Abstract
Myeloid ecotropic viral integration site 1 (Meis1) forms a heterodimer with Pbx1 that augments Hox-dependent gene expression and is associated with leukemogenesis and HSC self-renewal. Here we identified 2 independent actions of Meis1 in hematopoietic development: one regulating cellular proliferation and the other involved in megakaryocyte lineage development. First, we found that endogenous Mesp1 indirectly induces Meis1 and Meis2 in endothelial cells derived from embryonic stem cells. Overexpression of Meis1 and Meis2 greatly enhanced the formation of hematopoietic colonies from embryonic stem cells, with the exception of erythroid colonies, by maintaining hematopoietic progenitor cells in a state of proliferation. Second, overexpression of Meis1 repressed the development of early erythroid progenitors, acting in vivo at the megakaryocyte-erythroid progenitor stage to skew development away from erythroid generation and toward megakaryocyte development. This previously unrecognized action of Meis1 may explain the embryonic lethality observed in Meis1−/− mice that arises from failure of lymphatic-venous separation and can result as a consequence of defective platelet generation. These results show that Meis1 exerts 2 independent functions, with its role in proliferation of hematopoietic progenitors acting earlier in development from its influence on the fate choice at the megakaryocyte-erythroid progenitor between megakaryocytic and erythroid development.
Introduction
Meis1 was identified as a common site of proviral integration by the ecotropic virus in BXH-2 mice that promoted myeloid leukemias.1,2 Meis1 belongs to the TALE class of homeodomain transcription factors characterized by a 3-amino acid loop extension between the α-helices within its homeodomain. Meis1 interacts with other homeodomain proteins,3 in particular Pbx1,4 forming a heterodimer that recognizes DNA. The Meis1 protein contains a domain that recognizes wild-type Pbx proteins, but not chimeric Pbx1 proteins formed by translocations, such as the E2a-Pbx1 oncoprotein.5 The Meis1/Pbx dimer cooperatively associates with Hox homeodomain proteins, and in vitro interaction between Meis1, Hoxa9, and Pbx proteins can occur in the absence of DNA.6 In addition to its Pbx interaction motif,5 Meis1 also contains a carboxyl-terminal (C-terminal) region that is required for leukemia induction.7 This C-terminal region of Meis1 contains transcriptional activity regulated by protein kinase A that appears dependent on the coactivator of cAMP response element-binding protein.8 Thus, Meis1 appears to augment Hox transcription factor activity and can be regulated by extracellular signaling cues.
Initial analysis of Meis1 focused on its role in leukemic transformation. Acute myeloid leukemia induced by Hoxa9 was significantly accelerated by coexpression with Meis1, but not by coexpression with Pbx1b.9 A cellular action of Meis1 appeared to be the suppression of differentiation and the promotion of proliferation in a system of cytokine-driven Hoxa9-immortalized cells.10 A Hoxa9 chimeric fusion protein, NUP98-Hoxa9, independently induced a silent preleukemic phase of disease, which was accelerated by Meis1, suggesting that Meis1 augments the activities of the Hoxa9-dependent transformational event.11 Interactions between Meis1 and Hoxa9 also occur in a model of leukemia induced by rearrangements of the MLL/ALL1 gene, which represents approximately 20% of acute lymphoblastic leukemias and 5% to 6% of acute myeloid leukemias.12 In this setting, Meis1 is an essential, rate-limiting regulator of the development of MLL-dependent leukemias.12,13
Studies based on Meis1 overexpression initially suggested a role in regulation of proximodistal limb axis development.14 However, studies based on targeted disruption of Meis1 in mice observed more substantial defects in hematopoiesis, angiogenesis, and eye development.15–17 Complete elimination of Meis1 by gene targeting caused death between embryonic days 11.5 and 14.5.16 Although definitive myeloerythroid lineages are present in Meis1−/− embryos, the total numbers of colony-forming cells are significantly reduced. Similar defects were observed when Meis1 was targeted by a strategy that potentially generated a dominant negative protein, but in this case defects were also observed in the developing eye, with partially duplicated retinas and smaller lenses.15 This latter effect potentially could represent interference with the normal actions of Meis2, rather than Meis1, because Meis2 has been demonstrated to regulate the expression of Pax6, a pivotal regulator of eye development.18 Meis2 has been shown to maintain retinal progenitor cells in a state of rapid proliferation, at least in part through regulation of cell cycle machinery, including cyclin D1.19
Early embryonic lethality resulting from Meis1 deficiency was initially thought to result from hemorrhage secondary to vascular defects,15,16 but subsequent studies demonstrated that the absence of platelets in Meis1−/− embryos leads to a failure in separation of lymphatic vessels during embryonic angiogenesis,17 as platelets are critical in mediating separation of the venous and lymphatic systems.20,21 Further studies have recognized additional defects in heart development in Meis1−/− embryos.22
In summary, although Meis1 promotes leukemogenesis and normal hematopoiesis by modulating self-renewal of progenitor-like cells, the basis for this action as well as the defect in platelet development in Meis1−/− mice are still incompletely understood. In this study, we identified 2 distinct biologic actions of Meis1. We find that Meis1 promotes a proliferative state in hematopoietic progenitors, which may be related to the reported involvement of Meis1 in the development of leukemia. Second, we identify a previously unrecognized action of Meis1 at the megakaryocyte-erythroid progenitor (MEP) stage, in which Meis1 inhibits erythroid development and favors megakaryocyte development.
Methods
Mouse ES cell generation and EB differentiation
To generate inducible embryonic stem (ES) cell lines (A2lox.Mesp1, A2lox.Meis1, and A2lox.Meis2), individual cDNAs were amplified from embryoid body (EB) RNA using gene-specific primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and cloned into the p2lox targeting vector.23 Site specific recombination into A2lox ESCs was performed using cotransfected Cre recombinase, and all A2lox ESC lines were maintained as described.24 For differentiation, ESCs were plated in suspension in Petri dishes at 1.5 × 104 cells/mL in IMDM with 10% FCS, nonessential amino acids, L-glutamine, Na Pyruvate, penicillin-streptomycin, and 2-mercaptoethanol as described,24 and supplemented where indicated with SC-51322 (Enzo Life Sciences). Gene expression was induced by addition of doxycycline (250-500 ng/mL).
ES/OP9 coculture
Day 6 EBs were trypsinized to single-cell suspensions and plated on a monolayer of irradiated OP9-GFP cells25 at a density of 100 000 cells/mL in IMDM with 10% FCS, nonessential amino acids, L-glutamine, Na pyruvate, penicillin-streptomycin, 2-mercaptoethanol, and cytokines (100 ng/mL recombinant murine [rm] SCF, 40 ng/mL rm thrombopoietin [TPO], 40 ng/mL rm vascular endothelial growth factor, 5% Flt3 ligand conditioned medium, 10 ng/mL rm IL-3, and 20 ng/mL rm IL-6). All cytokines are from PeproTech.
FACS analysis and sorting
EBs or ES/OP9 cocultures dissociated by trypsin, or blood cells collected from mouse BM, spleen, or peripheral blood, were treated with Fc block (BD Biosciences PharMingen) on ice for 5 minutes and then stained with antibodies. Primary antibodies: PE α-Tie2 (1 μg/mL, TEK4), peridinin chlorophyll protein (PerCp)–Cy5.5 α-CD16/32 (1 μg/mL, 93), PE-Cy7 α-CD41 (1 μg/mL, eBioMWReg30), allophycocyanin (APC) α-Flk1 (1 μg/mL, Avas12a1), APC α-CD150 (1 μg/mL, mShad150), APC α-AA4.1 (1 μg/mL, AA4.1), APC α-CD42d (1 μg/mL, 1C2), APC-eFluor 780 α-c-kit (1 μg/mL, 2B8), APC-eFluor 780 α-CD45.2 (1 μg/mL, 104), eFluor 450 α-B220 (1 μg/mL, RA3-6B2), and eFluor 450 α-CD105 (1 μg/mL, MJ7/18; eBioscience), PE α-CD71 (1 μg/mL, C2), PE-Cy7 α-Sca1 (1 μg/mL, D7), PE-Cy7 α-Mac1 (1 μg/mL, M1/70), APC α-Gr1 (1 μg/mL, RB6-8C5; BD Biosciences PharMingen), V500 α-B220 (1 μg/mL, RA3-6B2, BD Hrizon), PE or APC α-hCD4 (1 μg/mL, Invitrogen). Data were acquired on a FACSCanto II (BD Biosciences) and analyzed using FlowJo Version 7.6.5 software (TreeStar). A2lox.Mesp1 cells were sorted based on Flk1 and Tie2 expression using a MoFlo cytometer (Dako North America). A2lox.Meis1 or A2lox.Meis2 cells were sorted on the FACSAria II (BD Biosciences) based on levels of CD41 expression.
Colony-forming assay
Day 6 EBs were dissociated by trypsin and added to MethoCult GF M3434 methylcellulose-based medium (StemCell Technologies) for hematopoietic colonies. Methylcellulose suspension cultures were supplemented with doxycycline where indicated. All colonies were counted on day 6 of methylcellulose culture. Megakaryocyte progenitors (MkPs) were examined using MegaCult-C collagen-based medium supplemented with 50 ng/mL rm TPO, 10 ng/mL rm IL-3, 20 ng/mL rm IL-6, and 50 ng/mL rm IL-11, following the protocol provided by the manufacturer (StemCell Technologies). Megakaryocytic colony formation was assessed after growth for 6 days by dehydrating, fixating, and staining the slides with acetylthiocholine iodide (Sigma-Aldrich) and Harris hematoxylin (Sigma-Aldrich) counterstain. The acetylthiocholine iodide-stained colonies were counted. Bright-field images were captured by a Nikon Eclipse TS100 microscope with an Optronics 60800 camera and imported into MagnaFire Version 2.0 software.
BrdU labeling
BrdU was added to ES/OP9 cocultures 3 hours before harvesting the cells for FACS analysis. The percentage of BrdU+ cells among CD41+ cells was assessed by cell-surface marker staining followed by cell permeabilization and APC α-BrdU antibody staining (BD Biosciences PharMingen).
Apoptosis assay
To analyze the degree of apoptosis of CD41+ cells, ES/OP9 cocultures were disaggregated and stained with CD41 antibody on ice for 30 minutes. After one wash in 1 × Binding Buffer (BD Biosciences), per 1 × 105 cells were stained with 5 μL PE–annexin V (BD Biosciences PharMingen) and 5 μL cell viability dye 7-amino-actinomycin D (7-AAD) for 15 minutes at room temperature. The proportion of cells in different quadrants was determined by FACS.
Retrovirus production and cell infection
Meis1a, Meis1b, and Meis2(2b) were subcloned from p2lox constructs into IRES-human CD4 (hCD4)–retrovirus26 with EcoRI. BM cells were collected from 8- to 10-week-old 129S6/SvEvTac mice (Taconic Farms), and c-kit+ progenitor cells were enriched with CD117 MicroBeads as described by the manufacturer (Miltenyi Biotec). c-kit+ BM progenitors were expanded in IMDM with 10% FCS, 100 ng/mL rm SCF, and 50 ng/mL rm TPO overnight, and then infected with retroviral supernatant in the presence of polybrene (2 μg/mL) by spin infection.
BM transplantation
Twenty-four hours after infection, c-kit–enriched BM progenitor cells were washed in 1 × PBS and transplanted by retro-orbital injection into 129S6/SvEvTac recipients that had been sublethally irradiated at 600 cGy.
Gene expression analysis
RNA from FACS-sorted cell populations was extracted using RNeasy kits (QIAGEN), and cDNA was synthesized using Superscripts III (Invitrogen). Quantitative RT-PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and a StepOne Plus Real Time PCR System (Applied Biosystems) with intron-spanning, gene-specific primers shown in supplemental Table 1. Large-scale gene expression analysis of A2lox.Mesp1 samples was done using Affymetrix MOE430_2.0 arrays as described.24 Data were normalized and modeled using DNA-Chip Analyzer/dChip. For A2lox.Meis1 and A2lox.Meis2 samples, gene expression analysis was done using Affymetrix Mouse Gene 1.0 ST arrays, and data were normalized and modeled using ArrayStar. The microarray datasets were deposited in the NCBI GEO database under accession numbers GSE34537 (A2lox.Mesp1 data), GSE34543 (A2lox.Meis1 data), and GSE34541 (A2lox.Meis2 data).
Statistical analysis
Paired Student t tests were performed to calculate P values and are indicated in the graphs.
Results
Identification of Meis1 and Meis2 as genes expressed by Mesp1-dependent endothelium
Previously, we reported that the transcription factor Mesp1 regulates the epithelial-mesenchymal transition and promotes the cell fates of cardiomyocytes, smooth muscle, and vascular endothelium from differentiating ES cells.24 Mesp1 also reduces the in vitro development of hematopoietic lineages from ES cells, although we did not establish whether this action occurred in vivo. Because HSCs were shown to derive from hemogenic endothelium27–29 and Mesp1 regulates development of endothelium, we wondered whether Mesp1 might influence gene expression related to hematopoietic development.
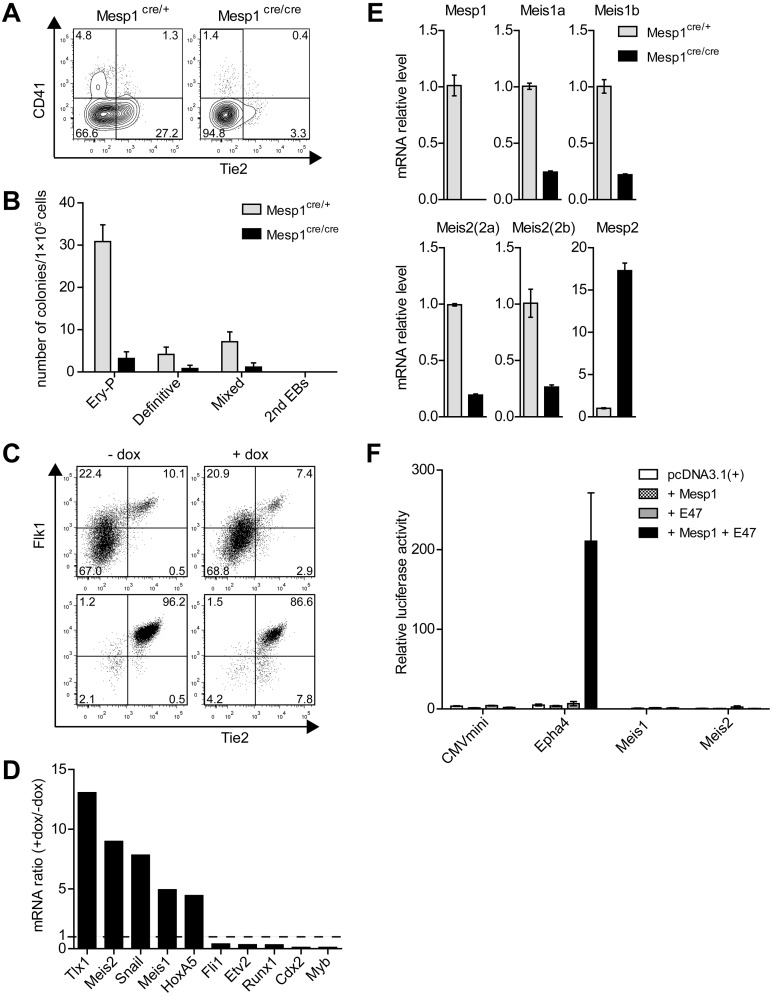
Previous fate mapping studies showed that Mesp1 is expressed in the precursors of the cardiovascular system, including endothelium, endocardium, myocardium, and epicardium,30 but did not carefully examine tracing of hematopoietic cells. We therefore carried out fate mapping using Cre recombinase expressed by the Mesp1 locus with a modified ROSA-GFP reporter locus (supplemental Figure 1A-B). Unexpectedly, we found that hematopoietic cells were efficiently labeled by Mesp1-Cre, with different efficiencies between individual animals ranging from 10% to as high as 99% of cells. Within a given animal, the same percentage of hematopoietic cells was labeled by ROSA-GFP across all hematopoietic lineages (supplemental Table 2). In particular, HSCs were labeled at the same frequency as multipotent progenitors, the MkPs, granulocyte-macrophage progenitors, and mature lineages derived from these progenitors. Although the basis for individual animal variation is still unclear, these results suggested that Mesp1 might be expressed in some fraction of hematopoietic progenitors, possibly hemogenic endothelium. Indeed, examination of Mesp1-deficient ES cells showed reduced numbers of developing Tie2+ endothelial cells and CD41+ hematopoietic progenitors (Figure 1A) as well as reduced hematopoietic development as assessed by methylcellulose assays (Figure 1B). Mesp1 lineage tracing previously identified that Mesp1 labels the endothelium of the dorsal aorta in the embryo. Because Mesp1 is not expressed in adult and mature hematopoietic lineages (supplemental Figure 1C), these results suggest a potential role of Mesp1 in optimal development of hemogenic endothelium.
Figure 1.
Mesp1 induces a subset of hematopoietic-associated transcription factors in ES cell–derived hemogenic endothelium. (A) ES cells derived from control heterozygous Mesp1cre/+ mice (Mesp1cre/+) or homozygous Mesp1-deficient Mesp1cre/cre mice (Mesp1cre/cre), as described previously,24 were differentiated as EBs for 6 days and analyzed by FACS. Shown are 2-parameter histograms for expression of CD41 and Tie2. Numbers indicate the percentage of cells in the indicated quadrant. (B) ES cells described in panel A were cultured as EBs for 6 days before plating in methylcellulose media with cytokines as described in “Methods.” Hematopoietic colonies were quantitated after 6 days of growth on methylcellulose based on morphologies. Data represent the average of 3 experiments. Error bars represent SD. (C) ES cells harboring a doxycyline (dox)–inducible Mesp1 gene (A2lox.Mesp1) were differentiated as EBs for 5 days in the absence (−) or presence (+) of dox from day 2 to day 4. Flk1+ Tie2+ cells composing between 5% and 10% of the population (Presort) were purified by cell sorting (Postsort). (D) Microarray analysis of transcription factors associated with hematopoietic development. Expression of the indicated genes is shown as a ratio of expression values by dox-treated endothelial cells relative to untreated cells. (E) Cells described in panel A were cultured as EBs for 5 days, and total RNA was isolated to detect the expression levels of the indicated genes by quantitative RT-PCR using primers described in supplemental Table 1. (F) The 293T cells were transfected with firefly luciferase reporter constructs containing a minimal CMV promoter (CMVmini), CMVmini with Epha4 enhancer (Epha4), or 1 kb upstream promoter/enhancer regions for Meis1 (Meis1) and Meis2 (Meis2). These were cotransfected along with expression vectors for Mesp1 and E47, either separately or together as indicated. Luciferase activity was normalized using cotransfected Renilla luciferase construct (prL-CMV). Shown is the normalized luciferase for the indicated constructs. Bars represent the SD of triplicate determinations.
To test this, we purified Flk1+ Tie2+ endothelium derived from ES cell differentiation as described24 (Figure 1C) and used microarray expression analysis to identify genes strongly induced by Mesp1 (Figure 1D). The transcription factor Snai1 was strongly induced by Mesp1, as we previously reported.24 Unexpectedly, Mesp1 also induced expression of Tlx1, required for spleen development,31 and both Meis1 associated with both hematopoietic and endothelial development15,16 and Meis2.19 However, Mesp1 did not globally induce genes associated with hematopoietic development because it inhibited expression of Fli1,32 Etv2,33 Runx1,34 Cdx2,35 and Myb36 (Figure 1D). We confirmed that Meis1 and Meis2 are indeed regulated by Mesp1 because both are reduced in expression in ES cells that are deficient for Mesp1 (Figure 1E). However, the regulation of Meis1 and Meis2 may be indirect because Mesp1 could be demonstrated to activate the Epha4 enhancer, but not the Meis1 or Meis2 promoter/enhancer regions (Figure 1F).
Meis1 and Meis2 promote hematopoietic colony formation from ES cells in vitro
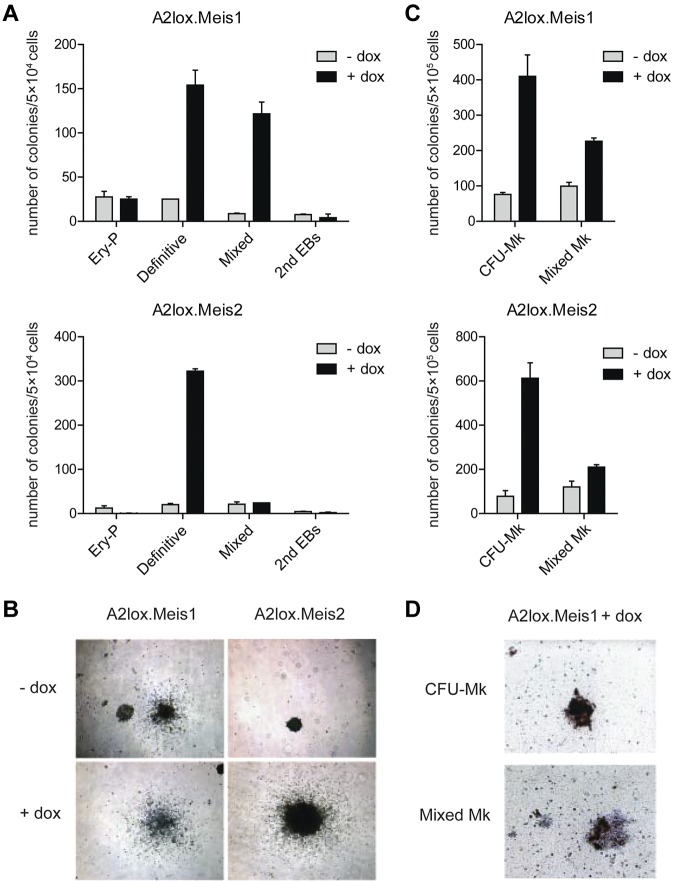
To determine the activity of these Mesp1-induced genes in differentiating endothelium, we generated ES cells with inducible Meis1 or Meis2 expression and examined their differentiation in vitro by colony formation assay (Figure 2). Induction of Meis1 caused a 5- to 6-fold increase in the numbers and size of definitive and mixed hematopoietic colonies (Figure 2A-B) but had little effect on primitive erythroid colonies. Meis2 also markedly increased numbers of definitive hematopoietic colonies (Figure 2A) and increased their size (Figure 2B). In collagen-based media containing TPO, induction of either Meis1 or Meis2 caused more than 4-fold increase in the numbers of pure megakaryocyte colonies and also increased the mixed megakaryocyte colonies (Figure 2C-D). To facilitate analysis of hematopoietic progenitors, we differentiated ES cells in liquid culture as EBs, and in coculture with OP9 cells,25 with or without cytokines (supplemental Figure 2A). We determined that doxycyline induced approximately a 4-fold increase in Meis1 expression over the endogenous Meis1 and more than 30-fold increase in Meis2 (supplemental Figure 2B). When examined in EBs, Meis1 caused only a slight increase in expression of CD41, a marker for the earliest hematopoietic progenitor37 on day 9 of differentiation, but this effect was lost by day 12 (supplemental Figure 2C). When examined in ES cells cocultured with OP9 cells alone, Meis1 produced a larger induction of CD41 on day 9, which again was lost by day 12. However, in ES cells cocultured with OP9 and cytokines, Meis1 caused a robust induction of CD41 in more than 50% of cells on day 9, and this effect persisted to day 12. Therefore, we used these last conditions to further study Meis1 and Meis2 in regulating hematopoietic differentiation.
Figure 2.
Meis1 and Meis2 increase the numbers of ES cell–derived definitive hematopoietic colonies in semisolid media. (A) ES cells with dox-inducible Meis1 (A2lox.Meis1) or Meis2 (A2lox.Meis2) were differentiated as EBs for 6 days before plating in methylcellulose media with cytokines as described in “Methods.” Hematopoietic colonies were quantitated after 6 days of growth on methylcellulose based on morphologies. Data represent the average of 3 experiments. Error bars represent SD. (B) Bright-field microscopy of definitive hematopoietic colonies derived from A2lox.Meis1 or A2lox.Meis2 ES cells with (+) or without (−) treatment with doxycycline (dox). Original magnification ×40. (C) A2lox.Meis1 or A2lox.Meis2 EBs were dissociated on day 6 after differentiation and plated in MegaCult-C media. After another 6 days, megakaryocyte colony formation was visualized by acetylthiocholine iodide and Harris hematoxylin counterstain, and CFU-Mk and mixed Mk colonies were quantitated. Data represent the average of 4 experiments. Error bars represent SD. (D) Bright-field microscopy of CFU-Mk and mixed Mk colonies derived from A2lox.Meis1 ES cells in the presence of dox. Original magnification ×100. CFU-Mk appeared brown because murine megakaryocytes express acetylcholinesterase, producing brown precipitate. Mixed Mk colonies were distinguished by the presence of nonmegakaryocytic cells within brown-staining cell clusters.
Meis1 increases most hematopoietic progenitors but inhibits early erythroid progenitors
Previous studies suggested that Meis1 augments the proliferative potential of various progenitor cells.10,19 Initially, we expected that Meis1 and Meis2 might increase all hematopoietic progenitors. Induction of Meis1 or Meis2 by doxycycline on day 7 and 8 increased the differentiation of CD71− CD41+ hematopoietic progenitors by 50% to 200% (Figure 3A). However, both factors decreased the number of early CD71+ erythroid progenitors, consistent with the observed decrease in numbers of primitive erythroid colonies in methylcellulose assays caused by Meis1 and Meis2 (Figure 2A).
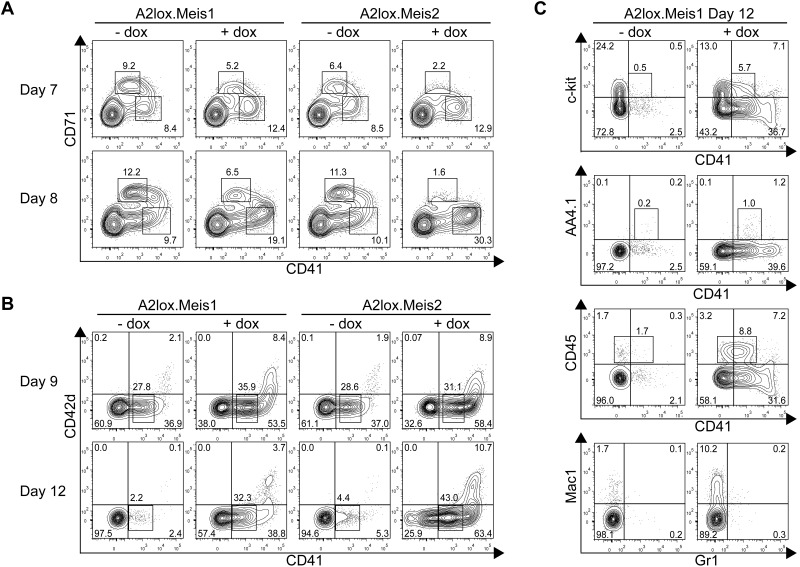
Figure 3.
Meis1 and Meis2 inhibit erythroid progenitor differentiation but increase CD41+ hematopoietic progenitor differentiation from ES cells cultured on OP9 with hematopoietic cytokines. (A) A2lox.Meis1 or A2lox.Meis2 ES cells were differentiated as EBs for 6 days before plating on OP9-GFP cell monolayers and cytokines, and treated with (+) or without (−) doxycycline (dox) every 2 days until day 12. On days 7 and 8, cells were analyzed by FACS for CD71 and CD41 expression. Data shown are for cells gated for negative expression of GFP to exclude OP9 cells from the analysis. Numbers indicate the percentage of cells within each quadrant. (B) Cells were treated as in panel A and analyzed on days 9 and 12 for expression of CD42d and CD41. (C) Cells were treated as in panel A and analyzed on day 12 for expression of c-kit, AA4.1, CD45, Mac1, and Gr1.
Meis1 and Meis2 caused a substantial increase of the numbers and maintenance of CD41+ hematopoietic progenitors on day 12 of differentiation (Figure 3B). Meis1 and Meis2 induced the formation of 2 populations of CD41+ cells. One population expressed intermediate levels of CD41 (CD41int), and the second expressed high levels (CD41hi). The CD41int cells induced by Meis1 and Meis2 were negative for expression of CD42d, a component of the von Willebrand factor receptor expressed by platelets. Notably, CD41hi cells coexpressed CD42d, suggesting that they are megakaryocytic precursors. CD41int cells expressed a low frequency of c-kit or AA4.1/CD93, an early hematopoietic progenitor marker, but expressed a higher frequency of CD45, a definitive hematopoietic marker (Figure 3C). Notably, CD41hi cells induced by Meis1 were largely negative for CD45, consistent with the lack of CD45 expression on megakaryocytic precursors. Meis1 induced the development of macrophages compared with control cultures (Figure 3C bottom panels). These results identify 2 distinguishable effects of Meis1 in differentiating ES cells. First, Meis1 increases the numbers of hematopoietic progenitors and maintains their persistence in culture. Second, and unexpectedly, Meis1 skews hematopoietic differentiation by suppressing erythroid while enhancing megakaryocytic progenitor differentiation.
Meis1 maintains proliferation of hematopoietic progenitors
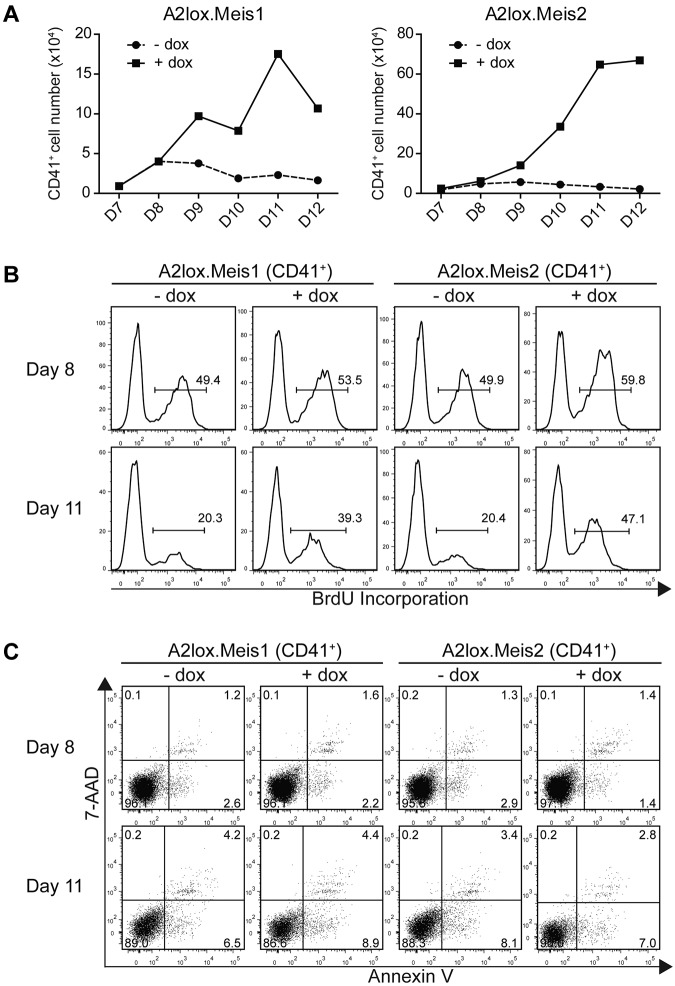
The actions of Meis1 noted in the preceding paragraph could result either from increased proliferation or decreased cell death of hematopoietic progenitors. To distinguish these possibilities, we determined the effect of Meis1 and Meis2 on the total number of CD41+ hematopoietic progenitors (Figure 4A). Both Meis1 and Meis2 markedly increased the total accumulation of CD41+ cells in culture, ranging from a 4-fold increase to more than a 10-fold increase in total cells (Figure 4A). This indicates that the increase in percentages of CD41+ populations seen earlier (Figure 3) represents an increase in total CD41+ cells. We next measured proliferation directly using BrdU incorporation in vitro (Figure 4B). Meis1 and Meis2 induced only slight increases in the rate of proliferation of CD41+ cells on day 8, 2 days after transfer into OP9 cultures (Figure 4B top panels). However, Meis1 and Meis2 caused the maintenance of cell proliferation on day 11 (Figure 4B). CD41+ cells, in which Meis1 or Meis2 was not induced, showed a marked decrease in proliferation at this time, with only 20% of cells incorporating BrdU with a 3-hour pulse. However, induction of Meis1 or Meis2 caused a rapid rate of proliferation to be maintained even on day 11 (Figure 4B bottom panels). This result is consistent with Meis1 and Meis2 being able to maintain progenitor cells in a proliferative state, as has been described previously.10,19
Figure 4.
Meis1 and Meis2 maintain the proliferation of CD41+ hematopoietic progenitors from ES cells cultured on OP9 with hematopoietic cytokines. (A) A2lox.Meis1 (left panel) or A2lox.Meis2 (right panel) ES cells were differentiated as in Figure 3A and analyzed for total cell numbers and CD41 expression on the indicated days. Absolute CD41+ cell numbers were determined from the product of total cell counts, and the percentage of CD41+ expressing cells was determined by FACS. (B) Cells differentiated as in panel A were pulsed with BrdU for 3 hours on day 8 or day 11 and analyzed by FACS for CD41 expression and BrdU incorporation. Numbers indicate the percentage of BrdU+ CD41+ cells. (C) Cells differentiated as in panel A were analyzed by FACS for staining with annexin V and 7-AAD to label apoptotic cells and with anti-CD41 antibody to label hematopoietic progenitors. Numbers indicate the percentage of CD41+ cells within each quadrant.
Increased cell numbers could conceivably arise from decreased apoptosis of CD41+ cells induced by Meis1. To test this, we stained differentiating ES cells in OP9 co-cultures with annexin V and 7-AAD (Figure 4C). First, CD41+ cells undergoing apoptosis (stained with annexin V but not 7-AAD) were rare in ES/OP9 cocultures, being less than 5% either with or without induction of Meis1 or Meis2. Second, the induction of Meis1 or Meis2 had no effect on the percentage of annexin V+ cells, indicating little effect of these factors on apoptosis of proliferating CD41+ progenitors.
Identification of gene targets of Meis1
Having identified 2 distinct effects of Meis1, we wished to determine their mechanisms. Meis1 might promote megakaryocyte-specific transcription factors or suppress genes associated with erythrocyte development. In regulating hematopoietic progenitor proliferation, Meis1 might directly induce genes associated with cell cycle, as reported for the induction of cyclin D19,38,39 or be involved in expressing growth factor receptors that could support progenitor proliferation. To distinguish these possibilities, we used microarray analysis to compare the various populations of cells developing in ES/OP9 cocultures in the presence and absence of Meis1 and Meis2 induction (supplemental Figure 3). To identify proximal gene targets of Meis1, we first needed to identify the critical time period in vitro during which Meis1 exerts its effects on proliferation and differentiation. Thus, we carried out a temporal analysis of Meis1's actions on hematopoietic cell development (supplemental Figure 3A). Optimal effects of Meis1 on inducing CD41 were observed when Meis1 was induced on either day 4 or day 6 of ES cell differentiation, but this effect was substantially reduced when induction of Meis1 was delayed until day 8 of differentiation (supplemental Figure 3B). Induction after day 8 led to essentially a loss of any activity of Meis1 induction on hematopoietic progenitor development.
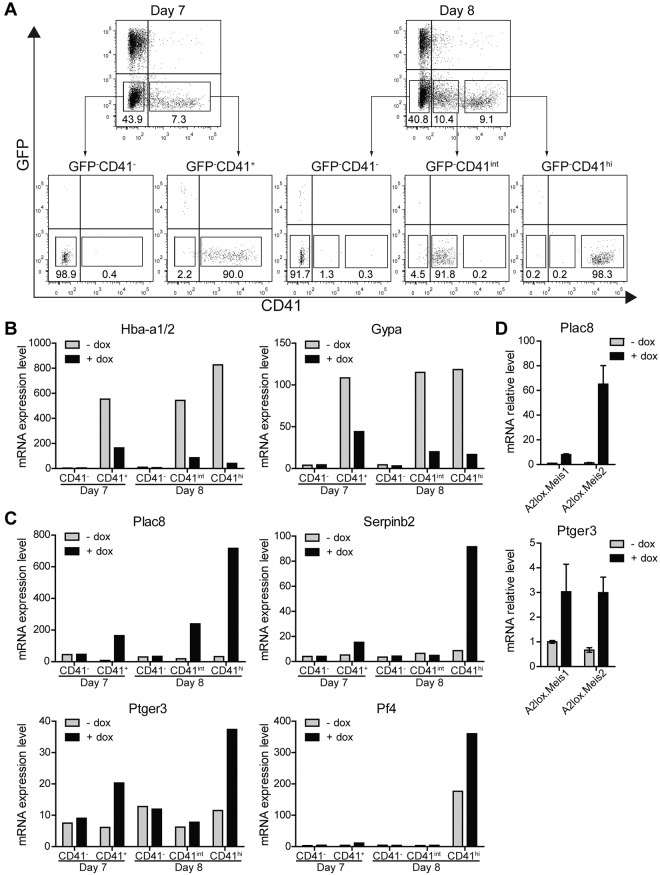
Therefore, to identify proximal targets of Meis1 in differentiating ES cells, we induced Meis1 on day 6 and purified populations of CD41− and CD41+ cells 24 hours after Meis1 induction (day 7), and CD41−, CD41int, and CD41hi populations 48 hours after induction of Meis1 (day 8; Figure 5A) and analyzed them by microarray (Figure 5B-C). Notably, induction of Meis1 and Meis2 substantially decreased a number of erythrocyte-specific genes, including hemoglobin α (Hba-a1/2) and glycophorin A (Gypa), both 24 hours and 48 hours after induction (Figure 5B). In addition, a number of other genes associated with erythroid development were repressed by the induction of Meis1 and Meis2 (supplemental Table 3). Expression of these erythroid genes was evident in CD41+ populations but absent from the CD41− populations, as expected. Thus, Meis1 appears to suppress erythrocyte gene expression within CD41+ hematopoietic progenitors, consistent with inhibition of CD71+ erythroid progenitors (Figure 3A) and reduced primitive erythroid colony formation (Figure 2A). This may be related to the requirement for Meis1 in platelet development, given that Meis1−/− embryos die of vascular defects secondary to the absence of platelets.15–17
Figure 5.
Identification of Meis1 and Meis2 target genes. (A) A2lox.Meis1 or A2lox.Meis2 ES cells were differentiated as in Figure 3A with or without treatment with doxycycline and were purified by cell sorting on day 7 or day 8 into the indicated populations based on levels of CD41 expression: CD41−, CD41+ (day 7) and CD41−, CD41int, and CD41hi (day 8). (B-C) Gene expression of A2lox.Meis2-derived populations with (+) or without (−) treatment by doxycyline (dox) was determined by microarray analysis. (B) Hemoglobin α (Hba-a1/2) and glycophorin A (Gypa) expression is shown for the indicated populations treated with (+) or without (−) dox. (C) Expression of placenta-specific 8 (Plac8), serine peptidase inhibitor, clade B, member 2 (Serpinb2), prostaglandin E receptor 3 (Ptger3), and platelet factor 4 (Pf4) is shown for the indicated populations and conditions. (D) Expression of Plac8 and Ptger3 by CD41hi day 8 cells treated with (+) or without (−) dox as described in panel A was determined by quantitative RT-PCR.
We also identified a few genes that were strongly induced by Meis1 and Meis2 (Figure 5C). Among these, placenta-specific 8 (Plac8) and serine peptidase inhibitor, clade B, member 2 (Serpinb2), were strongly induced primarily in CD41hi cells on day 8 but induced by lesser amounts on day 7. Notably, the prostaglandin E receptor 3 (Ptger3) was induced by Meis1 specifically in CD41+ cells on both days 7 and 8. We verified that the known target of Meis1, the platelet factor 4 (Pf4), was also induced by Meis1, and specific to CD41hi cells on day 8, consistent with its selective expression in megakaryocytes and platelets (Figure 5C). The induction of Plac8 and Ptger3 by Meis1 and Meis2 was confirmed by quantitative RT-PCR (Figure 5D).
Next, we turned to test the functional role of these 3 candidates in regulating hematopoietic cell progenitor proliferation. Among these candidates, Ptger3 appeared to be conceivably related to hematopoietic progenitor proliferation because its ligand prostaglandin E2 can expand HSCs in the setting of transplantation.40,41 However, Plac8 and Serpinb2 might conceivably regulate cell proliferation or survival. Plac8 reportedly supports cell proliferation and survival by modulating the Akt-Mdm2 pathway and p53 level,42 whereas Serpinb2 may enhance tumor cell survival.43 We tested Plac8 and Serpinb2 for their ability to influence accumulation of CD41+ cells in ES cell differentiation cultures using a doxycycline inducible system (supplemental Figure 4). However, induction of Plac8 or Serpinb2 alone or in combination showed no effect on the accumulation of CD41+ cells or on the expression of CD42d either on day 9 or day 12 in ES/OP9 cocultures. Plac8−/− mice are able to generate normal hematopoietic cell lineages, and only a slight influence on neutrophils function in an infectious model system has been reported.44 Conceivably, Plac8 might provide a proliferative advantage to hematopoietic progenitors that was obscured in the setting of the knockout. To test this notion, we generated mixed BM chimeras from wild-type and Plac8−/− donor mice (supplemental Figure 5). We compared the ability of Plac8−/− BM progenitors to compete with wild-type progenitors across a wide range of stages of hematopoietic development (supplemental Figure 5A). The ratio of chimerism observed for HSCs was similar to the ratio in all subsequent stages of myeloerythroid differentiation, indicating that loss of Plac8 does not influence progenitor proliferation relative to wild-type progenitors. Thus, Plac8 may be a target of Meis1 but does not appear to mediate its effects on hematopoietic progenitor proliferation.
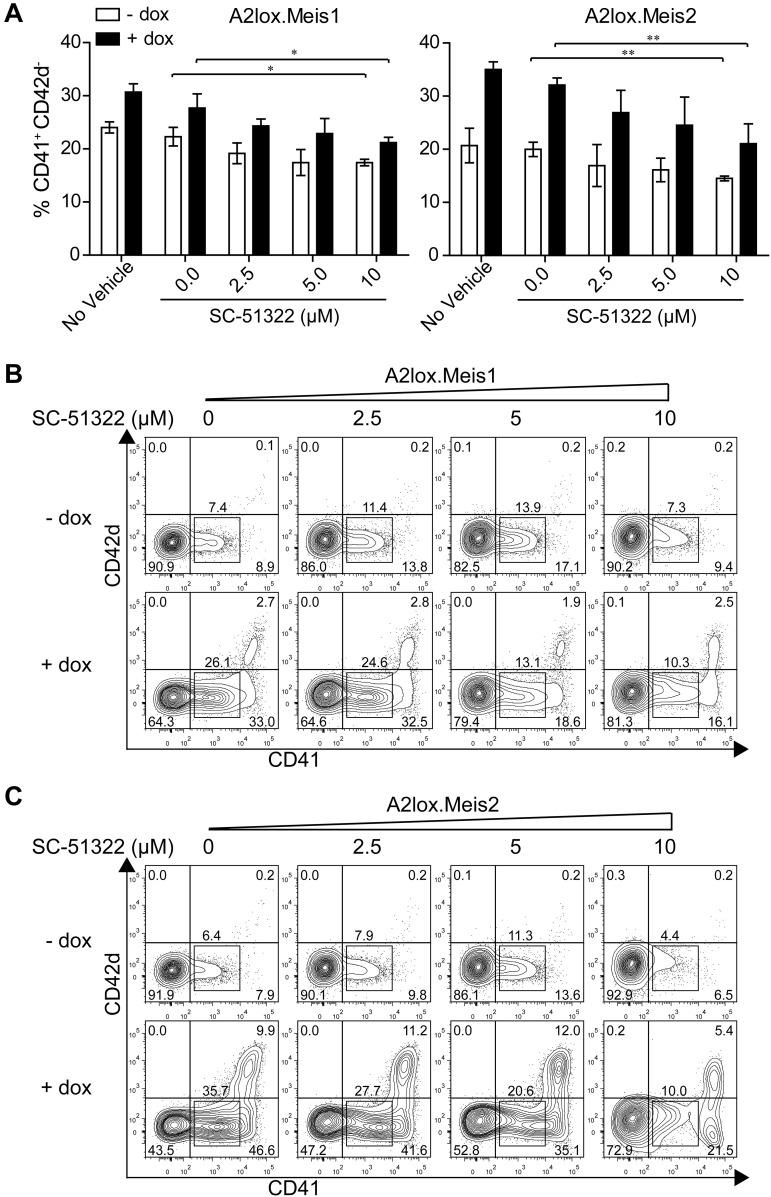
Meis1 induced expression of Ptger3 in CD41+ cells on day 7, and in CD41hi cells on day 8 (Figure 5C). Ptger3 is expressed in the MkP at high levels relative to other hematopoietic progenitors (supplemental Figure 8A). Meis1 expression is high in the MkP but is also expressed in other hematopoietic progenitors, except for erythroid precursors (supplemental Figure 8B). To test whether signaling through this receptor could mediate Meis1's effect on hematopoietic progenitor proliferation, we inhibited prostaglandin E receptor in the presence or absence of Meis1 induction in differentiating ES cells (Figure 6). In the absence of a prostaglandin E receptor antagonist SC-51322, both Meis1 and Meis2 induction caused a robust increase in the number of CD41int and CD41hi cells on day 12, as expected (Figure 6A-C). However, in the induced conditions, increasing doses of antagonist caused a statistically significant reduction in the CD41int population generated by overexpression of Meis1 and Meis2 (Figure 6B-C). The effect of this inhibition is more evident in conditions in which Meis1 or Meis2 is induced by doxycycline relative to the inhibition in the presence of endogenous levels of Meis1 or Meis2 (Figure 6A-C). This may indicate that the actions of Meis1 and Meis2 in promoting hematopoietic progenitor expansion may depend on signaling through the Ptger3. However, the inability of several shRNAs to significantly reduce endogenous Ptger3 expression in ES cells prevents definitive determination of the requirement for this candidate in mediating the effects of Meis1 (supplemental Figure 9).
Figure 6.
Meis1 and Meis2 induce CD41int hematopoietic progenitors through Ptger3 signaling pathway. (A) A2lox.Meis1 or Meis2 ES cells were differentiated as described in Figure 3A in the presence of the indicated concentration of SC-51322 added on day 6 and replenished daily. On day 9, cells were harvested and analyzed by FACS for expression of CD42d and CD41. Cells are gated as GFP-negative to exclude OP9 cells. Numbers indicate the percentage of CD41+ CD42d− cells developing in the indicated conditions from 3 independent experiments. Bars represent the SD. *P < .05 (paired Student t test). **P < .01 (paired Student t test). (B) A2lox.Meis1 ES cells were differentiated as in Figure 3A with the indicated concentration of SC-51322 added on day 6 and replenished each day until day 12. On day 12, cells were analyzed by FACS for expression of CD42d and CD41. Shown are data for GFP-negative cells to exclude analysis of OP9 cells. Numbers indicate the percentage of cells in the indicated gates. (C) A2lox.Meis2 ES cells were differentiated and analyzed as in panel A.
Isoform-specific repression of in vivo erythroid progenitor development
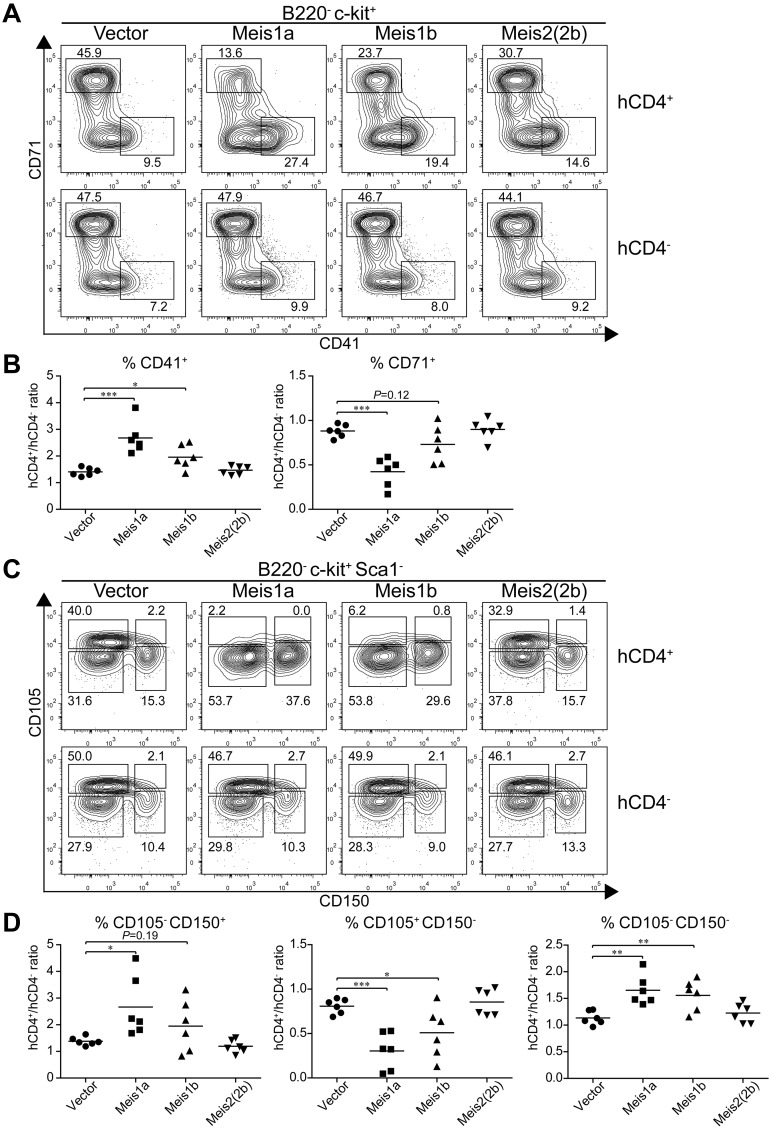
Because Meis1 and Meis2 have multiple isoforms,16 we wished to test their actions in regulating erythroid and megakaryocyte development. Meis1 has 2 naturally occurring isoforms, Meis1a and Meis1b (supplemental Figure 6A), formed by an alternative splicing of different terminal exons generating proteins with divergent C-terminal sequences. Meis2 has at least 6 reported isoforms generated through alternative splicing (supplemental Figure 6A). We first tested whether the alternative isoforms of Meis1 and Meis2 had similar effects on increasing CD41int hematopoietic progenitors (supplemental Figure 6B). Notably, Meis1a, Meis2(2a), and Meis2(4a) each substantially increased the number of CD41int hematopoietic progenitors on day 9 and day 12 relative to controls, indicating that these isoforms act similarly to Meis1b and Meis2(2b) tested earlier (Figure 3B). Interestingly, Meis1 and Meis2 appear to have different expression patterns in vivo. Meis1 is expressed in hematopoietic progenitors, whereas Meis2 is expressed in the nervous system.45 We find evidence for differential capacities of Meis1 and Meis2 isoforms to suppress erythroid differentiation and promote megakaryocytic progenitors in vivo (Figure 7). First, we used CD71 and CD41 expression to examine early erythroid progenitors from control BM or from BM transduced with retroviruses expressing Meis1a, Meis1b, or Meis2(2b) (Figure 7A). As a control, BM cells transduced with an empty retrovirus showed no difference in the extent of erythroid progenitor development compared with nontransduced BM within the same animal (Figure 7A left panel). As before, we found that expression of Meis1b substantially reduced the population of CD71+ erythroid precursors and increased the population of CD41+ CD71− MkPs. Notably, Meis1a caused a greater reduction in development of erythroid progenitors and a stronger enhancement of CD41+ MkPs compared with Meis1b. In contrast, Meis2(2b) appeared weaker than Meis1b in these actions.
Figure 7.
Meis1a and Meis1b inhibit the in vivo erythroid potential of the MEP. Stem/progenitor cell-enriched BM cells from donor mice were transduced with a control vector (IRES-hCD4) or a retroviral construct expressing Meis1a (Meis1a-IRES-hCD4), Meis1b (Meis1b-IRES-hCD4), or Meis2(2b) (Meis2(2b)-IRES-hCD4) as indicated. The transduced BM progenitor cells were transplanted into sublethally irradiated recipients and BM was analyzed 4 to 6 weeks later. (A) BM was analyzed for expression of hCD4, B220, c-kit, CD71, and CD41. Shown are 2-color contours of CD71 and CD41 expression for cells gated as B220− c-kit+. Numbers indicate the percentage of cells in the indicated gates. (B) The percentage of megakaryocytic progenitors (CD41+) or erythroid progenitors (CD71+) as in panel A was determined for both transduced (hCD4+) cells and nontransduced (hCD4−) cells within the same recipient. Shown is the ratio for each vector for megakaryocytic progenitors (CD41+) or erythroid progenitors (CD71+) as indicated. Each dot represents data from an individual recipient (n = 6). *P < .05. **P < .01. ***P < .001. (C) BM from recipients described in panel A was analyzed for expression of hCD4, B220, c-kit, Sca1, CD105, and CD150. Shown are 2-color contours of CD105 and CD150 expression for cells gated as B220− c-kit+ Sca1−. (D) The percentage of pre-MegE and MkP (CD105− CD150+), CFU-E (CD105+ CD150−), or granulocyte-macrophage progenitors and pre-GM (CD105− CD150−) progenitors was determined for both the transduced (hCD4+) and nontransduced (hCD4−) BM cells within the same recipient. Shown is the ratio of hCD4+ to hCD4− progenitors for each construct and cell population. Each dot represents data from one recipient mouse (n = 6). *P < .05. **P < .01. ***P < .001.
We repeated this analysis using CD105 and CD150 expression to distinguish earlier stages of erythroid and megakaryocyte differentiation. CD150+ CD105− population represents both the MEP and the committed MkP. As the MEP differentiates toward the erythrocyte lineage, CD105 expression is induced, followed by loss of CD150 expression. In this system, again BM cells transduced with an empty retrovirus showed very little effect on the inhibition of erythrocyte differentiation. In contrast, Meis1b substantially reduced the size of the CD105+ CD150− population of erythroid progenitors. Again, the Meis1a isoform was even more robust in extinguishing erythroid progenitor differentiation, and the Meis2(2b) isoform was much weaker than Meis1b or Meis1a.
Discussion
Meis1 was initially discovered as a common virus integration site for myeloid leukimias,1 and much subsequent analysis has associated its actions with leukemic transformation.9,13,46 However, the role of Meis1 in normal hematopoiesis is not well studied. Our study identified 2 distinct roles of Meis1 during normal hematopoiesis based on analysis of Mesp1-deficient ES cells and on analysis of overexpression of Meis1 and Meis2. First, we observed a robust action of Meis1 to maintain the proliferative state of early hematopoietic progenitors. Although previous study reported a reduction of HSC population in the fetal liver of Meis1−/− mice,15,16 suggesting Meis1 may play an important role in HSC self-renewal, there is no direct evidence that Meis1 acts on HSC proliferation. Our study demonstrated that Meis1 maintains a proliferative state in CD41+ hematopoietic progenitors by directly measuring their BrdU incorporation, which was associated with an increase of hematopoietic progenitors accumulating in ES cell/OP9 cocultures (Figure 4). Previous studies showed Meis1 interacts with Hoxa9 to accelerate leukemic transformation,9 and Hoxa9 also modulates Meis1 to influence normal hematopoiesis.47 However, we found that coexpression of Hoxa9 with Meis1 or Meis2 did not enhance proliferation of CD41+ hematopoietic progenitors derived from ES/OP9 cocultures (supplemental Figure 7), suggesting either that Hoxa9 is not a binding partner of Meis1 during in vitro hematopoietic differentiation of ES cells or that Hoxa9 is not present in limiting amounts in our system. Thus, our analysis extends our understanding of how Meis1 promotes HSC self-renewal, unifying its action with those described for Meis2 in regulating retinal progenitor cell proliferation during eye development.19,38
Second, our study shows that Meis1 actively suppresses erythroid progenitor differentiation while promoting MkP development. This previously unrecognized action is very likely to underlie the defect in platelet development that has been described as a basis for embryonic lethality in Meis1−/− mice.17 Our in vivo analysis of BM progenitors suggests that Meis1 regulated the early lineage decision choice at the stage of the MEP. In summary, our study demonstrates dual actions of Meis1, distinguishing its later action on lineage specification from its earlier role in HSC self-renewal.
In addition, our study identified novel gene targets of Meis1 to explain its actions during normal hematopoiesis. We identified 4 potential candidates, Plac8, Ptger3, Serpinb2, and Pf4, based on specific and robust induction by Meis1 in CD41+ hematopoietic progenitors. The platelet-specific gene Pf4/Cxcl4 is a chemokine-like protein and a known target of Meis1.48 We tested whether the other 3 candidates were sufficient or required for Meis1's actions. Plac8 reportedly supports cell proliferation and survival by decreasing p53 via the akt-Mdm2 pathway.42 However, we found it was neither sufficient to replace Meis1 for promoting hematopoietic cell progenitor proliferation nor necessary for maintaining in vivo HSC proliferation in a mixed BM chimera setting. Likewise, we found that Serpinb2 was insufficient either alone or in combination with Plac8 for maintaining the persistence of hematopoietic progenitors in ES/OP9 coculture as Meis1. Several reports have demonstrated that Meis1 can regulate progenitor cell proliferation through influencing the expression of cell cycle components, including cyclin D1 and cyclin D3.19,38,39 However, our gene expression analysis identified no cell cycle components to be substantially altered by Meis1 in CD41+ hematopoietic progenitors. Instead, our study suggests a novel mechanism by which Meis1 influences progenitor cell proliferation through inducing a receptor for the prostaglandin signaling pathway. We found that Ptger3, one of the 4 G-protein coupled receptors for prostaglandin E2, could potentially mediate increased hematopoietic progenitor proliferation because inhibition of prostaglandin signaling blocked Meis1's effect on proliferation of hematopoietic progenitors.
Our study also identified that Meis1 robustly repressed a number of erythroid-specific genes, including Hba-a1/2 and Gypa, consistent with an inhibitory role in erythroid differentiation. At present, we do not distinguish between direct or indirect actions of Meis1 on these target genes. Meis1 might directly recruit transcriptional corepressors to the target loci or could induce repressors that can inhibit expression of these erythroid-specific genes. In addition, other transcription factors, such as Klf1 and Fli1, have been reported to be critical regulators of erythroid and megakaryocyte development.49,50 It will be interesting to examine the transcriptional hierarchy of Meis1 and these other factors in erythroid cell and megakaryocyte development. A dissection of actions of Meis1 between early HSC proliferation and later erythroid and megakaryocyte lineage specification should help in understanding the important functions of Meis1 during normal hematopoiesis.
Supplementary Material
Acknowledgments
The authors thank Dr Raphael Kopan (Department of Developmental Biology, Washington University School of Medicine) for OP9-GFP cells and Dr Michael Kyba (Lillehei Heart Institute and Department of Pediatrics, University of Minnesota) for A2lox ES cells.
This work was supported by the Howard Hughes Medical Institute (K.M.M.) and an American Cancer Society postdoctoral fellowship (E.M.L.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.C. performed the majority of the experiments and analyzed data; E.M.L. and J.G.G. carried out the Mesp1 Flk1+Tie2+ sort microarray analysis; A.T.S., J.C.A., and W.K. helped with in vivo analysis based on bone marrow transplantation; J.C.A. generated Rosa26-CAG-STOP-eGFP+/+ reporter mice; T.L.M. bred the Mesp1Cre/+Rosa26-CAG-STOP-eGFP+/+ mice and carried out the initial fate mapping analysis in peripheral blood; and M.C., T. L.M., and K.M.M. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenneth M. Murphy, Department of Pathology and Immunology, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO 63110; e-mail: kmurphy@wustl.edu.
References
- 1.Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15(10):5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura T, Largaespada DA, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12(2):149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 3.Shen WF, Montgomery JC, Rozenfeld S, et al. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17(11):6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang CP, Jacobs Y, Nakamura T, et al. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17(10):5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A. 1997;94(26):14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen WF, Rozenfeld S, Kwong A, et al. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19(4):3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamo A, Krosl J, Kroon E, et al. Molecular dissection of Meis1 reveals 2 domains required for leukemia induction and a key role for Hoxa gene activation. Blood. 2006;108(2):622–629. doi: 10.1182/blood-2005-06-2244. [DOI] [PubMed] [Google Scholar]
- 8.Goh SL, Looi Y, Shen H, et al. Transcriptional activation by MEIS1A in response to protein kinase A signaling requires the transducers of regulated CREB family of CREB co-activators. J Biol Chem. 2009;284(28):18904–18912. doi: 10.1074/jbc.M109.005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroon E, Krosl J, Thorsteinsdottir U, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci U S A. 2001;98(23):13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kroon E, Thorsteinsdottir U, Mayotte N, Nakamura T, Sauvageau G. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 2001;20(3):350–361. doi: 10.1093/emboj/20.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlovsky K, Kalinkovich A, Rozovskaia T, et al. Down-regulation of homeobox genes MEIS1 and HOXA in MLL-rearranged acute leukemia impairs engraftment and reduces proliferation. Proc Natl Acad Sci U S A. 2011;108(19):7956–7961. doi: 10.1073/pnas.1103154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21(21):2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercader N, Leonardo E, Azpiazu N, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402(6760):425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 15.Hisa T, Spence SE, Rachel RA, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23(2):450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azcoitia V, Aracil M, Martinez A, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. 2005;280(2):307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Carramolino L, Fuentes J, Garcia-Andres C, et al. Platelets play an essential role in separating the blood and lymphatic vasculatures during embryonic angiogenesis. Circ Res. 2010;106(7):1197–1201. doi: 10.1161/CIRCRESAHA.110.218073. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev. 2002;16(16):2097–2107. doi: 10.1101/gad.1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heine P, Dohle E, Bumsted-O'Brien K, Engelkamp D, Schulte D. Evidence for an evolutionary conserved role of homothorax/Meis1/2 during vertebrate retina development. Development. 2008;135(5):805–811. doi: 10.1242/dev.012088. [DOI] [PubMed] [Google Scholar]
- 20.Bertozzi CC, Hess PR, Kahn ML. Platelets: covert regulators of lymphatic development. Arterioscler Thromb Vasc Biol. 2010;30(12):2368–2371. doi: 10.1161/ATVBAHA.110.217281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertozzi CC, Schmaier AA, Mericko P, et al. Platelets regulate lymphatic vascular development through CLEC-2-SLP-76 signaling. Blood. 2010;116(4):661–670. doi: 10.1182/blood-2010-02-270876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stankunas K, Shang C, Twu KY, et al. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ Res. 2008;103(7):702–709. doi: 10.1161/CIRCRESAHA.108.175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyba M, Perlingeiro RC, Hoover RR, et al. Enhanced hematopoietic differentiation of embryonic stem cells conditionally expressing Stat5. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11904–11910. doi: 10.1073/pnas.1734140100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsley RC, Gill JG, Murphy TL, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3(1):55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17(6):749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 26.Ranganath S, Ouyang W, Bhattarcharya D, et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161(8):3822–3826. [PubMed] [Google Scholar]
- 27.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464(7285):112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand JY, Chi NC, Santoso B, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464(7285):108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boisset JC, van Cappellen W, Andrieu-Soler C, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464(7285):116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 30.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127(15):3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 31.Roberts CW, Shutter JR, Korsmeyer SJ. Hox11 controls the genesis of the spleen. Nature. 1994;368(6473):747–749. doi: 10.1038/368747a0. [DOI] [PubMed] [Google Scholar]
- 32.Spyropoulos DD, Pharr PN, Lavenburg KR, et al. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol. 2000;20(15):5643–5652. doi: 10.1128/mcb.20.15.5643-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee D, Park C, Lee H, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2(5):497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Yabuuchi A, McKinney-Freeman S, et al. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc Natl Acad Sci U S A. 2008;105(22):7756–7761. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mucenski ML, McLain K, Kier AB, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65(4):677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 37.Ferkowicz MJ, Starr M, Xie X, et al. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130(18):4393–4403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- 38.Bessa J, Tavares MJ, Santos J, et al. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135(5):799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- 39.Argiropoulos B, Yung E, Xiang P, et al. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115(20):4071–4082. doi: 10.1182/blood-2009-06-225573. [DOI] [PubMed] [Google Scholar]
- 40.North TE, Goessling W, Walkley CR, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frisch BJ, Porter RL, Gigliotti BJ, et al. In vivo prostaglandin E2 treatment alters the bone marrow microenvironment and preferentially expands short-term hematopoietic stem cells. Blood. 2009;114(19):4054–4063. doi: 10.1182/blood-2009-03-205823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogulski K, Li Y, Rothermund K, et al. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene. 2005;24(51):7524–7541. doi: 10.1038/sj.onc.1208897. [DOI] [PubMed] [Google Scholar]
- 43.Tonnetti L, Netzel-Arnett S, Darnell GA, et al. SerpinB2 protection of retinoblastoma protein from calpain enhances tumor cell survival. Cancer Res. 2008;68(14):5648–5657. doi: 10.1158/0008-5472.CAN-07-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledford JG, Kovarova M, Koller BH. Impaired host defense in mice lacking ONZIN. J Immunol. 2007;178(8):5132–5143. doi: 10.4049/jimmunol.178.8.5132. [DOI] [PubMed] [Google Scholar]
- 45.Cecconi F, Proetzel G, Alvarez-Bolado G, Jay D, Gruss P. Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somitic mesoderm. Dev Dyn. 1997;210(2):184–190. doi: 10.1002/(SICI)1097-0177(199710)210:2<184::AID-AJA10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 46.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24(2):617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29(18):5181–5192. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okada Y, Nagai R, Sato T, et al. Homeodomain proteins MEIS1 and PBXs regulate the lineage-specific transcription of the platelet factor 4 gene. Blood. 2003;101(12):4748–4756. doi: 10.1182/blood-2002-02-0380. [DOI] [PubMed] [Google Scholar]
- 49.Doré LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118(2):231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Starck J, Cohet N, Gonnet C, et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol. 2003;23(4):1390–1402. doi: 10.1128/MCB.23.4.1390-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.