Summary
Background
The expression of microRNA-206 (miR-206) is high in skeletal muscle but low in most other tissues. The expression of miR-206 is increased in muscular dystrophy, suggesting its involvement in the pathogenesis of muscle diseases. To determine the role of miR-206 in muscle cell differentiation and explore a possible gene therapy vector, we constructed a miR-206 adenoviral expression vector (AdvmiR-206) and tested for transfection into C2C12 stem cells.
Material/Methods
A 355-bp PCR amplicon from C57B6 mouse skeletal muscle genomic DNA was inserted into the adenoviral shuttle vector pAdTrack-CMV, which was then co-transformed with the adenoviral backbone plasmid pAdEasy-1 into competent E. coli BJ5183 bacteria. The specificity and function of this recombinant adenoviral MiR-206 were studied in C2C12 cells by Northern blot, immunofluorescence, Western blot, and flow cytometry.
Results
Increased expression of miR-206 in AdvmiR-206 transfected C2C12 cells (P<0.001) and resulted in morphological and biochemical changes over time that were similar to serum deprivation, including elongated cells and increased myosin heavy chain proteins. Even in the absence of serum deprivation, miR-206 overexpression accounted for a 50% reduction of S-phase cells (P<0.01). Moreover, in untransfected C2C12 cells, the introduction of miR-206-specific antisense oligoribonucleotides inhibited the normal response to serum deprivation. Twenty-four hours after lipofection of antisense oligoribonucleotides, the number of elongated cells was reduced by half (P<0.01).
Conclusions
Collectively, these data support a role for miR-206 in myoblast differentiation. We foresee potential applications for the AdvmiR-206 vector in research and therapy.
Keywords: microRNA-206, myogenesis, C2C12 cells, adenoviral vector
Background
MicroRNAs are small non-coding RNAs that negatively regulate gene expression or promote mRNA degradation by binding to complementary sequences in the 3′ untranslated regions (UTRs) of target genes [1,2]. More than 30% of protein-coding genes are regulated by microRNAs [3], which have been implicated in diverse biological and pathological processes, including apoptosis, differentiation, metabolism, and carcinogenesis [4–8]. Hundreds of microRNAs have been identified to date. For the majority, the biological functions and relevant mechanisms are unknown [9].
During development, the miR-1 (miR-1-1, miR-1-2, and miR-206) and miR-133 families (miR-133a-1, miR-133a-2, and miR-133b) of microRNAs appear to be specifically involved in muscle cell differentiation [10,11]. Among them, the expression of miR-206 is more specifically for skeletal muscle [12,13]. The first study by Rao et al. showed that, during the myoblast-myotube transition in primary human myoblasts and mouse mesenchymal C2C12 stem cell lines; the expression of miR-206 was robustly induced, along with several other conserved muscle-specific microRNAs. This indicated that the induction of miR-206 was important for regulating the expression of muscle-specific proteins [14]. Later, Kim et al. reported a similar observation and identified the p180 subunit of DNA polymerase alpha and three other genes as direct targets of miR-206 [9]. The more recent studies showed that miR-206 regulates skeletal muscle satellite cell proliferation and differentiation by repressing Paired-box transcription factor Pax7 [15,16] and Pax 3 [17,18]. The function of miR-206 could be modulated by TGF-beta through regulation of histone deacetylase 4 [19]. Increasing studies in the field have been able to identify that there are more functions of miR-206 in muscular hypertrophy, atrophy, and dystrophy. For example, up-regulation of miR-206 was related to skeletal muscle hypertrophy [20]. Compared to the control mouse, the miR-206 expression was significantly elevated in the dystrophic tibialis anterior muscles of the MDX mouse. The MDX mouse was an inbred C57BL mouse strain that arose from a spontaneous mutation, and displayed a phenotype similar to human muscular dystrophy. There was a marked increase in the expression of miR-206 found in newly formed myotubes, which reflected active regeneration and efficient maturation of skeletal muscle fibres [21]. MiR-206 was induced in Duchenne muscular dystrophy patients during muscular regeneration [22]. Another study by Williams et al. showed that the induction of miR-206 expression delayed amyotrophic lateral sclerosis progression and promoted regeneration of neuromuscular synapses in mice [23].
More recently, a study by Nakasa et al. showed that a local injection of double-stranded miR-206 combined with two other microRNAs could accelerate muscle regeneration in a rat skeletal muscle injury model [24]. However, local injection of microRNA only provided a temporary induction of target genes. To investigate the possibility of a prolonged effect of miR-206, we constructed an adenoviral mouse miR-206 expression vector for transfection and tested it in vitro in C2C12 myoblasts. As a complementary approach, we measured miR-206 levels in serum-deprived C2C12 cells in the presence and absence of a specific antisense 2′-O-methyl-modified oligoribonucleotide.
Material and Methods
Animals and tissues
C57B6 male mice (n=3) aged 8 weeks were obtained from the Jackson Laboratories. The muscle, liver, spleen, kidney, heart, brain, lung, and intestine tissues were removed from mice that had been sacrificed by decapitation. All procedures were performed according to the humane and customary care and use of experimental animals, and followed a protocol approved by the Internal Animal Care and Use Committee of the Third Military Medical University.
The tissues were trimmed then homogenized with 1 ml RNAiso reagent (TaKaRa, Japan). After 5 minutes incubation, 0.2 ml CHCl3 was added into each sample, mixed and left at room temperature for 5 minutes. The samples were spin down at 12,000 g at 4°C for 15 minutes. The total RNAs in the upper phase were precipitated with 0.5 ml isopropanol then washed in 1 ml 75% ethanol. The RNAs were re-dissolved in DEPC-treated H2O.
Vector construction
Using the miRBase sequence database (http://microrna.sanger.ac.uk/sequence), we identified the nucleotide sequence for the pre-microRNA-206 gene for matching to the mouse genome database. We used Primer 5.0 software (Premier Biosoft International, USA) to design the following primers, which are complementary to the nucleotide sequences that flanked the miR-206 gene:
Forward Primer: 5′-CTAGCTAGCGTTTCTGGGAGTGTAG AATGGATG-3′
Reverse Primer: 5′-CGCGGATCCTGGGGAGCATAGTT GACCTG-3′
We amplified a 355-bp PCR product from C57B6 mouse skeletal muscle genomic DNA. The PCR amplicon was inserted into the adenoviral shuttle vector pAdTrack-CMV, which was then co-transformed with the adenoviral backbone plasmid pAdEasy-1 (a gift from Dr. Shi-Wu Dong; Anatomy Department of the Third Military Medical University, China) into competent E. coli BJ5183 bacteria. Optimal transfection conditions, based on high transduction efficiency and low toxicity, were determined by the percentage of green fluorescent protein (GFP)-positive cells across a range of virus concentrations (plaque forming unit; pfu).
Cell culture, transfection, and cytotoxicity
C2C12 myoblasts (ATCC #: CRL-1772) were maintained at sub-confluent densities in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Grand Island, NY) containing 20% heat-inactivated fetal bovine serum (FBS; GIBCO), penicillin G (100 IU/ml), and streptomycin (100 mg/ml). Myogenic differentiation was induced by changing the sub-confluent cells to DMEM containing 2% heat-inactivated horse serum (DM) [25]. For adenovirus transfection, the culture was maintained in a humid incubator at 37°C in 5% CO2. C2C12 cells were seeded at 2×105 cells per well on 6-well plates in DMEM containing 20% FBS. Twenty-four hours later, the cells were exposed to either AdvmiR-206 or AdvEGFP (control) for 1 hour. We measured the cell viability by following the manufacturer’s instructions of the CCK-8 assay method (Dojindo Laboratories, Mashiki, Japan),; in order to monitor the cellular toxicity induced by transfection, In short, C2C12 cells were seeded into 96-well plates at 2000 cells/well. After 24 hours, C2C12 cells were infected with the control/AdvmiR-206 adenovirus at a variety of MOIs, and five replicate wells were plated for each MOI. Cell viability was determined 3 days after adenoviral infection by absorbance at 450 nm.
For antisense inhibition studies, we identified a candidate sequence:
5′-CCACACACUUCCUUACAUUCCA-3′ for Phosphorothioated 2′-O-methyl modification [26]. The antisense inhibitor of miR-206 and the negative control oligonucleotides were synthesized by GeneChem Biotech Co (Shanghai, China). For antisense inhibition studies, C2C12 cells were cultured to 30% confluence and transfected with either 100 nM antimiR-206 or negative control RNA by Lipofectamine™ 2000 (Invitrogen, USA) according to the manufacturer’ instructions. Twenty four hours after transfection, the growth media of cells were substituted by differentiation media and induced myogenic differentiation for 48 h.
Northern blot
Total RNA was isolated from mouse tissues (brain, intestine, skeletal muscle, spleen, kidney, liver, cardiac muscle, lung) and cultured cells using the Trizol® reagent (Invitrogen, USA) according to the manufacturer’s instructions. Major RNA classes were separated on a 15% denaturing polyacrylamide gel and then transferred to a nylon membrane (GeneScreen Plus, Perkin-Elmer, Inc., Waltham, MA) using a Trans-Blot electrophoretic transfer apparatus (BioRad, USA). RNAs were cross-linked to the membrane by UV exposure (1200J, Queue, USA) and heat (80°C for 30 min). Oligonucleotide probes for microRNA-206 (anti-miR-206, 5′-ccacacacttccttacattcca) and a small RNA control (anti-U6, 5′-Atatggaacgcttca att) [26] were prepared by g-32P-ATP labeling according to the manufacturer’s instructions (Ji Kai Biotech Co., Shanghai, China). The membrane was pre-hybridized in ExpressHyb solution (Ambion, USA) at 65°C for at least 1 hour and hybridized with g-32P ATP labeled oligonucleotide probes in fresh ExpressHyb solution at 37°C for 12–24 hours. The blots were exposed to X-ray film at −70°C for 48–72 hours.
Immunofluorescence
Twenty-four hours after transfection, the cells were fixed in 4% paraformaldehyde-PBS for 10 min, permeabilized in 0.1% NP-40-PBS for 15 min, and then incubated overnight in a rabbit polyclonal primary antibody directed against the myosin heavy chain (anti-MHC; Santa Cruz Biotech, USA) at a 1:160 dilution in PBS with 3% BSA and 0.1% NP-40. After rinsing with PBS, the cells were incubated for 1 hour with secondary antibody (fluorescein or rhodamine-conjugated goat anti-rabbit IgG) at a 1:100 dilution (Zhong Shan Reagent Co., Beijing, China). Cells were washed once with DAPI working buffer, incubated with DAPI-methanol solution (1 μg/ml) for 5 minutes, and then washed once in methanol. After having been washed in PBS thrice, the cells were visualized with an immunofluorescence microscope.
Western blot for MHC protein detection
C2C12 cells were harvested and washed once in PBS followed by centrifugation at 500×g for 5 minutes. The supernatant was discarded and dissociated cell pellets were incubated in RIPA buffer (Roche) on ice for 10 minutes, and then centrifuged at 14,000×g at 4°C for 15 minutes. The supernatants were collected and protein concentrations were measured using the Bradford method. Forty micrograms of total protein from each sample were separated on 12% Tris-HCl PAGE gels and electrotransferred to PVDF membranes. Primary antibodies against MHC (Santa Cruz, rabbit polyclonal, 1:1000) and b-tubulin (Sigma, mouse monoclonal, 1:1000) were detected with HRP-conjugated anti-rabbit (Sigma) or HRP-conjugated anti-mouse antibodies (Sigma) and the ECLplus Western Blotting Detection Kit (Amersham).
Flow cytometric (FCM) analysis of cell cycle phase
Twenty-four hours after transfection, the cells were spun down, and the supernatant was discarded. Cell pellets were re-suspended in 0.25% trypsin-0.02% EDTA solution, spun down, washed in cold PBS (4°C), and fixed with 70% ethyl alcohol at −20°C. The final cell suspension was stored at −4°C for 24 hours, and then stained with 1 μg ml propidium iodide for analysis by flow cytometry.
Results
In vivo and in vitro expression of microRNA-206
We observed significant differences in the abundance of miR-206 across C57BL mouse tissues (Figure 1A). A very strong miR-206 signal was observed only in skeletal muscle, and there was not any detectable miR-206 signal observed in the brain, intestine, spleen, kidney, liver, or cardiac muscle. This finding confirmed the specificity of our g32P-labelled Northern blot probe.
Figure 1.
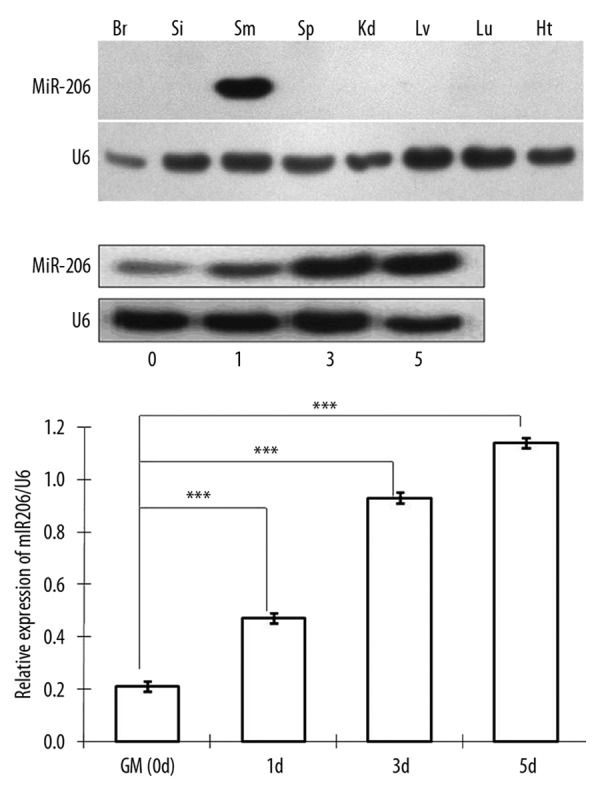
Skeletal muscle-specific expression of the miR-206 in different tissues of C57B6 mice and its temporal changes in a myogenesis model of C2C12 cells. (A) Northern blots were performed to detect miR-206 RNA in various mouse tissues. The expression of miR206 was specifically observed in skeletal muscle and U6 snoRNA was used as an internal control. Br: brain; Si: small intestine; Sm: skeletal muscle; Sp: spleen; Kd: kidney; Lv: liver; Lu: lung; Ht: heart. (B) During skeletal myogenesis of the C2C12 myoblast cell line, expression of miR-206 was measured by Northern blot at indicated times post-differentiation. U6 snoRNA was used as a control. (C) Quantitative assessment of microRNA levels was based on densitometry of exposed X-ray films. Expression of miR-206 was significantly higher 1, 3, and 5 days after medium exchange in comparison to baseline (*** P<0.001, one-way ANOVA, n=3).
Next, we used C2C12 mesenchymal stem cells to assess changes in miR-206 during muscle development. In this well-characterized in vitro model, medium containing 2% horse serum is used to induce myogenic differentiation. As shown in Figure 1, miR-206 levels increased significantly over time, and there were 5-fold higher levels by day 5 based on Northern blot analyses (Figure 1B,C) (P < 0.001, one way ANOVA, n=3).
Myoblast differentiation induced by AdvmiR-206 transfection in C2C12 cells
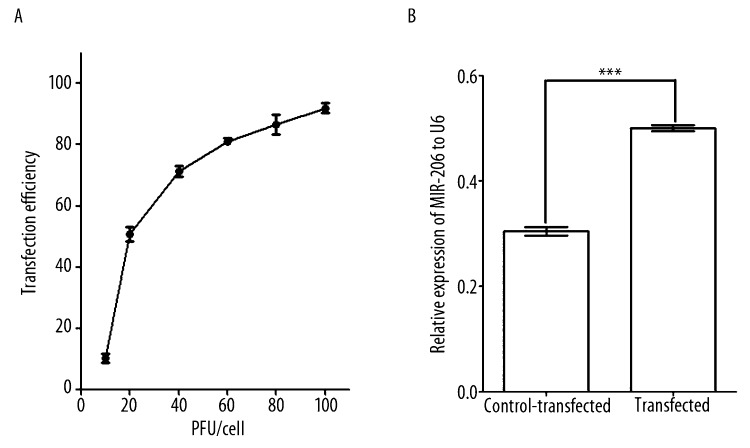
To explore the role of miR-206 in myogenesis, we constructed a vector that would direct the synthesis of miR-206 (AdvmiR-206) in transfected cells. Transfection was optimal at 30 pfu, as defined by a high transduction efficiency of 70% (Figure 2A), however, with low toxicity (data not shown). Forty-eight hours after transfection, visual inspection by fluorescence microscopy showed greater than 70% of cells were positive for green fluorescent protein (GFP). Transfected cells showed long-term viability, as evidenced by the presence of GFP-positive cells after 10 days (data not shown). Based on Northern blot analyses, miR-206 expression was 67% higher in AdvmiR-206 transfected C2C12 cells (0.50±0.01), compared to the control-transfected cells (0.30±0.01) (P<0.001, t-test, n=3) 48 hours post-transfection (Figure 2B).
Figure 2.
Adenovirus transfection and expression in C2C12 cells. (A) The efficiency of transfection was based on visual inspection of green fluorescence protein signal in cells at 48 hours. Transfection conditions were standardized at 30 pfu/cell and based on these results. (B) At baseline (24 hours), miR-206 expression was approximately 20% higher in AdvmiR-206 transfected cells compared to control-transfected cells (*** P<0.001, t-test, n=3).
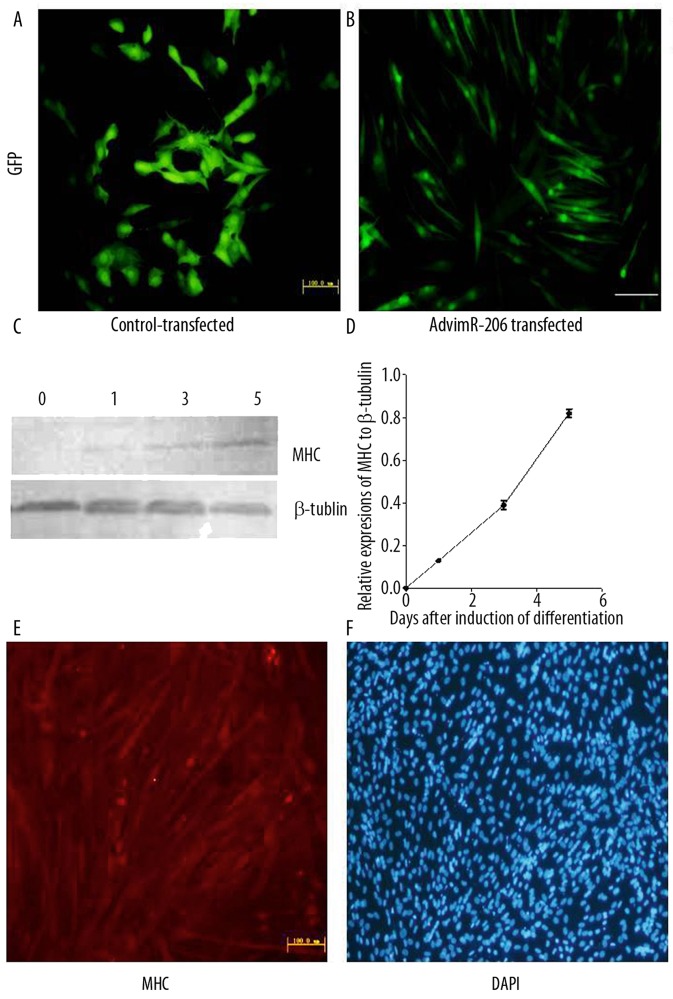
To test the functional consequences of miR-206 expression, we incubated transfected C2C12 cells in standard growth medium (DMEM). C2C12 cells typically do not differentiate under these conditions. However, when transfected with AdvmiR-206, they differentiated into muscle cells and gradually fused together to form grossly multinucleated myotubes by 5 days (Figure 3B). These morphological changes were not observed in control-transfected (i.e., AdvEGFP-transfected) cells (Figure 3A). Myosin heavy chain (MHC) is a marker of the terminal differentiation linked to myotube formation [25,27]. To confirm the positive correlation between MHC expression and the differentiation of C2C12 cells, we measured the MHC proteins levels in non-induced versus post-serum induced cells on day 1, 3, and 5. We identified a positive correlation between MHC expression and the number of days after induction of differentiation by Western blot (Figure 3C,D). There was no detectable expression of MHC in non-induced, undifferentiated C2C12 cells. However, by day 5 post-serum induction, a significant amount of MHC was observed in differentiated C2C12 cells. Based on those observations, positive expression of MHC was used as a marker for C2C12 cell differentiation. We found that MHC was expressed in AdvmiR-206 transfected C2C12 cells but not in AdvEGFP-infected control cells (Figure 3E), as determined by immunofluorescence histochemistry. To confirm the formation of multinucleated myotubes in miR-206 induced differentiation, all cells were also counter stained with DAPI (Figure 3F).
Figure 3.
Induction of myogenesis in AdvmiR-206 transfected C2C12 cells. (A,B) C2C12 cells were transfected with AdvmiR-206 or AdvEGFP control vector (30 pfu/cell), then maintained in standard growth medium for 5 days. C2C12 cells were GFP-positive with both vectors, which suggested a good viability of transfection. However, AdvmiR-206-transfected C2C12 cells showed the elongated morphology change. (C,D) Expression of myosin heavy chain (MHC) proteins was positively correlated to the differentiation process by Western blot. (E) As indicated by immunofluorescence, MHC was only detected in AdvmiR-206-transfected C2C12 cells. (F) Counterstaining with DAPI showed fused multi-nucleate myoblast cells post-differentiation in AdvmiR-206-transfected C2C12 cells.
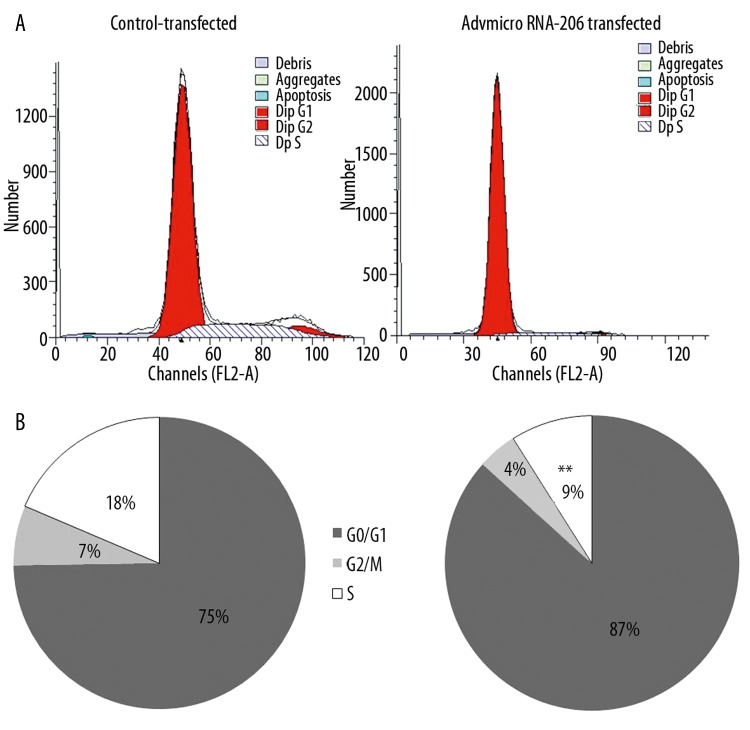
When cells undergo differentiation, they stop cell division, which can be assessed as a decrease in mitotic activity. By flow cytometry, we found that AdvmiR-206-transfected C2C12 cells were significantly less likely to be in S-phase in comparison to control-transfected cells: 9±1.4% vs. 18±0.3% (P<0.01, t-test, n=3) (Figure 4). As expected, there were no significant differences between the AdvEGFP control and non-infected groups (data not shown).
Figure 4.
Cell cycle alteration in AdvmiR-206-transfected cells vs. the control. AdvmiR-206-transfected cells showed cell cycle changes, which were consistent with myogenesis. Compared to control-transfected cells, there were fewer cells in S-phase, as determined by flow cytometry at 24 hours (** P<0.01, t-test, n=3). (A) A representative flowcytometry photograph. (B) Percentage of cell cycles stages distributed in the control vs. transfected groups.
Inhibition of myogenesis by antisense miR-206
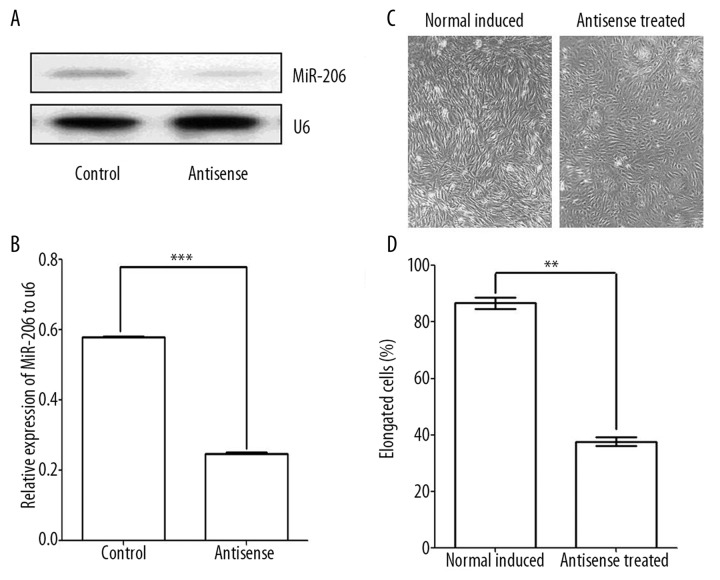
Our gain-of-function studies suggested a functional role for miR-206 in myogenesis. Therefore, we hypothesized that inhibition of endogenous miR-206 would decrease or delay differentiation in response to serum deprivation. Twenty-four hours after being transfer into differentiation medium, miR-206 levels were 2.5 times lower in C2C12 cells when a specific antisense oligoribonucleotide was present (P<0.001, t-test, n=3) (Figure 5A,B).
Figure 5.
Myogenesis in C2C12 cells with and without exposure to the microRNA-206 antisense probe. (A,B) The results of Northern blot analysis showing 2.5 times lower levels of miR-26 when cells were exposed to the antisense probe (*** P<0.001, t-test, n=3). (C,D) Anticipated morphological changes during differentiation were inhibited or delayed by antisense ribonucleotide. Cells exposed to miR-206 antisense showed half the number of elongated (differentiated) cells after 48 hours incubation in differentiation medium (** P<0.01, t-test, n=3).
To confirm anti-miR-206 oligoribonucleotide inhibition, C2C12 cells were observed under the light microscope, and elongated cells were counted. The percentage of elongated cells was significantly reduced in anti-miR-206 oligoribonucleotide transfected C2C12 cells in comparison to controls: 37±2% vs. 87±2% (P<0.01, t-test, n=3) (Figure 5C,D).
Discussion
Regulation of gene expression is complicated in multicellular organisms, particularly during development. MicroRNAs regulate gene expression by base-pairing to complementary sequences on target genes [28]. However, the exact function of most microRNAs is unknown. Early surveys suggested that miR-206 expression was specific to skeletal muscle [9,12]. In this study, we measured the signal of miR-206 RNA in various tissues of C57BL mice. Consistent with published reports, we indentified a very specific skeletal muscle expression. The above observation reinforced the concept that manipulating the expression and activity of myogenic microRNAs could lead to new therapeutic approaches for muscle diseases.
To gain insight into microRNA function in skeletal muscle, we used a well-characterized in vitro model of myogenesis: C2C12 mesenchymal stem cells. C2C12 cells differentiate along the myogenic pathway when they are transferred from growth medium into differentiation medium containing reduced serum [29]. Gain-of-function analyses have been one of the most fruitful approaches in mechanistic studies of specific microRNAs [5,30–33]. This strategy typically involves the transient transfection of cells with double-stranded RNA complexes that mimic mature functional microRNAs [34]. We know, however, that functional miRNAs are derived from pre-microRNA gene transcripts, which not only determines the final sizes but also the structure and function of mature microRNAs [28]. This knowledge shaped our strategy for the ectopic expression of microRNA-206. For construction of the adenovirus vector, we used a 355-bp PCR amplicon that incorporated the entire pre-miRNA-206 sequence. Overexpression of miR-206 was accomplished by sustained expression of the miR-206 precursor transcript in cells transfected with an adenovirus vector. We used an adenovirus vector because of its high transduction efficiency, which makes it easier to produce a high titer of recombinant virus. Because transfected genes are not integrated into the host chromosome, the risk of insertions or mutations is low. Therefore, adenoviruses are considered relatively safe for use in gene therapy [35]. Several research laboratories have now reported the successful transfer of genes using adenoviral vectors [8,36]. The therapeutic value of this technology with several other genes has been applied in clinical trials [35,37,38]. By using a microRNA-206 expressing adenovirus, we improved the low transfection efficiency associated with some other systems.
Using flow cytometry, we found that ectopic expression of miR-206 inhibited cell cycle changes associated with differentiation. Specifically, we found a 2-fold reduction in S-phase cells at 24 hours, which was consistent with other published data. For example, Kim et al. (2006) observed similar changes over a shorter time-course. The identification of DNA polymerase-alpha as a potential direct target for miR-206 suggests a role in the inhibition of DNA synthesis during differentiation [9]. However, we should not exclude the possibility of multi-mechanistic regulation. Increasing evidence shows that miR-206 might be involved in the regulation of development, differentiation, apoptosis, repair, and disease of skeletal muscle through multiple molecular mechanisms. For example, miR-206 directly targeted DNA polymerase alpha [9]. The miR-206 repressed histone deacetylase 4 (HDAC4) translations were implicated in the control of neuromuscular gene expressions [23]. The skeletal muscle differentiation gene Myo-D, activated the expression of miR-206, which in turn targeted two other downstream muscular differentiation genes: follistatin-like 1 and utrophin [39]. MiR-206 also downregulated expression of connexin43, a gap junction channel protein involved in myoblast fusion [9,26].
Our characterization would not be complete without a loss-of-function study [40]. For this, we constructed a 2-O-methyl-modified oligoribonucleotide that was complementary to miR-206. Other microRNAs transcribed from the miR-1 cluster are not likely be targeted as their specificity of the probe had been tested before [41–44]. MiR-206-specific antisense oligoribonucleotide blocked the normal in vitro response to serum depletion, which is consistent with a proposed role for miR-206 in differentiation. Like adenovirus-directed gene expression, antisense technology has great therapeutic potential because of its specificity and efficiency [40, 44]. MicroRNAs may eventually be used for the treatment or prevention of human diseases [45]. However, there are many obstacles to overcome.
Conclusions
We constructed an adenoviral miR-206 expression vector (AdvmiR-206) for transfection into C2C12 mesenchymal stem cells. Increased expression of miR-206 in these cells resulted in morphological and biochemical changes that were similar to serum deprivation-induced myogenesis. MiR-206 overexpression accounted for a 50% reduction of S-phase cells. MiR-206-specific antisense oligoribonucleotides inhibited the normal response to serum deprivation-induced myogenesis in untransfected C2C12 cells. Twenty-four hours post-transfection of antisense oligoribonucleotides, the number of elongated cells was reduced. These data support an important role for miR-206 in myoblast differentiation.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
The manuscript has been read and approved by all authors and each author believes the manuscript represents honest work.
Source of support: This work was supported by grants from National Key Basic Research and Development Plan of China (“973” Projects, No. 2005CB522605)
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–64. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 3.Berezikov E, Guryev V, van de Belt J, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol. 2008;110:206–15. doi: 10.1016/j.ygyno.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 5.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 6.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–65. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 7.O’Donnell KA, Wentzel EA, Zeller KI, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 8.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 9.Kim HK, Lee YS, Sivaprasad U, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779:682–91. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg I, Eran A, Nishino I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–21. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy JJ, Esser KA, Andrade FH. MicroRNA-206 is overexpressed in the diaphragm but not the hindlimb muscle of mdx mouse. Am J Physiol Cell Physiol. 2007;293:C451–57. doi: 10.1152/ajpcell.00077.2007. [DOI] [PubMed] [Google Scholar]
- 14.Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–26. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cacchiarelli D, Martone J, Girardi E, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–51. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Chen JF, Tao Y, Li J, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–79. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goljanek-Whysall K, Sweetman D, Abu-Elmagd M, et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc Natl Acad Sci USA. 2011;108:11936–41. doi: 10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirai H, Verma M, Watanabe S, et al. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347–65. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winbanks CE, Wang B, Beyer C, et al. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805–14. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 21.Yuasa K, Hagiwara Y, Ando M, et al. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct Funct. 2008;33:163–69. doi: 10.1247/csf.08022. [DOI] [PubMed] [Google Scholar]
- 22.Greco S, De Simone M, Colussi C, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. Faseb J. 2009;23:3335–46. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- 23.Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–54. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakasa T, Ishikawa M, Shi M, et al. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495–505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–66. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–71. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett AM, Tonks NK. Regulation of distinct stages of skeletal muscle differentiation by mitogen-activated protein kinases. Science. 1997;278:1288–91. doi: 10.1126/science.278.5341.1288. [DOI] [PubMed] [Google Scholar]
- 28.Ohler U, Yekta S, Lim LP, et al. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–22. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubo Y. Comparison of initial stages of muscle differentiation in rat and mouse myoblastic and mouse mesodermal stem cell lines. J Physiol. 1991;442:743–59. doi: 10.1113/jphysiol.1991.sp018817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennecke J, Hipfner DR, Stark A, et al. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 32.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–38. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 33.Xu P, Vernooy SY, Guo M, et al. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–95. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 34.Kwon C, Han Z, Olson EN, et al. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Relph KL, Harrington KJ, Pandha H. Adenoviral strategies for the gene therapy of cancer. Semin Oncol. 2005;32:573–82. doi: 10.1053/j.seminoncol.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Okubo Y, Bessho K, Fujimura K, et al. Expression of bone morphogenetic protein-2 via adenoviral vector in C2C12 myoblasts induces differentiation into the osteoblast lineage. Biochem Biophys Res Commun. 1999;262:739–43. doi: 10.1006/bbrc.1999.1281. [DOI] [PubMed] [Google Scholar]
- 37.Gabrilovich DI. INGN 201 (Advexin): adenoviral p53 gene therapy for cancer. Expert Opin Biol Ther. 2006;6:823–32. doi: 10.1517/14712598.6.8.823. [DOI] [PubMed] [Google Scholar]
- 38.Vattemi E, Claudio PP. Adenoviral gene therapy in head and neck cancer. Drug News Perspect. 2006;19:329–37. doi: 10.1358/dnp.2006.19.6.1015352. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg MI, Georges SA, Asawachaicharn A, et al. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krutzfeldt J, Poy MN, Stoffel M. Strategies to determine the biological function of microRNAs. Nat Genet. 2006;38(Suppl):S14–19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- 41.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–46. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369–76. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 44.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–89. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 45.Huang ZP, Neppl RL, Jr, Wang DZ. Application of microRNA in cardiac and skeletal muscle disease gene therapy. Methods Mol Biol. 2011;709:197–210. doi: 10.1007/978-1-61737-982-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]