The selection, extension, and elaboration of dendritic processes can be followed from shortly after birth when multiple small neurites are present until a single dendrite develops into its final shape weeks later. With further dilution of the virus, even mature Purkinje cells could be fully identified. Sagittal sections from mice injected with low-titer AAV8-triple-YFP (between 1.0×108 and 4.0×108 particles/hemisphere) displayed many well-isolated Purkinje cells, allowing the visualization of complete dendritic arbors (Fig. 12 Bii) down to the level of individual dendritic spines (Biii) in labeled cells (Video 1).
Figure 12. Neonatal AAV transgenesis offers a new window into Purkinje cell development and in vivo dynamics.
(A) High magnification images of sagittal sections through the cerebellum following P0 intraventricular injection with 2 µl of AAV8-triple-YFP (3.8×109 particles/hemisphere, top row) or AAV1-YFP (1.3×1010 particles/hemisphere, second row). Sparse fluorescent labeling provides a clear view of postnatal dendritic maturation from P2 until P14. (Bi) Low magnification view of a fixed sagittal section through the cerebellum. The images were taken from from an 8-week old mouse injected at P0 with AAV8-triple-YFP (1.0×108 particles/hemisphere) demonstrating sparse labeling of Purkinje cells. The surface of the brain is outlined and the border of the granule cell layer is dashed to provide structural landmarks. (Bii) High magnification projection of the Purkinje cell outlined in panel (Bi). (Biii) Magnified view of a single optical section resolves individual spines on a terminal dendrite. (C) Z-projections of a cerebellar Purkinje cell imaged in vivo by 2-photon microscopy. Top panel: Z-projection of the volume in the imaged orientation from 0 to 200 µm depth (220×60 pixels, 133×35 µm FOV). Bottom panel: Orthogonal view of the image stack displays the full dendritic arbor.
Based on our previous success imaging virally-labeled cortical neurons in vivo, and recognizing that the same sparse bright expression that made this possible in the cortex was present in the cerebellum, we tested whether Purkinje cell dendritic arbors could also be imaged in situ through a cranial window over the cerebellum of a P0-injected mouse. Remarkably, Purkinje cell dendritic arbors could be imaged in great detail by 2-photon microscopy and reconstructed in three dimensions from the image stack, despite the fact that cells were imaged from above with limited resolution in the Z-axis by this technique (Fig. 12C and Video 2). With practice, it should be possible to place the cranial window at an angle that offers even better resolution of the dendritic processes, and with it the potential for chronic imaging of these complex cells in vivo.
Discussion
We present neonatal intraventricular viral injection as an efficient and rapid method to genetically manipulate the rodent brain. We have optimized the intrinsic mosaic transduction pattern produced by this method to allow expression of multiple transgenes at any desired density and to readily identify the genetically modified cells by co-expressed fluorescent proteins. In the course of our study, we discovered that the timing of injection, the serotype selected for packaging, and the promoter chosen for expression each influence the pattern and cell types transduced.
Neonatal viral transduction has several advantages over other approaches commonly used for gene delivery to the CNS, such as germ-line transgenesis (Guo et al., 2002; Chakravarthy et al., 2008; Rotolo et al., 2008; Young et al., 2008) Zong et al., 2005; Lao et al., 2012), in utero and postnatal electroporation (Saito & Nakatsuji, 2001; Boutin et al., 2008; Chesler et al., 2008; LoTurco et al., 2009; De Vry et al., 2010), and in utero, intravenous, and adult stereotaxic viral injection (Hashimoto & Mikoshiba, 2003; 2004; Shen et al., 2004; Stott & Kirik, 2006; Rahim et al., 2009; Rahim et al., 2011). First, neonatal intraventricular injections are relatively easy to learn and implement compared to other methods. It takes only minutes to perform and can be done using inexpensive tools and cryoanesthesia. Second, the technique can be used either alone or in addition to other germ-line genetic manipulations, and generates animals with widespread transgene expression. Third, the procedure appears to cause little long-term damage to the brain; animals injected at P0 have normal neuroanatomy as adults. Most importantly, the speed and flexibility of AAV-based gene delivery affords ready access to a growing number of genetic tools for manipulating the nervous system (Arenkiel & Ehlers, 2009), including calcium indicators (Tian et al., 2009; Dombeck et al., 2010), light-activated channels (Banghart et al., 2004; Zhang et al., 2007), and shRNA libraries (Xia et al., 2004).
AAV may be unique in producing widespread transduction following intraventricular delivery. The pattern of transduction suggests that the virus follows the flow of the CSF through the subarachnoid space (Passini & Wolfe, 2001). At just 20–25 nm in diameter, the small size of AAV particles may facilitate their dissemination throughout the brain. In contrast, at 100+ nm in diameter, lentivirus injected at the same age transduced only the ventricular surface and choroid plexus (Watson et al., 2005). Although not yet empirically tested, the still larger herpes simplex virus (180–200 nm) might also be expected to show little transduction outside the ventricle. Size is clearly not the only factor influencing viral spread, as unlike AAV1, 2, 6, 8, and 9 (our data and (Passini & Wolfe, 2001; Passini et al., 2003; Broekman et al., 2006; Cearley et al., 2008), AAV5 transduction does not advance much beyond the injection site (Watson et al., 2005). The distribution of cellular receptors and their affinity for different AAV serotypes may also contribute to viral spread. AAV5, and to a lesser extent, AAV1 (Figure 6), appear to bind strongly at the ventricular surface, leaving fewer particles to enter the parenchyma.
Because of their varying receptor affinities, viral transgenesis also opens the possibility of harnessing serotype specificity to target distinct cellular populations. We demonstrate that AAV1 favors superficial layers of cortex, while AAV8 transfects more evenly across layers. AAV6 offers improved transduction of cerebellar Purkinje neurons, but works less well in the forebrain. Past work on neonatal AAV transduction has shown that serotype strongly biases which brain regions and cell types are targeted, with select capsid proteins preferring inhibitory neurons, astrocytes, or oligodendrocytes (Broekman et al., 2006); (Cearley et al., 2008; Nathanson et al., 2009). Although the precise mechanism of AAV transduction is not well understood, receptors for several serotypes have been identified, including the 37/67 kD laminin receptor (AAV8), platelet-derived growth factor receptor (AAV5), αVβ5 integrin (AAV2), hepatocyte growth factor receptor (AAV2), and fibroblast growth factor receptor (AAV2 and 3; reviewed in Akache et al. (Akache et al., 2006)). Specific sialic acid and heparan sulfate linkages also contribute to AAV tropism, and binding of several serotypes can be eliminated by enzymatic deglycosylation of cultured cells (AAV2–5). With over 100 AAV variants isolated to date, the repertoire of possible transduction patterns has yet to be fully exploited (Wu et al., 2006), and rational engineering of AAV glycoproteins and their cell-surface receptors promises even greater control in future (Wang et al., 2011).
Tapping into these natural serotype biases, we demonstrate that mosaic expression of multiple transgenes can be achieved in either overlapping or discrete cell populations by injecting two viruses together. Co-injection of the same serotype resulted in a high degree of co-infection. Conversely, different serotypes transduced largely non-overlapping populations. These natural preferences offer the possibility of expressing different transgenes pre- and post-synaptically. Luo et al. achieved a similar outcome with a Cre-based system that randomly excised one of two stop cassettes to differentially label neurons with red or yellow fluorescence (Zong et al., 2005). Our system now allows this dual mosaic labeling in wild-type mice (or used in addition to germ-line manipulations), and offers the flexibility to independently control the density of both labels.
The ability to express polycistronic transcripts from a single viral promoter also makes it easy to design AAV vectors that both genetically modify and fluorescently label the transduced cells, as we show through reliable co-expression of tTA or Cre with YFP or tdTomato. This method allows genetically manipulated and wild-type cells to be distinguished for morphological analysis, electrophysiological studies, or even fluorescence activated cell sorting (FACS) (Lobo, 2009; Yang et al., 2011). Although AAV has a relatively small packaging limit compared to other viral vectors (Natkunarajah et al., 2008; Karra & Dahm, 2010), the construct we used allowed 2.3 kb of cDNA to be inserted in addition to the 716 bp YFP coding sequence, 937 bp chick β-actin promoter and 600 bp post transcriptional regulatory element (WPRE). In theory, proteins up to 800 amino acids long could be incorporated into the construct and still allow fluorescent labeling of transgenic cells. Smaller promoters like synapsin-1 would further increase capacity (Shevtsova et al., 2005).
In addition to the size barrier, another perceived disadvantage of AAV particularly for developmental studies was the reported delay between injection and expression. Past work suggested that AAV-encoded fluorescent proteins could take up to a week to appear, with peak expression several weeks after onset (Sarra et al., 2002; McCarty et al., 2003; Natkunarajah et al., 2008). In contrast, we show that functional Cre recombinase was present within 2 days of injection, and by P7 the distribution of fluorescent reporter was similar to the adult. This timing is better aligned with the 24 hr onset reported by Pilpel et al following neonatal injection of AAV8 encoding a fluorescent label under control of the human synapsin promoter (Pilpel et al., 2009). Although in utero injections are still needed to manipulate embryonic development, the rapid onset of AAV expression makes neonatal injection an attractive alternative for postnatal studies.
Finally, we demonstrate that neonatal injection can be used to label neurons sparsely and brightly enough for in vivo two-photon imaging of neuronal morphology. Using a polycistronic cassette to triple expression of YFP, we attained fluorescence strong enough to observe detailed microstructure and stable enough to allow chronic imaging of dendritic structural plasticity over several months (M. Jiang and J.-Y. Kim, unpublished data). Past studies have used AAV-GFP virus for in vivo imaging following stereotaxic injection into mice and monkeys (Stettler et al., 2006; Lowery et al., 2009). Local injection has the benefit of eliminating background fluorescence from distant projection neurons, but at the cost of having less control over the density of labeled cells due to a sharp gradient in transduction from the site of injection. Neonatal transduction provides improved consistency in the expression pattern, and offers a serviceable alternative to Thy1-XFP lines (Feng et al., 2000), particularly when working with models that already require multiple transgenes or modified alleles. Viral transgenesis also provides access to neurons not labeled in the Thy1-XFP mice, notably Purkinje cells of the cerebellum, which in the past have required acute injection of synthetic dyes for morphological study in vivo (Gobel & Helmchen, 2007). Given the high plasticity of cerebellar circuitry and the progressive but poorly understood degeneration of Purkinje neurons in many inherited ataxias (Boyden et al., 2004; Carlson et al., 2009), chronic in vivo imaging of these arbors during motor learning and disease will likely grant new insight into cerebellar function and dysfunction. Combined with the potential to genetically manipulate the labeled neurons, neonatal viral transduction opens the possibility for experiments probing the relationship between targeted proteins, dendritic morphology, and neuronal function within single cells of the intact brain (O'Connor et al., 2009).
Although this technique has many advantages over past methods, several limitations should be noted. First, as mentioned above, the small packaging size of AAV limits the length and number of transgenes that can be co-expressed. In some situations this can be overcome by trans-splicing of co-injected viruses, but this may not be possible in every setting (Lai et al., 2005; Ghosh & Duan, 2007). Second, widespread transduction may not be ideal when more restricted expression is needed. Where available, spatial or cell-type specificity could be attained using Cre-dependent flex-signal viruses (Atasoy et al., 2008) with Cre-expressing transgenic lines (e.g., nagy.mshri.on.ca/). In other cases, selectivity might be achieved using an intersectional strategy of complementary elements introduced on co-injected viruses (Dymecki et al., 2010; Haubensak et al., 2010; Fujimoto et al., 2011). Third, the level of viral gene expression varies between cells due to differences in multiplicity of infection inherent in viral transgenesis. This fluctuation may complicate some studies of neuronal function, but may be lessened at extremes of high and low titers where infection can be maximized or dilution-limited to a single particle. Fourth, viral transduction may not target all neuronal subtypes. While the serotypes and promoters we tested expressed strongly in cortical pyramidal neurons, cerebellar Purkinje cells, olfactory granule neurons, and striatal interneurons, they produced very little expression in cortical interneurons and granule neurons of the dentate gyrus and cerebellum. Expression in these cell types might be attained using different serotypes and promoters, but must be tested empirically. Finally, there is a strict temporal window during which this technique can be used. Injections must be done within the first 12–24 hours after birth for AAV1, and within the first few days for AAV8. The timing of AAV injection may also limit which cell types can be transduced, as several neuronal populations are generated after birth. At the back end, however, expression of viral transgenes can be readily delayed using temporal control elements such as CreER and TTA.
By optimizing its natural mosaic transduction pattern, we discovered that neonatal viral transgenesis opens a wide range of experimental opportunities that were not possible with existing methods. Cell-autonomous and –extrinsic effects can now be readily distinguished. Purkinje neurons can now be easily manipulated and imaged in vivo. New constructs can be rapidly screened without germline transgenesis. The final advantage of the approach is the rising availability of compatible off-the-shelf viral preparations (e.g., Penn Vector Core and UNC Gene Therapy Center) and vectors (e.g., Addgene) that can be custom packaged into a variety of serotypes. These resources for viral manipulation complement a growing community of mouse repositories where newly characterized mutant strains can be purchased online (e.g., Jackson Laboratories, MMRRC, GENSAT, EMMA). As both the pattern and expression level of viral-delivered transgenes can depend on a number of factors including the transgene itself, the construct design (i.e., promoters and enhancers), the capsid serotype, the quality of the viral preparation, and the viral titer, each new application will require some optimization. However, the richness of viral manipulation and the rate at which it has recently advanced suggests that with additional experimentation a wide range of cell type specificities and novel applications are within reach.
Supplementary Material
Images were acquired at high resolution (0.16 µm per pixel in X–Y plane) in 0.5 µm optical slices with a 169×224×15 µm field of view.
Z-series of images acquired from the cerebellum by in vivo 2-photon microscopy were imported into NeuroLucida and used to reconstruct the complete dendritic arborization of this YFP-labeled Purkinje cell.
Acknowledgements
We thank Kazuhiro Oka and the Baylor College of Medicine Viral Vector Core for AAV production, Anna Gumpel, Carolyn Allen, Yuanyuan Zhang, and Bryan Song for mouse care, Bernard Lee and Bernard Kuecking from Zeiss for microscope support, Ben Arenkiel for sharing the EF1α-iCre-2A-tdTomato AAV vector, and Roy Sillitoe and Ben Arenkiel and for helpful comments on the manuscript. Grant support: American Health Assistance Foundation Alzheimer’s Disease Research Grant A2010097, National Institute of Aging R21 AG038856, and National Institutes of Health Office of the Director New Innovator Award DP2 OD001734.
Abbreviations
- AAV
adeno-associated virus
- CBA
chicken beta-actin
- EF1α
elongation factor 1α
- GFP
green fluorescent protein
- ITR
inverted terminal repeat
- p0
postnatal day 0
- tdTomato
tandem dimer Tomato
- TTA
tetracycline-responsive transactivator
- YFP
yellow fluorescent protein
Footnotes
Conflict of Interest: None
References
- Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenkiel BR, Ehlers MD. Molecular genetics and imaging technologies for circuit-based neuroanatomy. Nature. 2009;461:900–907. doi: 10.1038/nature08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin C, Diestel S, Desoeuvre A, Tiveron MC, Cremer H. Efficient in vivo electroporation of the postnatal rodent forebrain. PloS one. 2008;3:e1883. doi: 10.1371/journal.pone.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Katoh A, Raymond JL. Cerebellum-dependent learning: the role of multiple plasticity mechanisms. Annu Rev Neurosci. 2004;27:581–609. doi: 10.1146/annurev.neuro.27.070203.144238. [DOI] [PubMed] [Google Scholar]
- Broekman ML, Comer LA, Hyman BT, Sena-Esteves M. Adeno-associated virus vectors serotyped with AAV8 capsid are more efficient than AAV-1 or-2 serotypes for widespread gene delivery to the neonatal mouse brain. Neuroscience. 2006;138:501–510. doi: 10.1016/j.neuroscience.2005.11.057. [DOI] [PubMed] [Google Scholar]
- Carlson KM, Andresen JM, Orr HT. Emerging pathogenic pathways in the spinocerebellar ataxias. Current opinion in genetics & development. 2009;19:247–253. doi: 10.1016/j.gde.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearley CN, Vandenberghe LH, Parente MK, Carnish ER, Wilson JM, Wolfe JH. Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:1710–1718. doi: 10.1038/mt.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P, Jansen-West K, Beccard A, Ceballos-Diaz C, Levites Y, Verbeeck C, Zubair AC, Dickson D, Golde TE, Das P. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: evidence against inflammation as a driving force for amyloid deposition. FASEB J. 2010;24:548–559. doi: 10.1096/fj.09-141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Keck T, Roelandse M, Hartman R, Jeromin A, Perry S, Hofer SB, Mrsic-Flogel T, Levelt CN. Cre-dependent expression of multiple transgenes in isolated neurons of the adult forebrain. PloS one. 2008;3:e3059. doi: 10.1371/journal.pone.0003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler AT, Le Pichon CE, Brann JH, Araneda RC, Zou DJ, Firestein S. Selective gene expression by postnatal electroporation during olfactory interneuron nurogenesis. PloS one. 2008;3:e1517. doi: 10.1371/journal.pone.0001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A, Soriano P. Ephrin signaling in vivo: look both ways. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232:1–10. doi: 10.1002/dvdy.20200. [DOI] [PubMed] [Google Scholar]
- De Vry J, Martinez-Martinez P, Losen M, Temel Y, Steckler T, Steinbusch HW, De Baets MH, Prickaerts J. In vivo electroporation of the central nervous system: a non-viral approach for targeted gene delivery. Progress in neurobiology. 2010;92:227–244. doi: 10.1016/j.pneurobio.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Espinosa JS, Wheeler DG, Tsien RW, Luo L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron. 2009;62:205–217. doi: 10.1016/j.neuron.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fujimoto E, Gaynes B, Brimley CJ, Chien CB, Bonkowsky JL. Gal80 intersectional regulation of cell-type specific expression in vertebrates. Developmental dynamics : an official publication of the American Association of Anatomists. 2011;240:2324–2334. doi: 10.1002/dvdy.22734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Duan D. Expanding adeno-associated viral vector capacity: a tale of two vectors. Biotechnology & genetic engineering reviews. 2007;24:165–177. doi: 10.1080/02648725.2007.10648098. [DOI] [PubMed] [Google Scholar]
- Gobel W, Helmchen F. New angles on neuronal dendrites in vivo. J Neurophysiol. 2007;98:3770–3779. doi: 10.1152/jn.00850.2007. [DOI] [PubMed] [Google Scholar]
- Guo C, Yang W, Lobe CG. A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis. 2002;32:8–18. doi: 10.1002/gene.10021. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Mikoshiba K. Mediolateral compartmentalization of the cerebellum is determined on the"birth date" of Purkinje cells. J Neurosci. 2003;23:11342–11351. doi: 10.1523/JNEUROSCI.23-36-11342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Mikoshiba K. Neuronal birthdate-specific gene transfer with adenoviral vectors. J Neurosci. 2004;24:286–296. doi: 10.1523/JNEUROSCI.2529-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, Fanselow MS, Luthi A, Anderson DJ. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, Luo L. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hubener M, Keck T, Knott G, Lee WC, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci. 2010;33:409–440. doi: 10.1146/annurev.neuro.051508.135722. [DOI] [PubMed] [Google Scholar]
- Karra D, Dahm R. Transfection techniques for neuronal cells. J Neurosci. 2010;30:6171–6177. doi: 10.1523/JNEUROSCI.0183-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Miller VM, Levites Y, West KJ, Zwizinski CW, Moore BD, Troendle FJ, Bann M, Verbeeck C, Price RW, Smithson L, Sonoda L, Wagg K, Rangachari V, Zou F, Younkin SG, Graff-Radford N, Dickson D, Rosenberry T, Golde TE. BRI2 (ITM2b) inhibits Abeta deposition in vivo. J Neurosci. 2008;28:6030–6036. doi: 10.1523/JNEUROSCI.0891-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Yue Y, Liu M, Ghosh A, Engelhardt JF, Chamberlain JS, Duan D. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol. 2005;23:1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Z, Raju GP, Bai CB, Joyner AL. MASTR: A Technique for Mosaic Mutant Analysis with Spatial and Temporal Control of Recombination Using Conditional Floxed Alleles in Mice. Cell reports. 2012;2:386–396. doi: 10.1016/j.celrep.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK. Molecular profiling of striatonigral and striatopallidal medium spiny neurons past, present, and future. International review of neurobiology. 2009;89:1–35. doi: 10.1016/S0074-7742(09)89001-6. [DOI] [PubMed] [Google Scholar]
- LoTurco J, Manent JB, Sidiqi F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i120–i125. doi: 10.1093/cercor/bhp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery RL, Zhang Y, Kelly EA, Lamantia CE, Harvey BK, Majewska AK. Rapid, long-term labeling of cells in the developing and adult rodent visual cortex using double-stranded adeno-associated viral vectors. Developmental neurobiology. 2009;69:674–688. doi: 10.1002/dneu.20735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3'-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci U S A. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene therapy. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natkunarajah M, Trittibach P, McIntosh J, Duran Y, Barker SE, Smith AJ, Nathwani AC, Ali RR. Assessment of ocular transduction using single-stranded and self-complementary recombinant adeno-associated virus serotype 2/8. Gene therapy. 2008;15:463–467. doi: 10.1038/sj.gt.3303074. [DOI] [PubMed] [Google Scholar]
- O'Connor DH, Huber D, Svoboda K. Reverse engineering the mouse brain. Nature. 2009;461:923–929. doi: 10.1038/nature08539. [DOI] [PubMed] [Google Scholar]
- Park KM, Bowers WJ. Tumor necrosis factor-alpha mediated signaling in neuronal homeostasis and dysfunction. Cellular signalling. 2010;22:977–983. doi: 10.1016/j.cellsig.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Watson DJ, Vite CH, Landsburg DJ, Feigenbaum AL, Wolfe JH. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of beta-glucuronidase-deficient mice. J Virol. 2003;77:7034–7040. doi: 10.1128/JVI.77.12.7034-7040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Wolfe JH. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol. 2001;75:12382–12392. doi: 10.1128/JVI.75.24.12382-12392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel N, Landeck N, Klugmann M, Seeburg PH, Schwarz MK. Rapid, reproducible transduction of select forebrain regions by targeted recombinant virus injection into the neonatal mouse brain. Journal of neuroscience methods. 2009;182:55–63. doi: 10.1016/j.jneumeth.2009.05.020. [DOI] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Ahmadi S, Hoefer K, Buckley SM, Hughes DA, Nathwani AN, Baker AH, McVey JH, Cooper JD, Waddington SN. In utero administration of Ad5 and AAV pseudotypes to the fetal brain leads to efficient, widespread and long-term gene expression. Gene therapy. 2011 doi: 10.1038/gt.2011.157. [DOI] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Howe SJ, Buckley SM, Acosta-Saltos AD, Elston KE, Ward NJ, Philpott NJ, Cooper JD, Anderson PN, Waddington SN, Thrasher AJ, Raivich G. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene therapy. 2009;16:509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- Rotolo T, Smallwood PM, Williams J, Nathans J. Genetically-directed, cell type-specific sparse labeling for the analysis of neuronal morphology. PloS one. 2008;3:e4099. doi: 10.1371/journal.pone.0004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Developmental biology. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Sarra GM, Stephens C, Schlichtenbrede FC, Bainbridge JW, Thrasher AJ, Luthert PJ, Ali RR. Kinetics of transgene expression in mouse retina following sub-retinal injection of recombinant adeno-associated virus. Vision research. 2002;42:541–549. doi: 10.1016/s0042-6989(01)00230-9. [DOI] [PubMed] [Google Scholar]
- Shen JS, Meng XL, Maeda H, Ohashi T, Eto Y. Widespread gene transduction to the central nervous system by adenovirus in utero: implication for prenatal gene therapy to brain involvement of lysosomal storage disease. The journal of gene medicine. 2004;6:1206–1215. doi: 10.1002/jgm.630. [DOI] [PubMed] [Google Scholar]
- Shevtsova Z, Malik JM, Michel U, Bahr M, Kugler S. Promoters and serotypes: targeting of adeno-associated virus vectors for gene transfer in the rat central nervous system in vitro and in vivo. Experimental physiology. 2005;90:53–59. doi: 10.1113/expphysiol.2004.028159. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stettler DD, Yamahachi H, Li W, Denk W, Gilbert CD. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron. 2006;49:877–887. doi: 10.1016/j.neuron.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Stott SR, Kirik D. Targeted in utero delivery of a retroviral vector for gene transfer in the rodent brain. Eur J Neurosci. 2006;24:1897–1906. doi: 10.1111/j.1460-9568.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Faust SM, Rabinowitz JE. The next step in gene delivery: molecular engineering of adeno-associated virus serotypes. Journal of molecular and cellular cardiology. 2011;50:793–802. doi: 10.1016/j.yjmcc.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley interdisciplinary reviews. Systems biology and medicine. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- Watson DJ, Passini MA, Wolfe JH. Transduction of the choroid plexus and ependyma in neonatal mouse brain by vesicular stomatitis virus glycoprotein-pseudotyped lentivirus and adeno-associated virus type 5 vectors. Human gene therapy. 2005;16:49–56. doi: 10.1089/hum.2005.16.49. [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59:200–207. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008;11:721–728. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Images were acquired at high resolution (0.16 µm per pixel in X–Y plane) in 0.5 µm optical slices with a 169×224×15 µm field of view.
Z-series of images acquired from the cerebellum by in vivo 2-photon microscopy were imported into NeuroLucida and used to reconstruct the complete dendritic arborization of this YFP-labeled Purkinje cell.