Summary
Background
This study aimed to examine the relationship between XRCC1, p53 and MDR1 protein, along with polymorphisms of their genes and their prognostic values in breast cancer. The following clinical and pathological parameters were evaluated: histopathological type of tumor, grade, stage, Her2/neu expression, ER, PR positivity and involvement of regional lymph nodes.
Material/Methods
Expression of proteins was determined in 39 samples of breast cancer by immunohistochemistry. Nucleotide polymorphisms were analyzed by PCR-RFLP. For statistical analysis, chi-square test (Yates), Fisher’s exact test, and correlation test were used to analyze the data.
Results
The highest protein expression was immunohistochemically found in MDR1 protein, with 54% of samples testing positive. In addition, the evaluation of MDR1 expression revealed higher positive immunoreactivity in lobular (LIC) and other types of tumor in comparison to ductal (DIC) type. The expression of p53 and XRCC1 protein was equal, but lower compared to MDR1, both testing positive in 36% of all tissue samples. Comparison of XRCC1 protein and histopathological type of tumor revealed that DIC and LIC types were mostly XRCC1-negative, while other types, papillary and mucinous were more likely to be XRCC1-positive. Interestingly, when evaluating LIC samples separately, a negative correlation between the Her2/neu and expression of XRCC1 was detected. Apparently, all Her2/neu-positive samples were XRCC1-negative (6/86%). The correlation test indicated a negative correlation between Her2/neu-positive samples and XRCC1-negative specimens (r=1, p<0.05). Statistical analysis did not reveal a correlation of p53 expression with clinical and pathological parameters. Similarly, no statistically significant difference was found between the tested polymorphisms and protein expression.
Conclusions
We did not find statistically significant correlation between tested polymorphisms and their protein expression.
Keywords: XRCC1, p53, MDR1, polymorphisms, breast cancer
Background
Breast cancer is one of the most common human neoplasms, accounting for approximately one quater of all cancers in females [1]. Its occurrence is associated with the lifestyle in advanced economy countries, with the highest incidence in the Western countries. The frequency of breast cancer remains high, and its clinical courses are markedly variable. It is therefore of utmost importance to predict the biology of the tumor and subsequently the course of the disease in the individual patient in order to choose adequate therapy and patient surveillance. Surgery remains the principal therapeutic approach in breast cancer. Advanced cases require supplementary therapy involving pharmacotherapy and/or radiotherapy. The main reason for therapeutic failure in cases of invasive breast cancers involves resistance to diverse regimens of chemotherapeutics and radiotherapy.
X-ray repair cross-complementing protein 1 (XRCC1) is required for single-strand break repair in human cells. Several polymorphisms in this gene have been implicated in cancer risk and clinical prognostic factors. The XRCC1 gene is located on chromosome 19q13.2 and contains 17 exons [2]. The gene shows that 3 relatively common polymorphisms affect the amino acid sequence in codons 194(Arg/Trp), 280(Arg/His), and 399(Arg/Gln) [3]. Several reports indicate that the variant alleles of the repair gene polymorphisms may affect DNA repair function. DNA repair proficiency has also been proposed as a potential susceptibility factor for breast cancer [4]. On the other hand, cells defective in XRCC1 have increased sensitivity to ionizing radiation, UV, hydrogen peroxide, and mitomycin [5]. Identification of the factors that characterize the resistant cases would permit immediate treatment of the patients with alternative therapeutic approaches.
Many therapeutic agents act by damaging DNA, which may in certain contexts function as a signal to trigger apoptosis. p53, encoded by the Tp53 gene, is considered as one of the most commonly aberrant tumor suppressor genes in human cancer. It is a nuclear phosphoprotein capable of binding to specific DNA sequences and activating specific target genes. Normal p53 exerts its antiproliferative action by inducing reversible or irreversible cell cycle arrest, or apoptosis. Various mechanisms, such as Tp53 mutations, alteration of p53 regulators, or alteration of p53 target genes, often alter the normal function of p53 in cancer cells. The mutations can result in accumulation and overexpression of mutant p53 protein. In normal cells, the expression level of p53 protein is generally below the detection level of immunohistochemical methods. Mutations of p53 have been detected in more than 50% of human cancers [6]. Overexpression of p53 (detected immunohistochemically) has been found in breast, colon, lung, and brain cancers, and is usually associated with tumor progression and unfavorable prognosis [6]. This overexpression is thought to be due to either stabilization of mutant p53 or stabilization of wild-type p53 by interaction with other proteins that prevents its degradation. These events can lead to loss of the function of p53 as a transcription factor involved in the regulation of the cell cycle.
ABC transporters belong to the family of transporter proteins that contribute to drug resistance via ATP-dependent drug efflux pumps. P-glycoprotein (P-gp/MDR1), encoded by the MDR1 gene, is an ABC transporter normally involved in the excretion of toxins from the cells. It also confers resistance to certain chemotherapeutic agents such as anthracyclines, vinca alkaloids, colchicines, epipodophyllotoxins, and paclitaxel [7]. MDR1 is overexpressed in chemotherapy-resistant tumors (colon and kidney cancers). It is regularly upregulated after disease progression following chemotherapy in various malignancies such as leukemia and breast cancer. The results from a breast cancer meta-analysis indicated that MDR1 expression could be detected in 41% of breast cancer patients, with increased levels post-therapy [8]. The expression of MDR1 may indicate the presence of multidrug-resistant clones from which tumor recurrence and dissemination might arise [9].
The main aim of our study was to determine XRCC1, p53 and MDR1 gene polymorphisms in breast cancer patients and to compare them with related protein expressions in tumor tissue samples. In addition, the expression levels of XRCC1, p53 and MDR1 gene products correlated with histological type, stage and grade of tumors, and involvement of regional lymph nodes. Finally, the co-expression and relationship among XRCC1, p53, MDR1, Her2neu, ER, and PR proteins were assessed and evaluated as possible prognostic markers.
Material and Methods
Patients and clinical samples
Immunohistochemical analysis was performed retrospectively on tissue samples taken for routine diagnostic purposes. The cases were selected based on tissue availability and were not stratified for known preoperative or pathological prognostic factors. The patients gave their informed consent before their inclusion into the study.
Fragment samples from studied tumors were fixed in 9% buffered formalin and embedded in paraffin. In every case, hematoxylin and eosin stained preparations were subjected to histopathological evaluation by 2 pathologists. The stage of the tumors was assessed according to the TNM classification system [10]. Tumor grade was estimated according to Bloom-Richardson and the modification of Elston and Ellis [11].
In this study, we used 39 samples of breast carcinoma. The samples were obtained from the Department of Pathology, Pasteur Faculty Hospital, Košice, Slovak Republic.
Patients and tumor characteristics are summarized in Table 1.
Table 1.
Patients and tumor characteristics.
| Characteristics | No/% |
|---|---|
| All patients | 39/100% |
| Histopathological type | |
| DIC (ductal invasive c.) | 27/69% |
| LIC (lobular invasive c.) | 7/18% |
| Other | 5/13% |
| Histological grade | |
| G1 – good | 10/26% |
| G2 – moderate (worse) | 20/51% |
| G3 – poor | 9/23% |
| Histological stage | |
| 1 | 9/23% |
| 2 | 1/2.5% |
| 2A | 8/20.5% |
| 2B | 4/10.5% |
| 3A | 5/13% |
| 3B | 9/23% |
| 3C | 2/5% |
| 4 | 1/2.5% |
| HER2/neu | |
| Positive (3+) | 7/18% |
| Negative (0,1+) | 32/82% |
| Estrogen receptor status (ER) | |
| Positive | 26/67% |
| Negative | 13/33% |
| Progesteron receptor status (PR) | |
| Positive | 16/41% |
| Negative | 23/59% |
| Involvement of regional lymph nodes | |
| Positive | 23/59% |
| Negative | 16/41% |
Immunohistochemical detection of XRCC1, p53, MDR1, ER and PR
Thirty-nine samples of breast carcinoma were evaluated for expression of XRCC1, p53, MDR1, ER and PR. The indirect enzymatic immunohistochemical method was used to detect these proteins. Formalin-fixed, paraffin-embedded tissue blocks were cut (7 μm) and attached to the slides. Next, the slides were processed for immunohistochemistry.
The slides were deparaffinized, rehydrated and washed with phosphate-buffered saline containing 0.05% Tween-20 (PBS-Tween), pH 7.6. Endogenous peroxidase was quenched in 0.3% H2O2 (3% H2O2 for ER and PR) in methanol for 30 min (minutes) at room temperature and slides were washed in PBS-Tween. According to the analyzed protein, sections were pretreated in citrate buffer solution in the microwave oven. The slides stained for XRCC1, p53 and MDR1 were pretreated for 2×5 min for estrogen and progesterone receptors in Target Solution (DakoCytomation, Denmark) for 45 min. XRCC1, p53 and MDR1 staining procedure continued by blocking nonspecific staining with blocking buffer (milk buffer) for 30 min in a humidified chamber at room temperature. Blocking buffer was removed from slides by flipping the slide without rinsing, and the primary antibodies were applied overnight in a humidified chamber at 4°C. As primary antibodies monoclonal antibody to XRCC1 (Ab-3 clone 144, Lab Vision Corporation), monoclonal antibody to p53 (BP53-12, Alexis Biochemicals), monoclonal antibody to MDR1 (UIC2, Santa Cruz Biotechnology, Inc.), monoclonal antibody to mouse anti-human estrogen receptor á (clone 1D5, Dako Cytomation, Denmark) and monoclonal antibody to mouse anti-human progesterone receptor (clone 1A6, Immunotech, France) were used. Tissue sections were rinsed in PBS-Tween 20 (3×5 min), in the cases of ER and PR in Tris buffer, and subsequently incubated with the biotinylated secondary antibody (Link). After washing, the streptavidin-peroxidase (Label) was applied for 30 min at room temperature (Link and Label – Universal LSAB™ + KIT/HRP, DakoCytomation, Denmark). The sections were then visualized with 3,3′-diaminobenzidine tetrahydrochloride (DAB) at a concentration of 0.5 mg/ml in Tris buffer, pH 7.6, and 0.015% H2O2. ER and PR immunostained sections were visualized with aminoethylcarbazole (AEC). Sections were stream-rinsed with tap water, counterstained with hematoxylin for 2 min, washed in tap water, dried, mounted, and cover slipped.
Sections processed with omission of primary antibody served as a negative control of the immunohistochemical procedure.
The cases were diagnosed as XRCC1-, p53-, and MDR1-positive (11–90% = 2+, 91−100% = 3+) when expression was observed in more than 10% of tumor cells or in numerous cell clumps. The samples with no reaction (0% = −) or the reaction present in only individual tumor cells (<10% = 1+) were considered negative.
Immunohistochemical detection of HER2/neu protein
The HercepTestTM kit (Dako Cytomation, Denmark) was used for the immunohistochemical detection of HER2/neu protein. In relation to the agreement accepted worldwide, all steps of the detection strictly follow the guidelines published in the kit’s manual.
Statistical analysis
Statistical analysis of the results employed chi-square (967;2) test to estimate the significance of differences between the expression of 3 proteins (XRCC1, p53, MDR1), the polymorphisms of their genes, and clinicopathological parameters used. A p-value 0.05 or less was considered statistically significant.
DNA extraction
Blood samples were taken from anti-coagulated (Na2EDTA) peripheral venous blood, and genomic DNA was extracted using the standard phenol/chloroform extraction method. Due to the acquisition of new and better equipment in our laboratory, samples collected more recently were extracted using the Wizard® Genomic DNA Purification Kit (Promega Corporation, USA).
PCR-RFLP genotyping
The single nucleotide polymorphisms were analyzed using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assays. Primer sets (Sigma-Aldrich, Germany) and annealing temperatures used for the PCR-RFLP assay were as follows; (1) p53 Arg72Pro, 5′-TTT CACCCATCTACAGTCCC-3′ and 5′-ACCTAGGCTCAGGGCAACTGACCG-3′ (63°C); (2) MDR1 C3435T, 5′-TGTTTTCAGCTGCTTGATGG-3′ and 5′-AAGGCATGTATGTTGGCCTC-3′ (60°C); (3) XRCC1 Arg399Gln, 5′-TTGTGCTTT CTCTGTGTCCA-3′ and 5′-TCCTCCAGCCTTTTCTGATA-3′ (63°C).
The PCR program had an initial denaturation step of 3 min at 94°C followed by 35 cycles of 30 s (seconds) at 94°C, 30 s of annealing at 60°C or 63°C, depending on the primers, and 45 s at 72°C. The PCR products were digested at 37°C overnight (minimum 3 h [hours]) using BstUI, Sau3AI and MspI for p53 Arg72Pro, MDR1 C3435T and XRCC1 Arg399Gln, respectively. The obtained restriction fragments were separated by electrophoresis on a 3% agarose gel for 45 min at 140V and analyzed after staining with ethidium bromide under ultraviolet light (Table 2).
Table 2.
PCR-RFLP genotyping of the polymorphisms.
| SNP genotype | Fragments after digestion |
|---|---|
| p53 Arg72Pro (G>C) | |
| GG | 182-bp, 136-bp |
| GC | 318-bp, 182-bp, 136-bp |
| CC | 318-bp |
|
| |
| MDR1 C3435T (C>T) | |
| CC | 158-bp, (39-bp) |
| CT | 197-bp, 158-bp, (39-bp) |
| TT | 197-bp |
|
| |
| XRCC1 Arg399Gln (G>A) | |
| GG | 374-bp, 221-bp (20-bp) |
| GA | 615-bp, 374-bp, 221-bp (20-bp) |
| AA | 615-bp |
Results
Evaluation of immunohistochemical reaction
The expression of XRCC1 was seen as a very specific nuclear staining of positive cells (Figure 1). Fourteen (36%) of all samples (n=39) were XRCC1-positive. The rest of the cases (n=25) showed no XRCC1 immunoreaction (Table 3). The stromal background was inert in all cases.
Figure 1.
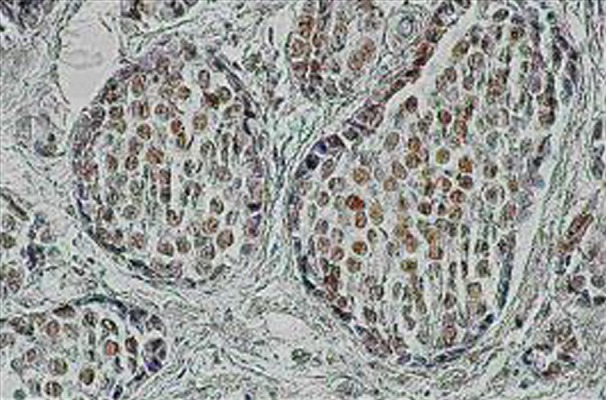
The expression of XRCC1 protein in cancer cells of lobular invasive carcinoma (LIC). Strong brown immunoreaction of cell’s nuclei (magnification: ×200).
Table 3.
Number/percentage of samples with XRCC1, p53 and MDR1 protein expression.
| Protein no 39 | Quantity of expression/% | No/% of negative | No/% of positive | |||
|---|---|---|---|---|---|---|
| (−) | (1+) | (2+) | (3+) | Samples | ||
| XRCC1 | 64% | 0% | 18% | 18% | 64% | 36% |
| p53 | 57% | 5% | 28% | 10% | 64% | 36% |
| MDR1 | 31% | 15% | 33% | 21% | 46% | 54% |
Immunostaining for p53 occurred in a nuclear localization and differed in intensity among individual cases (Figure 2). The number of p5- positive samples (14 [36%] of all cases, showed the same level as XRCC1. Twenty-five (64%) samples showed no p53-positivity (Table 3). The 25 samples that displayed no XRCC1 immunoreaction also showed no p53 immunopositivity.
Figure 2.
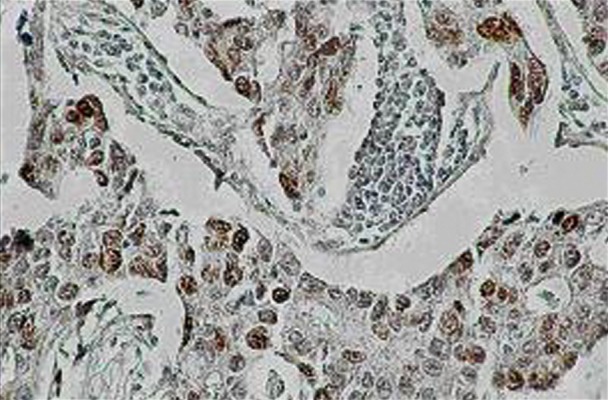
Strong reactivity of tumor cells of ductal invasive carcinoma (DIC), with the p53 monoclonal antibody as opposed to the inert stromal background. The p53 positive cells show clear brown nuclear staining (magnification: ×200).
Expression of MDR1 revealed a diffuse intracellular cytoplasmic localization (Figure 3). The intensity of reaction varied among individual cases. Positive immunostaining for MDR1 was found in 21 (54%) cases, while 18 (46%) samples were MDR1-negative (Table 3).
Figure 3.

Cancer cells of ductal invasive carcinoma with the expression of MDR1 (magnification: ×100).
In all 3 protein samples, we found no immunostaining in control of the immunohistochemical method (without primary antibody).
Relationship between XRCC1 and relevant clinical and pathological parameters
XRCC1 and histopathological type
The breast carcinoma samples were divided into 3 groups according the histopathological type: 1) Ductal invasive carcinoma – DIC (27 samples), 2) Lobular invasive carcinoma – LIC (7 samples) and 3) Other types (papillary and mucinous carcinoma) (5 samples).
Comparison of XRCC1 and histopathological type of tumor revealed that DIC and LIC types are mostly XRCC1-negative. Only other types of samples (papillary and mucinous) showed higher level of XRCC1-positivity. Nine (33%) of the DIC cases (n=27) were XRCC1-positive, whereas 18 (67%) showed no XRCC1 expression. Moreover, LIC samples (n=7) manifested XRCC1 in 1 (14%) case, while the rest of the specimens (n = 6; 86%) were XRCC1-negative. The results suggest a strong difference in samples of other types (n=5), since 4 (80%) samples expressed XRCC1-positivity and only 1 (20%) specimen exhibited no XRCC1 immunoreactivity (Table 4).
Table 4.
Number of patients with positive and negative expression of XRCC1 protein compared to different histopathological type of tumor and expression of Her2/neu in LIC samples.
| XRCC1+ | XRCC1− | ||
|---|---|---|---|
| Positive | Negative | ||
| Histo-pathol. type | |||
| DIC (n=27) | 9/33% | 18/67% | |
| LIC (n=7) | 1/14% | 6/86% | |
| Other (n=5) | 4/80% | 1/20% | |
| Correlation test | |||
| LIC Her2/neu positive (n=7) | 1/14% | 6/86% | R=1 (p<0.05) |
XRCC1 and LIC Her2/neu positive
Interestingly, the negative correlation between the Her2/neu and expression of XRCC1 was detected when evaluating LIC samples separately. Apparently, all LIC samples (n=7) were Her2/neu-positive, with only 1 sample expressing XRCC1. The obtained results represent the negative correlation between Her2/neu-positive samples and XRCC1-negative specimens, according to correlation testing (R=1, p<0.05) (Table 4).
The correlation of XRCC1 with histopathological type, grading, staging, Her2/neu, estrogen, progesterone receptor status and involvement of regional lymph nodes was not statistically significant (data not shown).
XRCC1 and histological grade
XRCC1-positive samples attained the highest level in grade 1 (n=4; 40%) and grade 2 (n=4; 20%). In grade 3 the level of XRCC1 positivity markedly decreased to 1 (11%) sample (Table 5). Nevertheless, statistical analysis did not reveal significant differences between the expression of XRCC1 and histological grade.
Table 5.
Number of patients with positive and negative expression of XRCC1 protein compared to different histological grade.
| XRCC1+ | XRCC1− | |
|---|---|---|
| Positive | Negative | |
| Grade | ||
| G1 (n=10) | 4/40% | 6/60% |
| G2 (n=20) | 4/20% | 16/80% |
| G3 (n=9) | 1/11% | 8/89% |
Relationship between MDR1 and relevant clinical and pathological parameters
MDR1 and histopathological type of tumor
The analysis in Table 6 shows the relationship between the expression of the MDR1 with the histopathological type of breast carcinoma. We found that, in comparison to DIC samples, the positive immunoreactivity score of MDR1 expression was higher in samples of LIC and other types. In cases of LIC (n=7), 5 samples (71%) showed positive immunoreactivity for MDR1 and 2 samples (29%) showed negative immunoreactivity for MDR1. Similarly, other types (papillary and mucinous, n=5) expressed MDR1 in 4 (80%) samples, while 1 (20%) sample manifested no MDR1 immunoreaction. Significantly lower MDR1 expression was found in DIC samples (n=27), where 15 samples (55%) showed negative expression of MDR1 protein and 12 samples (45%) exhibited MDR1-positivity.
Table 6.
Number/percentage of patients with positive and negative expression of MDR1 protein compared to different histopathological type of breast carcinoma.
| Protein | Total number | Histopathol. type | ||
|---|---|---|---|---|
| DIC | LIC | Other | ||
| MDR1 positive (+) | 21/100% | 12/45% | 5/71% | 4/80% |
| MDR1 negative (−) | 18/100% | 15/55% | 2/29% | 1/20% |
DIC – ductal invasive carcinoma, LIC – lobular invasive carcinoma, other types – papillary, mucinous.
Statistical analysis did not reveal any association between expression of MDR1 and the clinical parameters mentioned above (data not shown).
Relationship between p53 and relevant clinical and pathological parameters
Statistical analysis did not reveal a correlation between p53 expression and the following parameters: histopathological type of tumor, grade, stage, Her2/neu expression, ER-positivity, PR-positivity and involvement of regional lymph nodes (data not shown).
Despite the lack of statistical significance of the above results, some of the more interesting findings are depicted in tables.
The comparison of the XRCC1, p53, and MDR1 gene polymorphisms in terms of protein expression
The association of the variant genotypes of the XRCC1 Arg399Gln, p53 Arg72Pro and MDR1 C3435T polymorphisms with expression of relevant proteins in breast cancer tissues was further evaluated, as shown in Tables 7–9. The results revealed no statistically significant difference between the tested polymorphisms and protein expression.
Table 7.
IHC according to XRCC1 gene variants.
| Variable | IHC | p value | OR (95%CI) | |
|---|---|---|---|---|
| Positive (2+3) (n=9) | Negative (0+1) (n=9) | |||
| Allele | ||||
| G | 12 | 40 | 1.00 (Ref.) | |
| A | 6 | 20 | 1.000b | 1.00 (0.33–3.06) |
| Genotype | ||||
| GG | 5 | 13 | 1.00 (Ref.) | |
| GA | 2 | 14 | 0.405b | 0.37 (0.06–2.26) |
| AA | 2 | 3 | 0.621b | 1.73 (0.22–13.68) |
| GA+AA | 4 | 17 | 0.706b | 0.61 (0.14–2.74) |
(χ2-test);
(Fisher’s exact test).
Table 9.
IHC according to MDR1 gene variants.
| Variable | IHC | p value | OR (95%CI) | |
|---|---|---|---|---|
| Positive (2+3) (n=21) | Negative (0+1) (n=18) | |||
| Allele | ||||
| C | 22 | 16 | 1.00 (Ref.) | |
| T | 20 | 20 | 0.485a | 0.73 (0.30–1.78) |
| Genotype | ||||
| CC | 5 | 3 | 1.00 (Ref.) | |
| CT | 12 | 10 | 1.000b | 0.72 (0.14–3.79) |
| TT | 4 | 5 | 0.637b | 0.48 (0.07–3.35) |
| CT+TT | 16 | 15 | 0.702b | 0.64 (0.13–3.16) |
(χ2-test);
(Fisher’s exact test).
Discussion
We have described the expression of XRCC1, p53 and MDR1 proteins by immunohistochemistry and the polymorphisms of their genes by PCR-RFLP in 39 samples of breast carcinoma. The obtained results were subsequently compared to clinical and pathological parameters to discover the most probable mechanism of drug resistance in this disease. We were very careful in drawing general conclusions because of the relatively small number of analyzed samples. In this regard, our results supported the findings found in the literature conducted with large-size sets.
XRCC1
In case of XRCC1, we observed that if LIC samples were evaluated separately, a negative correlation between the Her2/neu and expression of XRCC1 was detected. Apparently, all Her2/neu-positive samples were XRCC1-negative. The correlation test revealed negative correlation between Her2/neu-positive samples and XRCC1-negative specimens (R=1, p<0.05). We were not able to compare this result with other studies because to our knowledge there are no published reports on the effect of XRCC1 in LIC samples on Her2/neu. Our findings indicated no relationship between XRCC1 protein and the histopathological type of tumor, which is in contrast with the results of Dufloth et al. [12], who found a relationship between the expression of XRCC1 and ductal type of breast carcinoma.
High proliferative activity is considered a high grade malignancy of breast carcinoma. In our results, the expression of XRCC1 decreases with grade. We partly agree with Dufloth et al [12], who hypothesized that patients who present the XRCC1 399(Arg/Gln) polymorphism could represent a subgroup of low grade cancers that are in general sporadic rather than hereditary. With reference to Costa et al. [13], XRCC1 Gln/Gln genotype also seems to be associated with less aggressive tumors, since this genotype correlated with well-differentiated tumors. Deficient efficiency of the XRCC1 protein has been described in XRCC1 Gln399 variant [14]. Moreover, the repair of more complex base lesion [15] by BER (base excision repair) pathway can potentially convert non-lethal lesions into lethal double-strand breaks [16]. Thus, deficiency in BER may actually reflect a well-differentiated nature of the tumor cells in less aggressive tumors, since less lethal lesions are produced.
The hormone receptors (estrogen-ER, progesterone-PR) participate in the carcinogenic process in the promotion phase, when expansion of mutated cells occurs. We wanted to know if there is an association between hormone receptor status and XRCC1 expression. The results did not show statistically significant association between XRCC1 and hormonal receptor status. These findings are in agreement with the results of Dufloth et al. [12], who also did not confirm the association between these 2 parameters. Furthermore, polymorphisms of genes that catalyze the balance of oestrogens, progesterones and androgens are part of the steroid hormone pathways and may alter the levels and/or effects of endogenous hormones [17]. In addition, our findings showed no statistically significant relation between XRCC1 and grade, stage of tumor, and involvement of regional lymph nodes.
P53
The inheritance of a Tp53 mutant allele results in a rare familial autosomal disorder, the Li-Fraumeni syndrome. It is characterized by a high incidence of multiple early cancers, including breast tumors. Many investigators have examined the value of p53 as an unfavorable prognostic marker in breast cancer. According to our results, 14 (36%) specimens demonstrated p53 positivity. Similarly, Vousden and Lu [18] and Borresen-Dale [19] indicated that between 20% and 40% of breast tumors have been shown to express a mutant p53. Generally, mutant p53 generally has an increased stability and accumulates in the nuclei of neo-plastic cells. Immunohistochemical detection of the amount of nuclear p53 has long been used as an indicator of p53 alteration, but this parameter appears highly dependent on the type of mutation [20]. Investigations of a “mutational hybrid” of p53 emphasized that wild-type p53 does not compensated for mutated p53; instead, it may be associated with a worse prognosis [21]. It is believed that mutation of p53 is a consequence of the inactive p53. However, it must be noted that not all p53 mutations are inactivating. Some mutant p53s display only partial loss of their DNA-binding activity [22]. In agreement with Montero et al. [23], disease-free survival curves showed that patients with p53-positive tumors had a significantly shorter disease-free survival compared to patients with p53-negative carcinomas. Tsutsui et al. [24] confirmed the findings of Montero et al. [23], and added that patients with p53-positive tumors had significantly worse overall survival compared to p53-negative patients.
Her2/neu amplification/overexpression is a marker of poor prognosis in breast cancer, and is one of the significant independent prognostic factors. Her2/neu+ tumors also containing p53 abnormalities (overexpression) tend to be hormone receptor- and bcl-2-negative, and have lymphoid infiltration and a high mitotic index [25]. With reference to our results, the majority of our samples were both Her2/neu- and p53-negative, which could predict good prognosis for this group of patients. Korkolis et al. [26] found a strong correlation between Her2/neu and p53 overexpression. Moreover, Chariyalertsak et al. [27] confirmed the significant correlation between p53 expression and Her2/neu (p<0.01). Erdem et al. [28] found that p53 protein expression correlated positively with Her2/neu expression. According to de Roos et al. [29], overexpression of Her2/neu and p53 was associated with local recurrence in patients treated for DIC and DCIS (ductal carcinoma in situ). In spite of previous observations, we cannot confirm any association between the expression of p53 protein and relevant clinical and pathological parameters.
MDR1
The unfavorable prognostic significance of MDR1 expression has been documented in several tumors, including breast cancer [30]. Most of the studies documented the negative prognostic significance in breast cancer with MDR phenotype treated with chemotherapy. Few of the studies have suggested that MDR1 participates in the resistance to hormonal therapy [31]. According to our results, the highest level among all studied proteins was observed in MDR1 protein, since 54% of samples demonstrated its positivity. These findings correspond with our previous findings and also with observations of other researchers. In a previous study we found that 57% of breast carcinomas show MDR1-positivity [32]. Other investigators have detected MDR1 expression in 55% [33] and 57% [34]. However, some authors present different levels of MDR1 in breast carcinoma. Faneyte et al [35] found no MDR1 expression in the tumor cells of 80 chemotherapy-naive tumors, while Yu et al. [36] detected only 41% of MDR1-positive breast carcinoma.
Next, we have shown that MDR1 immunoreactivity score is associated with the histopathological type of tumor. In cases of LIC, most of samples manifested MDR1-positivity, similar to other types (papillary and mucinous). Most DIC specimens were MDR1-negative. According to type of tumor, we could speculate about the relationship between the expression of MDR1 and appropriate therapy with regard to MDR1 substrate specificity (drug resistance mediated by this transporter). Finally, similar to results reported by Turgut et al. [37], we did not find any statistically significant difference between clinicopathological parameters and MDR1 of breast cancer patients.
Polymorphisms
Polymorphisms in breast cancer susceptibility genes with low-penetrance have been shown to contribute to breast tumorigenesis in combination with exogenous and endogenous exposures [38]. Nevertheless, confusion exists regarding the role of gene polymorphisms in cancer risk or overall prognosis and their role in drug response. According to the latest paper by Hosseini et al. [39], breast cancer might be significantly associated with 2 functional single-nucleotide polymorphisms (SNPs) in the 5,10-methylenetetrahydrofolate reductase (MTHFR) genes C677T and A1298C.
IL-6 can play an important role in pathogenesis of breast cancer. Its level in patients with breast cancer is higher than in the control group, irrespective of distribution of genotypes and frequency of the IL-6 (−174) C/G polymorphism [40]. On the other hand, genetic variants of TNF-related apoptosis-inducing ligand (TRAIL) might be associated with progression of breast cancer [41].
We tested immunohistochemical protein expression in relation to XRCC1 Arg399Gln, p53 Arg72Pro, and MDR1 C3435T polymorphisms.
To date, 12 observational studies have examined XRCC1 polymorphisms 194(Arg/Trp) and 399(Arg/Gln) in relation to breast cancer risk, with inconsistent results [42]. With regard to Sterpone et al. [43], a significant association was found between breast cancer occurrence and XRCC1 399(Arg/Gln) polymorphism in Caucasian women. Conversely, Saadat [44] found no significant association between XRCC1 haplotypes and risk of breast cancer among Caucasian subjects. Similarly, another 2 studies confirmed the association between XRCC1 399(Arg/Gln) polymorphism and breast cancer risk among African-Americans, but not among Caucasians [45,46], indicating that this polymorphism may be linked to another biologically effective mutation. Some of these studies suggested that associations between XRCC1 polymorphisms and breast cancer risk are stronger in women who have higher exposure to various antioxidants, including smoke [47]. The former study observed an association of breast cancer risk with all XRCC1 polymorphisms aside from 194(Arg/Trp) and 399(Arg/Gln) [48]. They observed an association between increased breast cancer risk and the 280(Arg/His) polymorphism in their study population (n=250 cases). Polymorphisms 194(Arg/Trp) and 399(Arg/Gln) were shown to affect increased mutagen sensitivity after bleomycin treatment, a radiation-mimicking agent that induces double-strand breaks in DNA [49]. The polymorphism in coding region of the XRCC1 gene at codon 399(Arg399Gln) could alter the XRCC1 function. Previous studies have demonstrated the significant association of Gln allele with a higher level of DNA adducts and glycophorin A mutations in erythrocytes [50,51], increased sister chromatid exchange frequencies [52,53], and higher sensitivity to ionizing radiation [54]. The present study did not find significant correlation between XRCC1 Arg399Gln polymorphism, and protein expression was found (p=0.405–1.000, χ2-test or Fisher’s exact test). In contrast to our results, Cheng et al. [55] found that the protein expression was significantly higher in patients with a Gln allele (ArgGln or GlnGln) than in patients with the ArgArg genotype in locally advanced cervical carcinoma.
Recent studies have suggested that genetic polymorphisms in the TP53 pathway influence tumor formation, progression, and response to therapy. A number of polymorphisms have been identified in the TP53 gene. The codon 72 polymorphism (G>C) in exon 4 of the p53 gene, which is carried by 20±40% of the population, leads to an arginine-to-proline substitution. The polymorphism Arg72Pro is the most frequent in Caucasian populations and its frequency varies from the equator to higher latitudes, suggesting a selection pressure upon 2 forms of p53 protein, Arg72Pro and Pro47Ser [56]. In the Turkish population the frequency of Arg72Pro genotype was 41.4% in patients [57]. Bisof et al [58] provided evidence of the association of the TP53 gene polymorphisms Arg72Pro and PIN3 (+16bp) with sporadic breast cancer in the Croatian population.
We did not observe any significant difference in p53 protein expression between particular alleles and genotypes of this polymorphism (p=0.437–1.000, χ2-test or Fisher’s exact test). Similarly, Akkiprik et al. [59] did not find an association between certain genotypes of p53 Arg72Pro polymorphism and the protein over-expression in breast cancer patients. On the other hand, several authors confirmed that the Pro variant is less efficient in suppressing cell transformation and slower in inducing apoptosis compared to the Arg variant [60].
Recent studies have reported the association of a silent C3435T polymorphism in exon 26 of MDR1 with altered mRNA and protein expression or MDR1 function [61,62]. C3435T polymorphism in the Caucasian population in the CC genotype was found in 46 (20.8%) patients and in 35 (31%) control subjects [61,62]. According to Čižmáriková et al. [63], the C allele was found in 45.2% of breast cancer patients and 54.9% of healthy donors, while the T allele was found in 54.8% of patients and 45.1% of controls. Similarly, results from Turgut et al. [37] indicated that the T allele was more frequently found in breast cancer patients compared to controls. Our results did not confirm the relationship between the tested polymorphism and protein expression (IHC) in breast cancer tissue (p=0.485–1.000, χ2-test or Fisher’s exact test). Although the original study by Hoffmeyer et al. [61] showed a borderline-significant association (p=0.056), other studies have failed to replicate this finding [64–66].
Conclusions
The most remarkable findings of this work suggest that the MDR1 protein has the highest protein expression when compared to XRCC1 and p53 proteins. Moreover, our results indicated a negative correlation between XRCC1 and Her2/neu in LIC samples, and we found a lack of relationship between tested polymorphisms and protein expression.
Table 8.
IHC according to p53 gene variants.
| Variable | IHC | p value | OR (95%CI) | |
|---|---|---|---|---|
| Positive (2+3) (n=14) | Negative (0+1) (n=25) | |||
| Allele | ||||
| A1 | 9 | 12 | 1.00 (Ref.) | |
| A2 | 19 | 38 | 0.437a | 0.67 (0.24–1.86) |
| Genotype | ||||
| A1A1 | 2 | 2 | 1.00 (Ref.) | |
| A1A2 | 5 | 8 | 1.000b | 0.63 (0.07–5.97) |
| A2A2 | 7 | 15 | 0.591b | 0.47 (0.05–4.03) |
| A1A2+A2A2 | 12 | 23 | 0.609b | 0.52 (0.07–4.18) |
(χ2-test);
(Fisher’s exact test).
Acknowledgements
We thank Ivana Čigašová-Perunská and Diana Gagyiová for their skillful technical assistance.
Abbreviations
- XRCC1
X-ray repair cross-complementing protein 1
- MDR1
multidrug resistance 1 genes
- DIC
ductal invasive carcinoma
- LIC
lobular invasive carcinoma
- PCR-RFLP
polymerase chain reaction-restriction fragment length polymorphism assays
- AEC
aminoethylcarbazole
- DAB
3,3′-diaminobenzidine tetrahydrochloride
Footnotes
Conflict of interest
The authors declare no conflict of interest in relation to the article.
Source of support: This work was supported by grant VEGA No. 1/0388/08, VEGA No. 1/0050/11 awarded by the Ministry of Education. It was also partially supported by the Agency of the Slovak Ministry of Education for the Structural Funds of the EU, under projects ITMS: 26220120039
References
- 1.Jemal A, Thomas A, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Lamerdin J, Montegomery M, Stilwagen S, et al. Genomic sequence comparison of the human and mouse XRCC1 DNA repair gene regions. Genomics. 1995;25:547–54. doi: 10.1016/0888-7543(95)80056-r. [DOI] [PubMed] [Google Scholar]
- 3.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exit at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8. [PubMed] [Google Scholar]
- 4.Helzlsouer KJ, Harris EL, Parshad R, et al. DNA repair proficiency: potential susceptibility factor for breast cancer. J Natl Cancer Inst. 1996;88:754–55. doi: 10.1093/jnci/88.11.754. [DOI] [PubMed] [Google Scholar]
- 5.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 6.Greenblatt MS, Bennet WP, Hollstein M, et al. Mutation of p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- 7.Gayet L, Dayan G, Barakat S, et al. Control of P-glycoprotein activity by membrane cholesterol amounts and their relation to multidrug resistance in human CEM leukemia cells. Biochemistry. 2005;44:4499–509. doi: 10.1021/bi048669w. [DOI] [PubMed] [Google Scholar]
- 8.Trock BJ, Leonessa F, Clarke R. Multidrug resistance in breast cancer: a meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst. 1997;2:917–31. doi: 10.1093/jnci/89.13.917. [DOI] [PubMed] [Google Scholar]
- 9.Chung HC, Rha SY, Kim JH, et al. P-glycoprotein: the intermediate end point of drug response to induction chemotherapy in locally advanced breast cancer. Breast Cancer Res Treat. 1997;42:65–72. doi: 10.1023/a:1005739525196. [DOI] [PubMed] [Google Scholar]
- 10.Sobin LH, Wittekind C. TNM classification of malignant tumours. 5. New York: Wiley-Liss Inc; 2002. [Google Scholar]
- 11.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–10. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 12.Dufloth RM, Arruda A, Heinrich JKR, et al. The investigation of DNA repair polymorphisms with histopathological characteristics and hormone receptors in a group of Brazilian women with breast cancer. Gen Mol Res. 2008;7:574–82. doi: 10.4238/vol7-3gmr376. [DOI] [PubMed] [Google Scholar]
- 13.Costa S, Pinto D, Pereira D, et al. XRCC1 Arg399Gln and RAD53 5′UTR G135C polymorphisms and their outcome in tumor aggressiveness and survival of Portuguese breast cancer patients. Breast Cancer Res Treat. 2008;109:183–85. doi: 10.1007/s10549-007-9637-1. [DOI] [PubMed] [Google Scholar]
- 14.Qu T, Morii E, Oboki K, et al. Micronuclei in EM9 cells expressing polymorphic forms of human XRCC1. Cancer Lett. 2005;221:91–95. doi: 10.1016/j.canlet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland BM, Bennet PV, Sutherland JC, et al. Clustered DNA damages induced by x rays in human cells. Radiat Res. 2002;157:611–16. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Ward JF. Radiation mutagenesis: the initial DNA lesions responsible. Radiat Res. 1995;142:362–68. [PubMed] [Google Scholar]
- 17.de Jong MM, Nolte IM, te Meerman GJ, et al. Genes other than BRCA1 and BRCA2 involved in breast cancer susceptibility. J Med Genet. 2002;39:225–42. doi: 10.1136/jmg.39.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nature Reviews Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 19.Borresen-Dale AL. TP53 and breast cancer. Hum Mutat. 2003;21:292–300. doi: 10.1002/humu.10174. [DOI] [PubMed] [Google Scholar]
- 20.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 21.Baumbusch LO, Myhre S, Langerod A, et al. Expression of full-lenght p53 and its isoform Δp53 in breast carcinomas in relation to mutation status and clinical parameters. Mol Cancer. 2006;5:47. doi: 10.1186/1476-4598-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedlander P, Haupt Y, Prives C. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Molecular and Cellular Biology. 1996;16:4961–71. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montero S, Guzmán C, Vargas C, et al. Prognostic value of cytosolic p53 protein in breast cancer. Tumour Biol. 2001;22:337–44. doi: 10.1159/000050636. [DOI] [PubMed] [Google Scholar]
- 24.Tsutsui S, Ohno S, Murakam S. Prognostic value of p53 protein expression in breast cancer, an immunohistochemical analysis of frozen sections in 514 Japanese women. Breast Cancer. 2001;8:194–201. doi: 10.1007/BF02967508. [DOI] [PubMed] [Google Scholar]
- 25.Ménard S, Fortis S, Castiglioni F, et al. Her2 as a prognostic factor in breast cancer. Oncology. 2001;61:67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- 26.Korkolis D, Ardavanis A, Yotis J, et al. Her2/neu overexpression in breast cancer: an immunohistochemical study including correlation with clinicopathologic parameters, p53 oncoprotein and cathepsin-D. Anticancer Res. 2001;21:2207–12. [PubMed] [Google Scholar]
- 27.Chariyalertsak S, Chariyalertsak S, Cheirsilpa A, et al. Prognostic importace of p53 and c-erbB-2 oncoproteins overexpression in patiens with breast cancer. J Med Assoc Tha. 1998;81:698–704. [PubMed] [Google Scholar]
- 28.Erdem O, Dorsun A, Coşkun U, et al. The prognostic value of p53 and c-erbB-2 expression, proliferative activity and angiogenesis in node-negative breast carcinoma. Tumori. 2005;91:46–52. doi: 10.1177/030089160509100109. [DOI] [PubMed] [Google Scholar]
- 29.de Roos MA, de Bock GH, de Vries J, et al. p53 overexpression is a predictor of local reccurence after treatment for both in situ and invasive ductal carcinoma of the breast. Surg Res. 2007;140:109–14. doi: 10.1016/j.jss.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 30.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 31.Linn SC, Giaccone G, van Diest PJ, et al. Prognostic relevance of P-glycoprotein expression in breast cancer. Ann Oncol. 1995;6:697–85. doi: 10.1093/oxfordjournals.annonc.a059284. [DOI] [PubMed] [Google Scholar]
- 32.Rybárová S, Hodorová I, Hajduková M, et al. Expression of MDR proteins in breast cancer and its correlation with some clinical and pathological parameters. Neoplasma. 2006;53:128–35. [PubMed] [Google Scholar]
- 33.Rudas M, Filipits M, Taucher S, et al. Expression of MRP1, LRP and Pgp in breast carcinoma treated with preoperative chemotherapy. Breast Cancer Res Treat. 2003;81:149–57. doi: 10.1023/A:1025751631115. [DOI] [PubMed] [Google Scholar]
- 34.Li EX, Li Y, Yang J, et al. Influence of P-glycoprotein expression on chemotherapeutic response of metastatic breast carcinoma. Ai Zheng. 2002;21:430–32. [PubMed] [Google Scholar]
- 35.Faneyte IF, Kristel PM, van de Vijver MJ. Multidrug resistence associated genes MRP1, MRP2 and MRP3 in primary and anthracycline exposed breast cancer. Anticancer Res. 2004;24:2931–39. [PubMed] [Google Scholar]
- 36.Yu P, Xiao NX, Chen YP. Expression of P-glycoprotein and lung resistence protein in brest carcinoma and its relationship with prognosis. Ai Zheng. 2003;22:1339–42. [PubMed] [Google Scholar]
- 37.Turgut S, Yaren A, Kursunluoglu R, et al. MDR1 C3435T polymorphism in patients with breast cancer. Arch Med Res. 2007;38:539–44. doi: 10.1016/j.arcmed.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Rothman N, Wacholder S, Caporaso NE, et al. The use of common genetic polymorphisms enhance the epidemiologic study of environmental carcinogens. Biochim Biophys Acta. 2001;1471:C1–10. doi: 10.1016/s0304-419x(00)00021-4. [DOI] [PubMed] [Google Scholar]
- 39.Hosseini M, Houshmand M, Ebrahimi A. MTHFR polymorphisms and breast cancer risk. Arch Med Sci. 2011;1:134–37. doi: 10.5114/aoms.2011.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jabłonowska E, Kołacinska A, Kuydowicz J, et al. Interleukin-6 and the IL-6 (–174) C/G polymorphism in breast pathologies and in HIV-infected patients. Arch Med Sci. 2010;6:860–65. doi: 10.5114/aoms.2010.19292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildiz Y, YaylIm-Eraltan I, Arikan S, et al. Is there any correlation between TNF-related apoptosis-inducing ligand (TRAIL) genetic variants and breast cancer? Arch Med Sci. 2010;6:932–36. doi: 10.5114/aoms.2010.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duell EJ, Millikan RC, Pittman GS, et al. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:217–22. [PubMed] [Google Scholar]
- 43.Sterpone S, Mastellone V, Padua L, et al. Single-nucleotide polymorphisms in BER and HRR genes, XRCC1 haplotypes and breast cancer risk in Caucasian women. J Cancer Res Clin Oncol. 2010;4:631–36. doi: 10.1007/s00432-010-0791-1. [DOI] [PubMed] [Google Scholar]
- 44.Saadat M. Haplotype analysis of XRCC1 (at codons 194 and 399) and susceptibility to breast cancer, a meta-analysis of the literatures. Breast Cancer Res Treat. 2010;124(3):785–91. doi: 10.1007/s10549-010-0895-y. [DOI] [PubMed] [Google Scholar]
- 45.Duell EJ, Millikan RC, Pittman GS, et al. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:217–22. [PubMed] [Google Scholar]
- 46.Nexo BA, Vogel U, Olsen A, et al. A specific haplotype of single nucleotide polymorphisms on chromosome 19q13.2-3 encompassing the gene RAI is indicative of post-menopausal breast cancer before age 55. Carcinogenesis. 2003;24:899–904. doi: 10.1093/carcin/bgg043. [DOI] [PubMed] [Google Scholar]
- 47.Shen J, Gammon MD, Terry MB, et al. Polymorphisms in XRCC1 modify the association between polycyclic aromatic hydrocarbon-DNA adducts, cigarette smoking, dietary antioxidants, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:336–42. doi: 10.1158/1055-9965.EPI-04-0414. [DOI] [PubMed] [Google Scholar]
- 48.Moullan N, Cox DG, Angele S, et al. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol Biomarkers Prev. 2003;12:1168–74. [PubMed] [Google Scholar]
- 49.Wang Y, Spitz MR, Zhu Y. From genotype to phenotype: correlatin XRCC1 polymorphisms with mutagen transporter. DNA Repair (Amst.) 2003;2:901–8. doi: 10.1016/s1568-7864(03)00085-5. [DOI] [PubMed] [Google Scholar]
- 50.Lunn RM, Langlois RG, Hsieh LL, et al. Xrcc1 polymorphisms: effects on aflatoxin b1-dna adducts and glycophorin a variant frequency. Cancer Res. 1999;59:2557–61. [PubMed] [Google Scholar]
- 51.Matullo G, Guarrera S, Carturan S, et al. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int J Cancer. 2001;92:562–67. doi: 10.1002/ijc.1228. [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Rahman SZ, Soliman AS, Bondy ML, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159:79–86. doi: 10.1016/s0304-3835(00)00537-1. [DOI] [PubMed] [Google Scholar]
- 53.Duell EJ, Wiencke JK, Cheng TJ, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damag in human blood mononuclear cells. Carcinogenesis. 2000;21:965–71. doi: 10.1093/carcin/21.5.965. [DOI] [PubMed] [Google Scholar]
- 54.Hu JJ, Smith TR, Miller MS, et al. Genetic regulation of ionizing radiation sensitivity and breast cancer risk. Environ Mol Mutagen. 2002;39:208–15. doi: 10.1002/em.10058. [DOI] [PubMed] [Google Scholar]
- 55.Cheng XD, Lu WG, Ye F, et al. The association of XRCC1 gene single nucleotide polymorphisms with response to neoadjuvant chemotherapy in locally advanced cervical carcinoma. J Exp Clin Cancer Res. 2009;28:91. doi: 10.1186/1756-9966-28-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckman G, Birgander R, Sjalander A, et al. Is p53 polymorphism maintained by natural selection? Hum Hered. 1994;44:266–70. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 57.Kara N, Karakus N, Ulusoy AN, et al. P53 codon 72 and HER2 codon 655 polymorphisms in Turkish breast cancer patients. DNA Cell Biol. 2010;29:387–92. doi: 10.1089/dna.2009.0995. [DOI] [PubMed] [Google Scholar]
- 58.Bisof V, Salihovic MP, Narancic NS, et al. TP53 gene polymorphisms and breast cancer in Croatian women: a pilot study. Eur J Gynaecol Oncol. 2010;31:539–44. [PubMed] [Google Scholar]
- 59.Akkiprik M, Sonmez O, Gulluoglu BM, et al. Analysis of p53 gene polymorphisms and protein over-expression in patients with breast cancer. Pathol Oncol Res. 2009;15:359–68. doi: 10.1007/s12253-008-9129-6. [DOI] [PubMed] [Google Scholar]
- 60.Dumont P, Leu JI, Della Pietra AC, et al. The codon 72 polymorphic variant of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 61.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci, USA. 2000;97:3473–78. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimchi-Sarfaty Ch, Oh JM, Kim IW, et al. A“silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–28. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 63.Čižmáriková M, Wagnerová M, Schonova L, et al. MDR1 (C3435T) polymorphism: relation to the risk of breast cancer and therapeutic outcome. Pharmacogenomics J. 2009;10:62–69. doi: 10.1038/tpj.2009.41. [DOI] [PubMed] [Google Scholar]
- 64.Owen A, Goldring C, Morgan P, et al. Relationship between the C3435T and G2677T(A) polymorphisms in the ABCB1 gene and P-glycoprotein expression in human liver. Br J Clin Pharmacol. 2005;59:365–70. doi: 10.1111/j.1365-2125.2005.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siegmund W, Ludwig K, Giessmann T, et al. The effects of the human MDR1 genotype on the expression of duodenal P-glycoprotein and disposition of the probe drug talinolol. Clin Pharmacol Ther. 2002;72:572–83. doi: 10.1067/mcp.2002.127739. [DOI] [PubMed] [Google Scholar]
- 66.Yoshikawa T, Kanazawa H, Tanaka J, et al. Gene polymorphism of epidermal growth factor receptor and airway hyperresponsiveness in young allergic subjects without respiratory symptoms. Med Sci Monit. 2010;16(4):CR163–71. [PubMed] [Google Scholar]


