Abstract
Purpose
To evaluate the feasibility of prospectively guiding 4-dimensional (4D) magnetic resonance imaging (MRI) image acquisition using triggers at pre-selected respiratory amplitudes to achieve T2 weighting for abdominal motion tracking.
Methods and Materials
A respiratory amplitude based triggering system was developed and integrated into a commercial turbo spin echo (TSE) MRI sequence. Initial feasibility tests were performed on healthy human subjects. Four respiratory states, the middle and the end of inhalation and exhalation, were used to trigger 4DMRI image acquisition of the liver. To achieve T2 weighting, the echo time (TE) and repetition time (TR) were set to 75ms and 4108ms, respectively. Single-shot acquisition, together with parallel imaging and partial k-space imaging techniques, was used to improve image acquisition efficiency. 4D MRI image-sets composed of axial or sagittal slices were acquired.
Results
Respiratory data measured and logged by the MRI scanner showed that the triggers occurred at the appropriate respiratory levels. Liver motion could be easily observed on both 4DMRI image data-sets by sensing either the change of liver in size and shape (axial) or diaphragm motion (sagittal). Both 4DMRI image data-sets were T2 weighted as expected.
Conclusions
This work demonstrated the feasibility of achieving T2 weighted 4DMRI images using amplitude based respiratory triggers. With the aid of the respiratory amplitude based triggering system, the proposed method is compatible with most MRI sequences and therefore has the potential to improve tumor-tissue contrast in abdominal tumor motion imaging.
Keywords: 4D imaging, MRI, T2 weighting, abdominal motion tracking
INTRODUCTION
For abdominal tumors treated with radiation therapy, a large treatment margin is normally used to account for uncertainties in tumor delineation and tumor motion. Accurate tumor delineation and quantitative breathing motion characterization can minimize treatment margins and subsequent normal organ dose. Many methods (1–5) are available to minimize the impact of breathing motion, including breath hold and treatment machine gating. While breath hold can, in principle, reduce or eliminate breathing motion, it requires that patients are able to reproducibly hold their breath, which greatly limits its clinical application (6, 7). Treatment machine gating offers the opportunity to activate the radiation beam only when the target volume is in the planned position. While this reduces treatment temporal efficiency, in reality, the total beam-on-time is only a small fraction of the overall treatment time. In addition, some linear accelerator manufacturers are providing high dose-rate beam modalities that may further improve linear accelerator gating efficiency. In order to accurately select a gating window and determine the subsequent treatment margin, tumor breathing motion must be quantitatively determined during the simulation process.
Four dimensional (4D) imaging (8–10) enables accurate motion assessment. 4D Computed Tomography (4DCT) (11–13) is the current standard. It has good spatial resolution and can be acquired quickly. However, CT has poor soft tissue contrast which leads to increased uncertainty in tumor delineation. In addition, 4DCT involves unwanted ionizing radiation. Magnetic Resonance Imaging (MRI), which does not rely on ionizing radiation, provides excellent tumor and soft tissue contrast. Therefore, it is appropriate for abdominal motion measurements. 4DMRI image sets can be generated using rapid repetition of 3D volumetric imaging (3D+t) (14), or 2D images of a given slice acquired continuously from all respiratory phases and repeating the process for all slices (2D+t+1D) (15). For the 2D+t+1D case, 2D images need to be retrospectively assigned to appropriate respiratory states using internal (16, 17) or external surrogates (18). Due to a lack of practical and robust respiratory triggering systems to prospectively guide 4DMRI image acquisition, most existing 4DMRI methods are respiratory phase based, which necessitate fast imaging techniques. Among a variety of rapid imaging sequences, spoiled gradient echo (GRE) (19) and balanced steady-state free precession (SSFP) (20) sequences are particularly effective and used dominantly in existing 4DMRI methods. Spoiled GRE (T1 weighted) and balanced SSFP (T2/T1 weighted) sequences use short repetition time (TR) to achieve fast imaging, but at a cost of tumor-tissue contrast. The weightings provided by those 4DMRI techniques offer relatively poor tumor-tissue contrast even with the aid of Gadolinium-based contrast agents.
In this work, a novel respiratory amplitude based triggering system was developed to guide 4DMRI image acquisition. By using triggers at pre-selected respiratory amplitudes, it was possible to acquire MRI images at different respiratory states in different respiratory cycles. This eliminated the restriction on the TR and thus made more MRI sequences, especially those with better tumor-tissue contrast, compatible with 4D imaging. To show the feasibility of the proposed work, T2-weighted abdominal 4DMRI image-sets were obtained to track the respiration-induced motion.
METHODS AND MATERIALS
Respiratory phase guided 4DMRI acquisition
The acquisition scheme for a traditional respiratory phase guided 4DMRI scan is illustrated in Figure 1. The respiratory cycle (as determined via external monitor) is divided into multiple bins according to phase (or time). A 2D MRI image is acquired within each bin. Image acquisition starts from the first slice, acquires images at all respiratory phases continuously within one respiratory cycle, and then moves on to the next slice until all slices are covered. Since internal organ motion is better correlated to respiratory amplitude than respiratory phase, phase based guidance may not represent the best solution. Under regular breathing, where there is a good correlation between respiratory amplitude and phase, the image acquired at a given respiratory phase can be mapped accurately to the corresponding respiratory amplitude and therefore internal organ position. Respiration, however, is irregular in most cases. Variable breathing patterns will disrupt the correlation between respiratory amplitude and phase, causing images being registered to the wrong respiratory amplitudes and, therefore, internal organ position. The acquisition scheme used in phase based 4DMRI can limit the misregistration to slices acquired during the breathing pattern change, rather than all slices after the time point of the breathing pattern change. This practice, however, limits the longest TR that can be used in 4D imaging, which equals the duration of the respiratory bin, and therefore compatible MRI sequences. As of today, respiratory phase guided 4DMRI can only provide T1 weighting or T2/T1 weighting.
Figure 1.
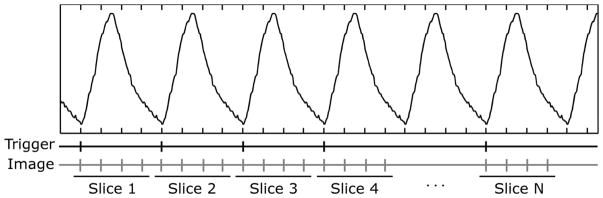
Image acquisition scheme for traditional 4DMRI methods. Each vertical bar corresponds to one 2D MRI image at a specific respiratory phase. Images at all respiratory phases from a single slice position are acquired continuously within one respiratory cycle. When image acquisition is finished for one slice, it moves to the next slice until all slices are covered.
Respiration amplitude guided 4DMRI
MRI weightings offered by traditional 4DMRI methods are less optimal for tumor delineation. Clinically, T2 weighted and diffusion weighted MRI have better tumor-tissue contrast. However, those sequences require long TR, which is not compatible with existing respiratory phase based 4DMRI methods. To use long TR, image acquisition of different respiratory states for each slice needs to be separated into multiple respiratory cycles to allow the magnetization to completely relax. To address this challenge, an amplitude based triggering system was developed to guide 4DMRI image acquisition.
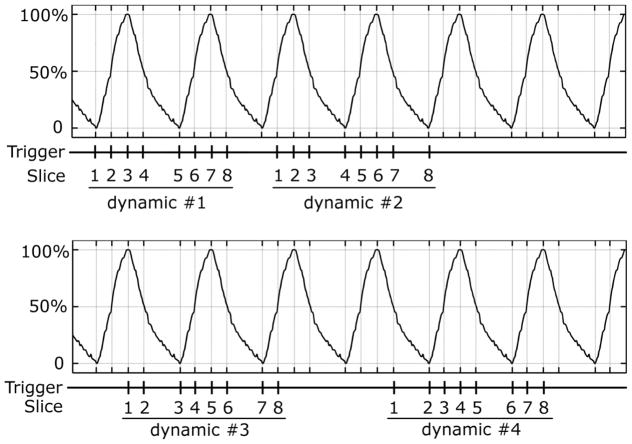
Figure 2 illustrates the acquisition scheme. The respiratory cycle is divided into multiple respiratory states based on respiratory amplitude and stage (i.e., inhalation↑ or exhalation↓), rather than respiratory phase. For example, in the 4-state case, the 4 respiratory states are 0%, 50%↑, 100%, and 50%↓. When the respiratory signal reaches a specific state, it triggers image acquisition for a specific slice and a 2D MRI image is obtained, corresponding to the given slice and respiratory state. A complete 4DMRI image acquisition requires N dynamic scans, where N equals the number of respiratory states. In each dynamic scan, one image is acquired from each slice. Slices are assigned to different respiratory states sequentially. For example, to acquire a 4DMRI image-set that has 4 respiratory states and 8 slices, the image acquisition includes 4 dynamic scans in a single MRI scan. No operator interaction is needed between dynamic scans. In dynamic scan #1, slices 1 through 8 are acquired in respiratory states 0%, 50%↑, 100%, 50%↓, 0%, 50%↑, 100% and 50%↓, respectively. In dynamic scan #2, each slice is assigned to a different respiratory state. Slices 1 through 8 are acquired in respiratory states 50%↑, 100%, 50%↓, 0%, 50%↑, 100%, 50%↓ and 0%, respectively. Similarly, in dynamic scan #3, slices 1 through 8 are acquired in respiratory states 100%, 50%↑, 0%, 50%↓, 100%, 50%↑, 0% and 50%↓, respectively, and in dynamic scan #4, slices 1 through 8 are acquired in respiratory states 50%↑, 0%, 50%↓, 100%, 50%↑, 0%, 50%↓ and 100%, respectively. After the 4th dynamic scan, 4 images are acquired from each slice, corresponding to the 4 respiratory states.
Figure 2.
Illustration of the respiratory amplitude based 4DMRI acquisition method. The respiratory cycle is divided into 4 states: 0%, 50%↑, 100%, and 50%↓. The whole acquisition includes 4 dynamic scans. In each dynamic scan, each slice is assigned to a specific respiratory state and one image is acquired for that slice. For the next dynamic scan, each slice is associated to a different respiratory state and another image is acquired for each slice. This process continues until images from all four respiratory states are acquired for every slice.
MRI pulse sequence implementation
The proposed triggering algorithm was implemented into the standard turbo spin echo (TSE) sequence using a commercial pulse sequence programming environment (Philips Healthcare, Andover, MA, version R2.5.3). The respiratory waveform used in the triggering system was acquired through the standard external respiratory bellow pressure monitor. Implementation of the proposed triggering system consisted of two stages: the preparation stage and the acquisition stage. In the preparation stage, the scanner spent 8 seconds acquiring respiratory signal. During this stage, no images were acquired. For the majority of human subjects, 8 seconds was sufficient to encompass a complete respiratory cycle. The respiratory cycle was analyzed in real-time to determine the minimum and maximum respiratory signals. During the acquisition stage, the respiratory cycle was divided into a user-defined number of respiratory states based on the respiratory range obtained in the preparation stage. Since the respiration included both inhalation and exhalation, the number of respiratory states was set to be an even number so that half states occurred during inhalation and half during exhalation.
For each respiratory state, the corresponding respiratory amplitude (or triggering level TL) and stage (or triggering stage TS) can be described as
| (1) |
where i is an integer identifying the ith respiratory state, N represents the number of respiratory states, Rmin and Rmax are the minimum and maximum respiratory amplitudes obtained during the preparation stage.
Instead of sending triggers at precise triggering levels, an acceptance range was used. Whenever the respiratory signal fell within the triggering level ± 5% of (Rmax−Rmin), a trigger was sent to start image acquisition. During image acquisition, the MRI sequence looped through dynamic scans. In each dynamic scan, the sequence looped over slices sequentially. Each slice was assigned to a specific respiratory state as described by
| (2) |
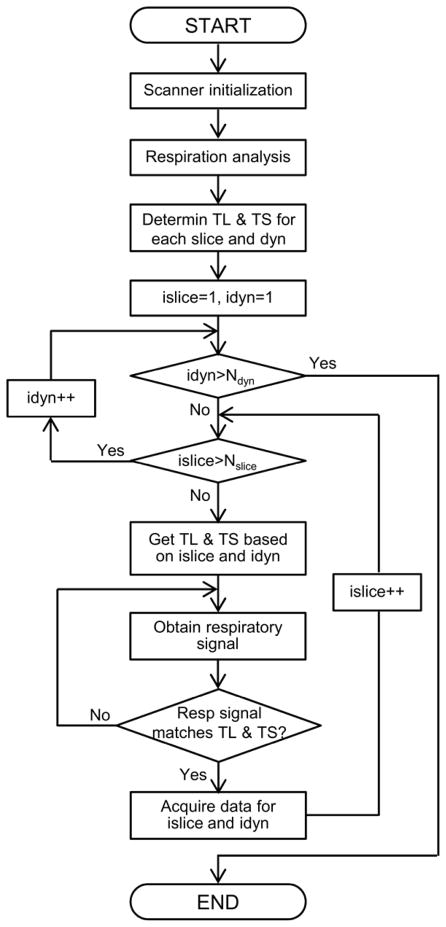
where m represents the mth slice, n represents the nth dynamic scan, N is the total number of respiratory states and % means the modulus operation. Each respiratory state was associated with a pre-defined triggering amplitude and triggering stage according to Eq. (1). For triggering purposes, a loop structure was placed in the sequence immediately before the image acquisition for each individual slice. The respiratory signal was continuously acquired in this loop at a rate of 100 samples/second and compared to the triggering amplitude and stage associated with a specific slice. Once a match was found, the MRI sequence broke out of the loop and started the image acquisition for that specific slice. After images from all slices and all respiratory states had been acquired, they were rearranged so that images from all slices at each respiratory state were assembled into a 3D volume at that respiratory state. 3D volumes at all respiratory states were obtained and stored for further evaluation. Illustration of the MRI pulse sequence implementation is shown in Figure 3.
Figure 3.
MRI pulse sequence implementation for the proposed respiratory amplitude based 4DMRI method. Ndyn, Nslice, idyn and islice represent the number of dynamic scans, the number of slices, the current dynamic scan and the current slice, respectively. TL, TS and Resp stand for triggering level, triggering state and respiratory, respectively.
Human subjects
Five healthy adult human subjects were scanned to test the feasibility of the proposed respiration amplitude guided 4DMRI acquisition. Three of them were scanned during the pulse sequence developmental process and two subjects were scanned with the finalized sequence. The study was approved by the Institutional IRB office.
MRI scan protocol
During the scan, the respiratory signal was acquired using the standard respiratory monitoring device utilizing an air-filled cushion and a pressure sensor. The respiratory signal was displayed on the console computer. During the time that k-space was sampled, markers were displayed along with the respiratory signal on the console computer to indicate the active image acquisition period on subject’s respiratory waveform. The digitized respiratory waveform for the whole scan and active acquisition markers were logged into a file for post-scan analysis. Sequence testing was performed on a clinical 1.5T MRI scanner (Philips Achieva). Four respiratory states (0%, 50%↑, 100% and 50%↓) were used to show the feasibility of the proposed 4DMRI method. Images were acquired in both axial and sagittal orientations.
For the 4D image set composed of axial slices, the in-plane field of view (FOV) was 375mm×264mm, in-plane spatial resolution was 1.5mm×1.5mm. Phase encoding direction was in the anterior-posterior direction. 32 5mm-thick slices with no inter-slice gaps were acquired to provide a spatial coverage of 160mm in the superior-inferior direction. The single-shot TSE sequence was used in image acquisition. Parallel imaging (acceleration factor=2) and partial k-space imaging (k-space coverage=0.685) techniques were used to reduce total acquisition window. TE was 75ms and TR was 4108ms to provide T2 weighting. The echo spacing was 4.4ms and total acquisition window was 270ms. To minimize inter-slice cross-talk, 32 slices were divided into 4 packages in an interleaved way, e.g. package 1 included slice 1, 5, 9, 13, 17, 21, 25 and 29. Slices in each package were acquired in ascending order, as were packages. 4 dynamic scans were included for a complete 4DMRI image data-set with no delay between dynamic scans. In each dynamic scan, each slice was associated to one of the 4 respiratory states. The total nominal scan time without considering the respiratory triggering was 65 seconds, while the total scan time for acquiring a complete 4DMRI image set was around 3 minutes. For the 4D image set composed of sagittal slices, imaging parameters were the same as those used in the acquisition of axial slices except that readout direction was changed to superior-inferior.
RESULTS
Figure 4 shows the respiratory triggering for all 8 slices in a representative package (package #2). Vertical bars represented the acquisition window for each slice. In different dynamic scans, each slice was associated with different respiratory states to ensure that images at 4 respiratory states were acquired. Due to partial k-space image acquisition, sampling of the k-space origin did not occur precisely at the acquisition window midpoint. In our cases, the TSE factor was 62 and the acquisition of k=0 was at the 19th echo (approximately 1/3 of the acquisition window).
Figure 4.
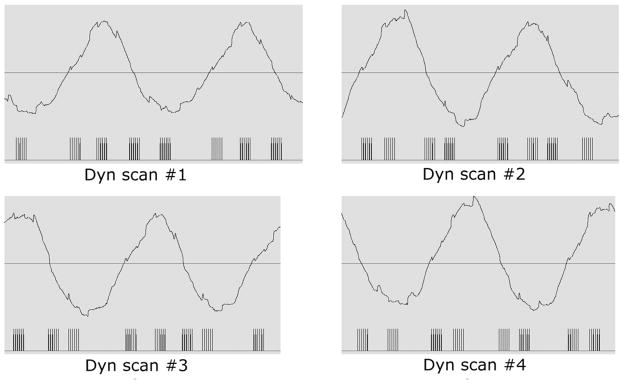
Respiratory triggering example for a representative package that includes 8 slices. The digitized respiratory waveform and triggering locations were logged by the MRI scanner and restored by a physiological data viewer included in the scanner pulse sequence programming environment. Vertical bars represent the acquisition window for each slice.
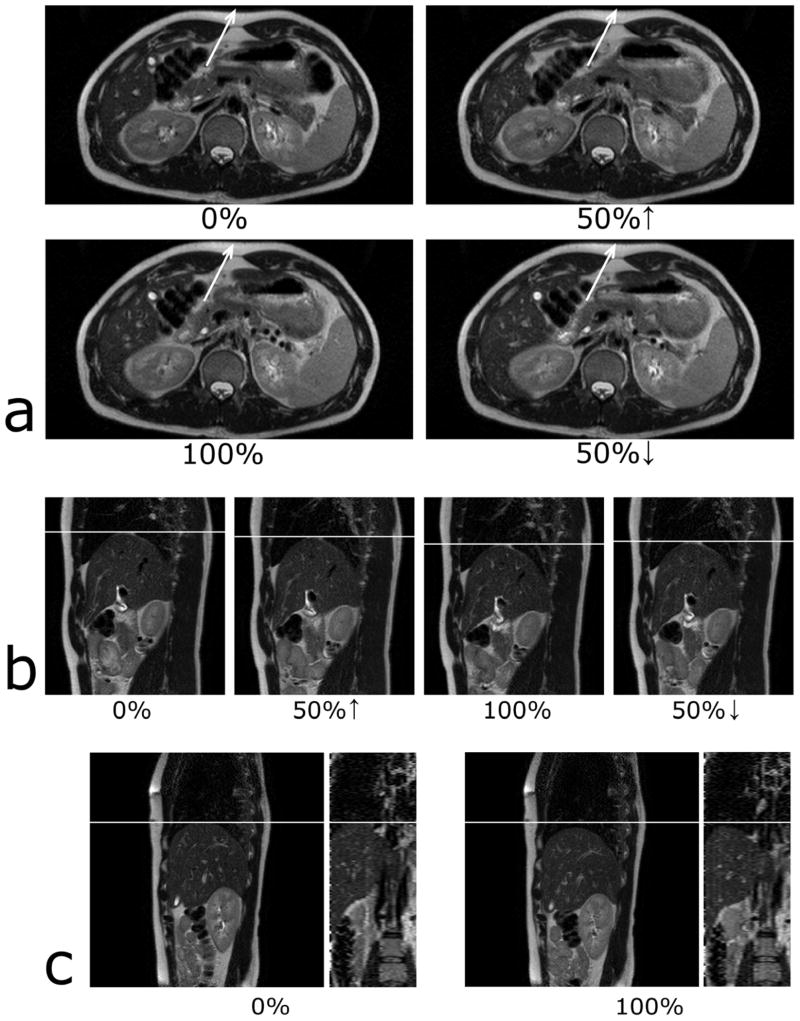
Figure 5a and 5b show T2-weighted images from four respiratory states for a representative axial slice and a sagittal slice, respectively. Respiratory motion is illustrated by the abdominal wall movement (5a) and the diaphragm motion (5b). Figure 5c shows sagittal and reconstructed coronal images at the end of exhalation (0%) and inhalation (100%). All images from Figure 5 came from the same human subject. The coronal image cuts the sagittal image through the center pixel (144th pixel out of 288) whereas the sagittal image intercepts the coronal image near the liver dome (24th slice out of 32). No abrupt positional changes occur in the reconstructed coronal view suggesting images were at the same respiratory state. All images exhibited the desired T2 weighting.
Figure 5.
T2-weighted images from four respiratory states for a representative axial slice (a) and for a representative sagittal slice (b). Sagittal and reconstructed coronal images at the end of exhalation (0%) and inhalation (100%) are shown in c. Respiratory motion can be noted by examining the abdominal wall movement (a) and the liver-lung boundary (b & c).
DISCUSSION
In this work, a respiratory amplitude based triggering system was successfully developed to guide 4DMRI image acquisition. Since image registration to appropriate respiratory states was prospectively guided by the proposed triggering system, separating image acquisition at different respiratory states into different respiratory cycles was possible, allowing magnetization sufficient time to relax (long TR). The proposed 4DMRI method was more flexible in the selection of MRI sequences and was theoretically compatible with most MRI sequences, including those with better tumor-tissue contrast, e.g. T2 weighted sequences. Therefore, it had great potential in monitoring abdominal tumor motion.
Compared to phase based 4DMRI methods, the amplitude based 4DMRI is more robust to irregular breathing. Phase based 4DMRI methods are susceptible to breathing irregularities in both amplitude and period since they all interrupt the correlation between respiratory phase and amplitude. The amplitude based 4DMRI method, however, is less sensitive to breathing irregularity, which can be illustrated in Figure 6. When a sudden change occurs in breathing amplitude (6α) or breathing frequency (6β) such that the expected respiratory state does not occur, or happens before the end of image acquisition from the previous slice, the MRI scanner remains idle until the expected respiratory state occurs again.
Figure 6.
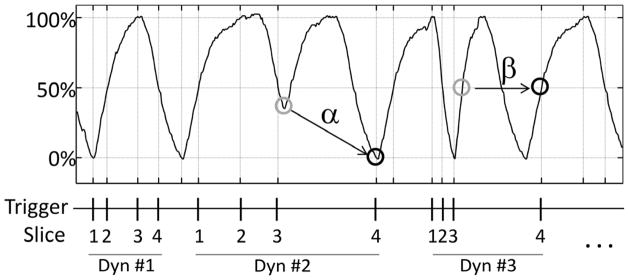
The proposed 4DMRI method is robust to breathing irregularities. When there is a sudden change in amplitude (α) resulting in the absence of a respiratory state, or an abnormal rapid breathing (β) causing a premature occurrence of the respiratory state before the end of image acquisition from the previous slice, the MRI scanner will remain idle until the next occurrence of the expected respiratory state is detected.
The external respiratory monitor, instead of internal navigator, was used as the surrogate in this work because of two reasons: 1) the external respiratory monitor comes with virtually all MRI scanners for triggered scans whereas the internal navigator method is only available in the recent MRI scanner as an advanced software option, and 2) there is no interference between the external respiratory monitor and image acquisition whereas the internal navigator method works only with sequences utilizing low flip angles. For T2-weighted TSE sequences using 90° excitation pulse, if the imaging slice overlaps with the navigator sampling region, magnetization will be saturated after image acquisition, thus altering the subsequent navigator signal.
Although four respiratory states were used in this study, the number of respiratory states is not limited to four. The maximum number of respiratory states is practically determined by both the individual’s respiratory pattern and the image acquisition time for each shot (or segment). Normally, inhalation is shorter than exhalation. For healthy volunteers, inhalation lasts around 1.0 – 2.0 seconds. With the single-shot technique, image acquisition time for a 2D image is about 0.3 seconds. Therefore, it is possible to fit 3 respiratory states during inhalation, or 6 in the whole respiratory cycle. In this study, we conservatively select 4 respiratory states (2 in inhalation and 2 in exhalation) to test the feasibility of the proposed 4DMRI method. To accommodate more respiratory states, the multi-shot technique can be utilized, wherein the long echo train of the single-shot technique is divided into several segments, acquires one segment at a time and combines afterwards to generate one 2D image. Since a segment rather than the whole k-space is acquired for each slice at a time, the acquisition time per segment can be significantly smaller than that per slice, allowing more respiratory states to be fit into the respiratory cycle.
One limitation of the proposed 4DMRI method is that it is susceptible to breathing pattern change between the preparation and acquisition stages. When breathing is deep in the preparation stage but shallow in the acquisition stage, image acquisition can be frequently halted, significantly decreasing acquisition efficiency. When breathing is shallow in the preparation stage but deep in the acquisition stage, the respiratory motion at the end of exhalation and inhalation is not sampled as the corresponding triggers are not set to the full range of the respiration. To address this challenge, possible solutions include optimizing acceptance range for variable breathers, using respiratory coaching, and/or increasing the duration of preparation stage for better respiratory waveform estimation. Another limitation is the elongated scan time under irregular breathing. The proposed 4DMRI method is able to handle mild irregular breathing or highly irregular breathing in period fairly well, which should not dramatically increase the total scan time. In this feasibility study, more than 95% of the triggers occurred as expected. However, for patients with highly irregular breathing, the total scan time could become unreasonably long due to frequently scanner halts. Those patients might not benefit from the proposed 4DMRI technique.
CONCLUSION
A triggering system based on respiratory amplitude has been developed to guide 4DMRI image acquisition. It is compatible with most MRI sequences and therefore has the flexibility of adopting MRI sequences with better tumor-tissue contrast in 4D imaging to monitor abdominal tumor motion. In this work, the feasibility of acquiring T2 weighted 4DMRI images was demonstrated on healthy volunteers.
SUMMARY.
In this work, a respiratory amplitude based triggering system was developed to guide 4DMRI image acquisition, making it possible to incorporate MRI sequences with better tumor-tissue contrast, e.g. T2 weighting, into 4D imaging to improve abdominal tumor motion tracking.
Acknowledgments
This work is supported by NIH grant 1R21CA167092-01A1.
Footnotes
Conflicts of Interest Notification: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shirato H, Shimizu S, Kunieda T, et al. Physical aspects of a real-time tumor-tracking system for gated radiotherapy. Int J Radiat Oncol Biol Phys. 2000;48:1187–1195. doi: 10.1016/s0360-3016(00)00748-3. [DOI] [PubMed] [Google Scholar]
- 2.Hanley J, Debois MM, Mah D, et al. Deep inspiration breath-hold technique for lung tumors: the potential value of target immobilization and reduced lung density in dose escalation. Int J Radiat Oncol Biol Phys. 1999;45:603–611. doi: 10.1016/s0360-3016(99)00154-6. [DOI] [PubMed] [Google Scholar]
- 3.Wong JW, Sharpe MB, Jaffray DA, et al. The use of active breathing control (ABC) to reduce margin for breathing motion. Int J Radiat Oncol Biol Phys. 1999;44:911–919. doi: 10.1016/s0360-3016(99)00056-5. [DOI] [PubMed] [Google Scholar]
- 4.Rosenzweig KE, Hanley J, Mah D, et al. The deep inspiration breath-hold technique in the treatment of inoperable non-small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2000;48:81–87. doi: 10.1016/s0360-3016(00)00583-6. [DOI] [PubMed] [Google Scholar]
- 5.Kubo HD, Hill BC. Respiration gated radiotherapy treatment: a technical study. Phys Med Biol. 1996;41:83–91. doi: 10.1088/0031-9155/41/1/007. [DOI] [PubMed] [Google Scholar]
- 6.Dawson LA, Brock KK, Kazanjian S, et al. The reproducibility of organ position using active breathing control (ABC) during liver radiotherapy. Int J Radiat Oncol Biol Phys. 2001;51:1410–1421. doi: 10.1016/s0360-3016(01)02653-0. [DOI] [PubMed] [Google Scholar]
- 7.Plathow C, Ley S, Zaporozhan J, et al. Assessment of reproducibility and stability of different breath-hold maneuvres by dynamic MRI: comparison between healthy adults and patients with pulmonary hypertension. Eur Radiol. 2006;16:173–179. doi: 10.1007/s00330-005-2795-9. [DOI] [PubMed] [Google Scholar]
- 8.Keall P. 4-dimensional computed tomography imaging and treatment planning. Semin Radiat Oncol. 2004;14:81–90. doi: 10.1053/j.semradonc.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Citrin D, Camphausen K, et al. Advances in 4D medical imaging and 4D radiation therapy. Technol Cancer Res Treat. 2008;7:67–81. doi: 10.1177/153303460800700109. [DOI] [PubMed] [Google Scholar]
- 10.Biederer J, Dinkel J, Remmert G, et al. 4D-Imaging of the lung: reproducibility of lesion size and displacement on helical CT, MRI, and cone beam CT in a ventilated ex vivo system. Int J Radiat Oncol Biol Phys. 2009;73:919–926. doi: 10.1016/j.ijrobp.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Vedam SS, Keall PJ, Kini VR, et al. Acquiring a four-dimensional computed tomography dataset using an external respiratory signal. Phys Med Biol. 2003;48:45–62. doi: 10.1088/0031-9155/48/1/304. [DOI] [PubMed] [Google Scholar]
- 12.Low DA, Nystrom M, Kalinin E, et al. A method for the reconstruction of four-dimensional synchronized CT scans acquired during free breathing. Med Phys. 2003;30:1254–1263. doi: 10.1118/1.1576230. [DOI] [PubMed] [Google Scholar]
- 13.Keall PJ, Starkschall G, Shukla H, et al. Acquiring 4D thoracic CT scans using a multislice helical method. Phys Med Biol. 2004;49:2053–2067. doi: 10.1088/0031-9155/49/10/015. [DOI] [PubMed] [Google Scholar]
- 14.Plathow C, Klopp M, Schoebinger M, et al. Monitoring of lung motion in patients with malignant pleural mesothelioma using two-dimensional and three-dimensional dynamic magnetic resonance imaging: comparison with spirometry. Invest Radiol. 2006;41:443–448. doi: 10.1097/01.rli.0000208222.03256.ba. [DOI] [PubMed] [Google Scholar]
- 15.von Siebenthal M, Szekely G, Gamper U, et al. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol. 2007;52:1547–1564. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- 16.von Siebenthal M, Cattin P, Gamper U, et al. 4D MR imaging using internal respiratory gating. Med Image Comput Comput Assist Interv. 2005;8:336–343. doi: 10.1007/11566489_42. [DOI] [PubMed] [Google Scholar]
- 17.Tokuda J, Morikawa S, Haque HA, et al. Adaptive 4D MR imaging using navigator-based respiratory signal for MRI-guided therapy. Magn Reson Med. 2008;59:1051–1061. doi: 10.1002/mrm.21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, Chang Z, Wang Z, et al. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: a feasibility study. Med Phys. 38:6384–6394. doi: 10.1118/1.3658737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekihara K. Steady-state magnetizations in rapid NMR imaging using small flip angles and short repetition intervals. IEEE Trans Med Imaging. 1987;6:157–164. doi: 10.1109/TMI.1987.4307816. [DOI] [PubMed] [Google Scholar]
- 20.Carr HY. Steady-state free precession in nuclear magnetic resonance. Phys Rev. 1958;112:1693–1701. [Google Scholar]