Abstract
To characterize the repertoire of T lymphocytes in chronically hepatitis C virus (HCV)-infected patients with and without mixed cryoglobulinaemia (MC). T cell receptor (TCR) variable (V) β clonalities in portal tracts isolated from liver biopsy sections with a laser capture microdissection technique in 30 HCV-positive MC patients were studied by size spectratyping. Complementarity-determining region 3 (CDR3) profiles of liver-infiltrating lymphocytes (LIL) were also compared with those circulating in the blood. The representative results of TCR Vβ by CDR3 were also obtained from liver tissues and peripheral blood lymphocytes (PBL) of 21 chronically HCV-infected patients without MC. LIL were highly restricted, with evidence of TCR Vβ clonotypic expansions in 23 of 30 (77%) and in 15 of 21 (71%) MC and non-MC patients, respectively. The blood compartment contained TCR Vβ expanded clones in 19 (63%) MC and 12 (57%) non-MC patients. The occurrence of LIL clonalities was detected irrespective of the degree of liver damage or circulating viral load, whereas it correlated positively with higher levels of intrahepatic HCV RNA. These results support the notion that TCR Vβ repertoire is clonally expanded in HCV-related MC with features comparable to those found in chronically HCV-infected patients without MC.
Keywords: hepatitis C virus, laser capture microdissection, mixed cryoglobulinaemia, T cell repertoire
Introduction
The persistence of a virus is evidence of its ability to escape the host's immune surveillance through the exploitation of several strategies, including integration into the host genome 1, silencing of viral gene expression 2, inhibition of antigen processing and presentation 3,4, synthesis of proteins homologous to known immune regulatory molecules 5, mutations that either inhibit viral responsiveness to anti-viral cytokines or preclude recognition by neutralizing antibodies 3 or else modify residues that are critical for recognition by the major histocompatibility complex 6 or T cell receptor (TCR) 7.
Hepatitis C virus (HCV) infection is characterized by a striking tendency to become chronic in a high proportion of patients. A 60–80% range of persistent infection has been documented by detection of HCV RNA 8. Infection with HCV results in liver disease of variable severity, which is thought to arise from an immune-mediated reaction 9. This is because the virus is not cytopathic, and histopathology of the HCV-infected liver frequently shows a large infiltration of inflammatory cells containing virus-specific human leucocyte antigen (HLA) class I-restricted cytotoxic T lymphocytes (CTL) 10. Some have argued that HCV-specific CTL are sequestered preferentially in the liver, where they can cause persistent hepatic damage without complete virus eradication 11.
Histopathology of HCV-infected liver is characterized by varying degrees of portal tract and parenchymal infiltration of T lymphocytes 12. The immune response is initiated following interaction between the TCR on T lymphocytes and antigenic peptides associated with major histocompatibility molecules on the antigen-presenting cells 13. TCR consists of four polypeptides (α, β, γ, δ) that form two types of heterodimer (αβ and γδ). Each polypeptide has variable (V), joining (J) and constant (C) regions and β and γ chains also have diversity (D) regions 14. TCR diversity is generated by the rearrangement of V, D, J and C regions. The random insertion of non-germline-encoded nucleotides at the junctions of these rearranged segments provides additional diversity, and is the main site of antigen recognition 15.
Restrictions of the T cell immune response as evidenced by oligoclonal T cell expansions are found preferentially in the memory T cell population of healthy individuals 16, and are possibly the result of the increasing number of antigens encountered during life.
The size and diversity of HCV-specific T cell populations in the liver and their relevance for the outcome of HCV infection are still poorly known 17. Currently, whether unsuccessful immune response is targeted against fewer antigens or consists of smaller effector T cell populations is one of the major questions being investigated. HCV mutations involving a TCR contact residue diminish T cell recognition significantly and fail to prime naive T cells effectively 18.
Studies in chimpanzees and humans infected with HCV have shown that broadly reactive HCV-specific T cell responses, established early after infection, are associated with prompt clearance of the virus 19. Conversely, diversity of clonal TCR usage is considered a factor in the development of escape mutations and correlates with a poor response to interferon (IFN)-α therapy 20.
HCV infection can be detected in mixed cryoglobulinaemia (MC) 21, a chronic immune complex-mediated disease with underlying skewing of the B cell repertoire 22,23. Morphologically, MC is characterized by bone marrow and liver multi-focal infiltrates of monoclonal B cells 21,23. The liver, indeed, is considered an ‘ectopic’ lymphoid organ, in that B cells bearing antigen-specific receptors are stimulated to proliferate and differentiate 24. Intrahepatic B cell clonotypes contribute to the formation of intraportal lymphoid nodules 25. The B cell repertoire in MC is somewhat restricted and the occurrence of B cell clonal expansions influences profoundly the clinical expression of HCV-infection 23,26. B cell clonal expansions, which involve mainly rheumatoid factor (RF)-secreting cells, represent the cogent molecular mechanism underlying the production of cryoprecipitating immune complexes.
To assess whether a limited TCR diversity exists in MC patients with HCV infection, a comprehensive analysis of the TCR Vβ repertoire of liver-infiltrating lymphocytes (LIL) and peripheral blood lymphocytes (PBL) was performed and compared with the T cell population in HCV-positive patients without MC.
Materials and methods
Patients
In this study, 30 HCV-positive patients with MC and 21 chronically HCV-infected patients without MC were enrolled. Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. MC diagnosis was based on the following criteria: (a) detection of serum cryoglobulins, as described previously 27, and (b) clinical symptoms comprising the triad purpura–weakness–arthralgia. All patients were anti-HCV- and HCV RNA-positive. Other potential causes of hepatitis (i.e. alcoholic liver disease, toxic or metabolic aetiology, chronic hepatitis B, autoimmune hepatitis) were ruled out. None of the patients had been treated previously with anti-viral therapy or steroids and immunosuppressive drugs. Liver biopsy specimens were obtained percutaneously from all patients. Histological activity and fibrosis grading were defined according to Ishak and Zimmerman 28. Blood samples from 30 healthy subjects were also considered for PBL TCR Vβ rearrangements. Healthy subjects were voluntary blood donors selected after responding to a panel of questions comprising a medical history. The healthy individuals were 16 males and 14 females, aged 55–62 years. All answered questions intended to identify and then exclude recipients of a previous transfusion, those who had had jaundice or signs of hepatitis and people having experienced high-risk sexual behaviour between 1 and 4 weeks preceding the intended donation. All samples were screened for HBV, HIV and HCV.
Isolation of PBL
PBL from patients and control subjects were isolated by Ficoll/Hypaque (Pharmacia, Freiburg, Germany) density gradient centrifugation 22.
Measurement of intrahepatic HCV RNA
Percutaneous liver biopsies were performed as part of the diagnostic evaluation. A tissue portion (∼2 mg) was used for HCV RNA measurement, whereas the remaining part was put into a RNase and DNase-free microtube and frozen immediately in liquid nitrogen until sectioning, as described elsewhere 22.
HCV RNA in tissue was measured by signal amplification with a branched DNA probe assay (Quantiplex, Chiron, Emeryville, CA, USA). Cold guanidine-HCl homogenizing solution [8 M guanidium thyocyanate, 50 mM Tris-HCl pH 7·5, 25 mM ethylenediamine tetraacetic acid (EDTA), 8% (v/v) 2-mercaptoethanol (ME) containing 3 M sodium acetate] was added to the frozen tissue and homogenized with a pellet pester mixer. Sarkosyl (10%) was added and mixed gently. Tubes were then centrifuged to sediment particulates. Supernatants were removed and added to the tubes containing poly A (10 mg/ml). Ethanol was then added and mixed thoroughly. Tubes were placed at −20°C overnight and centrifuged for 20 min at 4°C. Supernatants were aspirated, and the pellets were dried down with a Speedvac Rotatory Vacuum device (Eppendorf-Netheler-Hinz, Hamburg, Germany). After solubilization in nuclease-free H2O, HCV RNA was measured. Duplicate samples were added to the wells in which lysis, hybridization, capture and signal amplification occurred. A standard curve was constructed with reference sera obtained from Acrometrix OptiQuant HCV RNA Panel (Egret Court, Benicia, CA, USA). Results were expressed as pg HCV RNA per gram of bioptic tissue [conversion factor 0·52 obtained by dividing HCV RNA molecular mass by the number of viral copies/ml (3·13 × 106 g/mol/6·023 × 1023 copies/mol) × 105].
Isolation of portal tracts from liver biopsy sections
Portal tract structures were isolated by the laser capture microdissection (LCM) technique, as described elsewhere 29. Briefly, frozen liver tissue specimens were cut as a series of 5-μm sections mounted on slides coated with a thermoplastic membrane (Leica Microsystems, Wetzlar, Germany). Portal tracts were dissected selectively by focal melting of the membrane with an ultraviolet laser beam by Leica SVS LMD System (Leica Microsystems). Dissected microsamples were dropped into a cap tube under microscopic inspection. Microsamples were lysed in 50 μl lysis buffer and tubes were centrifuged. Then, pellets were washed with 70% ethanol and air-dried. DNA was isolated via QIAshedder columns and DNeasy Kit from Qiagen (Hilden, Germany). Portal tracts from liver biopsy sections of five patients with non-alcoholic steatosis (three men and two women, aged 55–61 years), and five with near-normal liver obtained during cholecystectomy (one man and four women, aged 51–60 years) were employed as controls.
Analysis of TCR Vβ repertoire by polymerase chain reaction (PCR) and capillary electrophoresis
DNA-based PCR approaches were established for detection of TCR Vβ gene rearrangements 30. The PCR approach for the TCR Vβ gene was designed as a multiplex PCR for the detection of Vβ–Jβ and incomplete Dβ–Jβ rearrangements. With 23 family-specific Vβ primers designated to recognize mainly functional Vβ gene segments, two specific Dβ primers and 13 specific Jβ primers, theoretically most complete Vβ–Jβ, and all incomplete Dβ–Jβ gene rearrangements, can be detected.
Each 25-μl PCR reaction contained 200 ng DNA, 12·5 μl 2× Taq PCR Master Mix (Qiagen) and 0·5 μmol/l of each primer. PCR used one cycle at 94°C for 3 min followed by 40 cycles at 95°C for 60 s, 60°C for 30 s and 72°C for 30 s, with a final 10-min extension at 72°C and a 4°C hold. The PCR product was diluted 1:10 in water and 1 μl was mixed with 12 μl of deionized formamide and 0·5 μl Gene Scan 400 HD Rox size Standard (Applied Biosystems, Foster City, CA, USA). The mixture was injected onto ABI PRISM 310 [5-s injection, 15 Kv, GSSTR POP 4 (1 ml) D module, 60°C, 24 min run-time]. Data were analysed using Genescan (Applied Biosystems), which records the fluorescence intensities in each peak. Run variations of the run-off are the results of different CDR3 lengths, reflecting an imprecise V–D–J joining process. The graphs representing CDR3 size patterns were standardized at 100% for the highest peak and the data used to generate these graphs could be used to determine the intensity of each peak expressed as relative fluorescence units and to evaluate the background. Dominant (oligoclonal) peaks were defined arbitrarily as a predominant length of CDR3 with a peak area of fluorescence corresponding to at least 40% of the sum of fluorescence intensities of all the peaks in a given family 31.
To eliminate the risk of false-positive results due to ‘background’ amplification of similar rearrangements in polyclonal cell populations, heteroduplex analysis was used. Homo- and heteroduplexes resulting from denaturation at 94°C and renaturation at a lower temperature were separated in non-denaturing polyacrylamide gels 32. TCR Vβ primer sequences depicted in Table 1 were labelled with 5′ FAM (6-carboxyfluorescein) and 5′ HEX (6-hexachlorofluorescein). Amplifiable DNA was confirmed in all samples by β-globin primers generating a 268 base pairs (bp) product (5′-HEX-CAACTTCATCCACGTTCACC-3′ and 5′-GAAGAGCCAAGGAGAGGTAC-3′).
Table 1.
Sequences of Vβ, Jβ and Dβ primers for PCR analyses of TCR Vβ gene rearrangements
| PCR analysis of TCR Vβ gene rearrangements | |||||
|---|---|---|---|---|---|
| Vβ family primers | Dβ primers | ||||
| Vβ 2 | 5′-AACTATGTTTTGGTATCGTCA-3′ | Dβ 1 | 5′-GCCAAACAGCCTTACAAAGAC-3′ | ||
| Vβ 4 | 5′-CACGATGTTCTGGTACCGTCAGCA-3′ | Dβ 2 | 5′-TTTCCAAGCCCCACACAGTC-3′ | ||
| Vβ 5/1 | 5′-CAGTGTGTCCTGGTACCAACAG-3′ | ||||
| Vβ 6a/11 | 5′-AACCCTTTATTGGTACCGACA-3′ | Jβ primers | |||
| Vβ 6b/25 | 5′-ATCCCTTTTTTGGTACCAACAG-3′ | Jβ 1·1 | 3′-GTGGGTCTAAGTGTCAACATCCATTC-5′ | ||
| Vβ 6c | 5′-AACCCTTTATTGGTATCAACAG3′ | Jβ 1·2 | 3′-CTGGTCCAATTGGCAACATCCATTC-5′ | ||
| Vβ 7 | 5′-CGCTATGTATTGGTACAACGA-3′ | Jβ 1·3 | 3′-TTCAACCGAGTGACAACATCCATTC-5′ | ||
| Vβ 8a | 5′-CTCCCGTTTTCTGGTACAGACAGAC-3′ | Jβ 1·4 | 3′-CTTGGGTCGAGAGACAGAACCCATAC-5′ | ||
| Vβ 9 | 5′-CGCTATGTATTGGTATAAACAG-3′ | Jβ 1·5 | 3′-CTGAGCTGAGAGGTAGGATCCATTC-5′ | ||
| Vβ 10 | 5′-TTATGTTTACTGGTATGCTAAGAAGC-3′ | Jβ 1·6 | 3′-GTCCGAGTGACACTGTCCATAC-5′ | ||
| Vβ 11 | 5′-CAAAATGTACTGGTATCAACAA-3′ | Jβ 2·1 | 3′-AGTGGCACGATCCATTCTTCC-5′ | ||
| Vβ 12a/3/13a/15 | 5′-ATACATGTACTGGTATCGACAAGAC-3′ | Jβ 2·2 | 3′-TCCGACTGGCATTGACCCATTC-5′ | ||
| Vβ 13b | 5′-GGCCATGTACTGGTATAGACAAG-3′ | Jβ 2·3 | 3′-ACTGTCACGAGCCATTCGCCC-5′ | ||
| Vβ 13c/12b/14 | 5′-GTATATGTCCTGGTATCGACAAGA-3′ | Jβ 2·4 | 3′-AGAGTCACGACCCATTCGACC-5′ | ||
| Vβ 16 | 5′-TAACCTTTATTGGTATCGACGTGT-3′ | Jβ 2·5 | 3′-CACGAGCCACACGCGC-5′ | ||
| Vβ 17 | 5′-GGCCATGTACTGGTACCGACA-3′ | Jβ 2·6 | 3′-TCCGACTGGCACGACCCGCTC-5′ | ||
| Vβ 18 | 5′-TCATGTTTTACTGGTATCGGCAG-3′ | Jβ 2·7 | 3′-GTCCGAGTGCCAATGTCCATTC-5′ | ||
| Vβ 19 | 5′-TTATGTTTATTGGTATCAACAGAATCA-3′ | ||||
| Vβ 20 | 5′-CAACCTATACTGGTACCGACA-3′ | ||||
| Vβ 21 | 5′-TACCCTTTACTGGTACCGGCAG-3′ | ||||
| Vβ 22 | 5′-ATACTTCTATTGGTACAGACAAATCT-3′ | ||||
| Vβ 23/8b | 5′-CACGGTCTACTGGTACCAGCA-3′ | ||||
| Vβ 24 | 5′-CGTCATGTACTGGTACCAGCA-3′ | ||||
PCR: polymerase chain reaction; TCR: T cell receptor; V: variable; J: joining; C: constant; D: diversity.
Statistical analyses
Values were expressed as mean and range and evaluated by linear regression analysis. Between-group differences were analysed by Kruskall–Wallis test for non-parametric data.
Results
The patients' characteristics are shown in Table 2. In the cryoglobulinaemic group, 28 had type II MC, immunoglobulin (Ig)Mk being the monoclonal component in all. Type III MC was demonstrated in the remaining two patients. All patients were shown to harbour productive HCV infection by repeated confirmation of HCV viraemia. The number of females was higher in the MC group. Liver disease was characterized histologically in all. Obvious cirrhosis was demonstrated in three patients of both groups. A moderate prevalence of genotype 2 was noted in MC patients. Liver enzymes were elevated in all. MC patients had measurable cryocrit (3·5 ± 1·8%).
Table 2.
Epidemiological, virological and histological data of 30 chronically HCV-infected patients with and 21 without mixed cryoglobulinaemia
| Patients | ||
|---|---|---|
| Mixed cryoglobulinaemia | ||
| Parameters | With | Without |
| n | 30 | 21 |
| Age, years | 66·4 (56–74) | 64·7 (51–76) |
| Sex (female/male) | 22/8 | 8/13 |
| HCV RNA positives, n (%) | 30 (100) | 21(100) |
| HCV genotypes, n (%) | ||
| 1 | 12 (40) | 12 (57) |
| 2 | 18 (60) | 9 (43) |
| Liver histology, n (%) | ||
| Chronic active hepatitis | 27 (90) | 18 (83) |
| Cirrhosis | 3 (10) | 3 (17) |
| Cryoglobulinaemia, n (%) | ||
| II | 28 (93) | |
| III | 2 (7) | |
| Cryocrit (%) | 3·5 ± 1·6 | |
| Purpura, n (%) | 27 (90) | |
| Arthralgia, n (%) | 22 (76) | |
| Weakness, n (%) | 24 (80) | |
HCV: hepatitis C virus.
DNA-based PCR approaches for TCR Vβ gene rearrangements were designed as multiplex PCR for detection of complete Vβ–Jβ and incomplete Dβ–Jβ rearrangements. TCR Vβ gene rearrangements were carried out in the liver tissues and in PBL collected in step with the liver biopsy. The polyclonal feature, which consisted of seven to 11 peaks in a Gaussian pattern with a 3-nucleotide interval, was observed in seven (23%) and six (29%) liver tissues and in 11 (37%) and nine (43%) PBL of MC and non-MC patients, respectively. In all but one PBL from healthy subjects (97%) and in all control liver tissues, a polyclonal TCR Vβ feature was demonstrated.
Restricted TCR Vβ patterns which contained a single peak, or a few dominant peaks, were observed in multiple variable segments. Quantitation of DNA fragments was defined by measurement of laser-light-induced fluorescence. The fluorescence intensity of the amplicons was expressed as an area under the curve (AUC) in relative fluorescence units. Clonal deviation of the CDR3 profile was taken when the AUC percentage exceeded the value of the reference panel by 3 standard deviations (s.d.).
A discrepancy was found between liver and PBL in terms of the frequency of TCR Vβ gene rearrangements and expansions. These findings comprised T cell clonal expansions in 23 (77%) and 15 (71%) liver tissues and in 19 (63%) and 12 (57%) PBL in MC and non-MC patients, respectively (Table 3).
Table 3.
Frequency of Vβ T cell clonal expansions in 30 chronically HCV-infected patients with and 21 without mixed cryoglobulinaemia
| Patients | |||
|---|---|---|---|
| Mixed cryoglobulinaemia | |||
| Vβ T cell clonalities | With | n (%) | Without |
| Polyclonal | |||
| Liver | 7 (23) | 6 (29) | |
| PBL | 11 (37) | 9 (43) | |
| Mono/oligoclonal expansion | |||
| Liver | 23 (77) | 15 (71) | |
| PBL | 19 (63) | 12 (57) | |
HCV: hepatitis C virus; PBL: peripheral blood monuclear cells; V: variable.
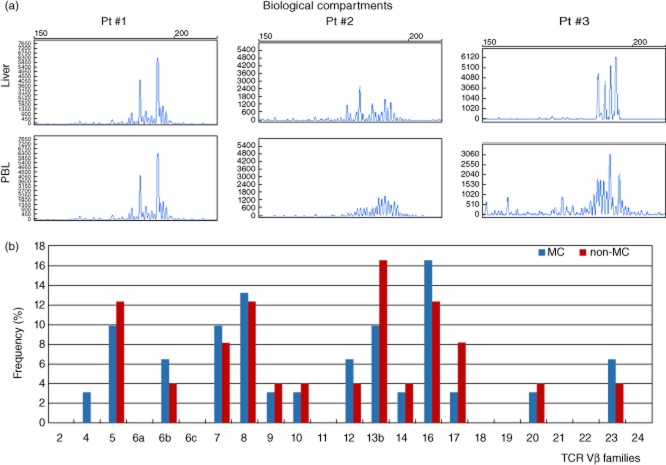
Spectratyping profiles showed that identical expanded T cell populations were recognized in the two biological compartments in eight (27%) and six (28·5%) MC and non-MC patients. Conversely, in 10 (33·3%) MC and five (23·8%) non-MC patients, T cell clonalities in the liver were demonstrated to be different from those found in PBL (Fig. 1a).
Fig. 1.
In (a), examples of variable (V)β T cell clonotypes analysed by laser capture microdissection (LCM)-based technique in portal infiltrates of liver biopsy samples and in corresponding peripheral blood lymphocytes (PBL). In patient 1, oligoclonal T cell receptor (TCR) Vβ complementarity-determining region 3 (CDR3) spectratype had an identical pattern in the two biological compartments. In patients 2 and 3 different spectratype features were demonstrated, being oligoclonal in the liver and polyclonal in the PBL in the former and oligoclonal with different peaks in the latter. In (b), frequency of TCR Vβ families are depicted in both mixed cryoglobulinaemia (MC) and non-MC groups.
In five (17%) MC and three (14%) non-MC patients, TCR Vβ gene rearrangements were demonstrated only in the liver, but not in the peripheral blood. In one (5%) non-MC patient, circulating T cell clonotypes occurred without evidence of intrahepatic TCR gene rearrangements. Frequencies of Vβ families determined in the liver and in the PBL of MC and non-MC patients were reported in Fig. 1b. No significant differences were noted between the two groups, with Vβ5, Vβ7, Vβ8, Vβ14 and Vβ16 families among the most frequent.
Demonstration of TCR gene rearrangements is related strictly to the detectability threshold which, indeed, is intrinsic to the current assays. The sensitivity levels of PCR assays in our hands reached the detection of one clonally rearranged cell in 100 non-rearranged control cells tested by diluting Jurkat cells in normal samples. However, the inability to detect TCR gene rearrangements includes many other reasons, such as atypical rearrangements, or rearrangements of non-functional genes leading to the incapability of PCR primers to anneal appropriately. Another factor responsible for attenuating the efficiency of PCR assays is the dilution of antigen-specific lymphocytes among the liver-resident T cell population. It can be assessed that primers currently used amplify CDR3 of T cells, but the products of a small clonal population may be obscured by polyclonal T cells in the sample. This drawback is particularly critical when considering total DNA extracted from the entire bioptic sample, which contains a mixture of relevant and irrelevant nucleic acids targets that probably interfere with the molecular analyses of cells of interest. In previous experiments in our laboratory, patterns of TCR Vβ clonalities obtained from total DNA extracted from the entire liver tissue were compared with those obtained from DNA extracted from microdissected intraportal inflammatory cells in 10 patients with HCV-related chronic active hepatitis. The results showed that despite polyclonal features obtained on total DNA, mono/oligoclonal patterns of TCR Vβ expansions were demonstrated by LCM-based analysis in two liver samples.
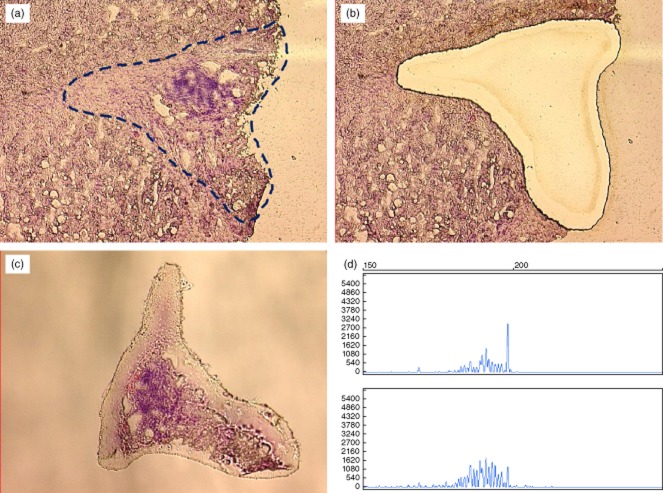
To overcome this critical point, LCM was then used to separate inflammatory cells precisely from the surrounding contaminating cells, as depicted in Fig. 2.
Fig. 2.
Example of laser capture microdissection (LCM)-based analysis: (a) section of liver biopsy showing enlarged portal tract with heavy inflammatory infiltrate and lymphoid aggregate, with appearance of a lymphoid follicle; (b) cleared area remaining in the context of tissue section after complete dissection of the portal structure; (c) dissected portal tract in the cap tube containing lysis buffer for molecular procedure; (d) analysis performed with genomic DNA obtained from microdissected portal tract in the upper square and on DNA obtained from entire liver sample in the lower square. Spectratyping profile shows an oligoclonal distribution in the former and plyclonal feature in the latter.
In Table 4, the relationship between the occurrence of expanded T cell clonalities and epidemiological, virological and laboratory parameters in MC and non-MC patients was summarized. The mean age and duration of HCV infection were not dissimilar between the groups. Interestingly, in spite of comparable average levels of circulating viral load, a significantly higher concentration of intrahepatic HCV RNA was demonstrated in patients with TCR Vβ gene rearrangements than in those without in both groups of patients.
Table 4.
Comparison of clinical, immunological, virological and histological parameters in mixed cryoglobulinaemia patients with and without intrahepatic TCR Vβ gene rearrangements
| HCV chronic infection | ||||
|---|---|---|---|---|
| Mixed cryoglobulinaemia | ||||
| With | Without | |||
| Vβ T cell gene rearrangements | ||||
| Parameters | With | Without | With | Without |
| Patients, n | 23 | 7 | 15 | 6 |
| Epidemiological | ||||
| Age (years) | 66·0 ± 4·7 | 64·3 ± 9·6 | 60·5 ± 12 | 61·3 ± 11·7 |
| Duration of HCV infection (years) | 18·5 ± 3·6 | 17·3 ± 2·5 | 20·8 ± 6·6 | 18·4 ± 5·9 |
| Virological | ||||
| HCV RNA | ||||
| Circulating (IU/ml) | 1 224 130 ± 803 350 | 1 090 606 ± 909 300 | 1 440 090 ± 660 449 | 1 360 880 ± 707 412 |
| Intrahepatic (pg/g) | 1120 ± 704* | 406 ± 216 | 1010 ± 860* | 396 ± 284 |
| Genotype | ||||
| I | 9 | 3 | 8 | 4 |
| II | 14 | 4 | 7 | 2 |
| Biochemical | ||||
| ALT (IU/ml) | 70 ± 24 | 78 ± 19 | 101 ± 31 | 98 ± 16 |
| Liver histology | ||||
| Chronic active hepatitis without cirrhosis (%) | 21 (91) | 6 (86) | 13 (87) | 5 (83) |
| Activity index | 6·7 ± 1·5 | 4·8 ± 0·9 | 7·1 ± 2·2 | 5·5 ± 1·6 |
| Stage | 2·6 ± 0·8 | 2·9 ± 0·6 | 3·0 ± 1·0 | 3·1 ± 0·96 |
| Cirrhosis (%) | 2 (9) | 1 (14) | 2 (13) | 1 (17) |
ALT: alanine aminotransferase; HCV: hepatitis C virus; TCR: T cell receptor; V: variable.
Discussion
As shown by spectratyping profiles of the TCR repertoire, MC patients with chronic HCV infection are characterized by clonally expanded T cells in either the liver or the peripheral blood. Almost 80% of examined livers and more than 60% of circulating blood samples showed rearrangements of the TCR Vβ gene. Similar features were found in HCV-infected patients without MC, in that T cell clonal expansions occurred in more than 70% of liver tissues and in almost 60% of peripheral lymphocytes.
Comparison of the T cell repertoire between liver and peripheral blood is of obvious importance if one assumes that T cell clonalities found more frequently in the liver than in the blood are likely to be relevant regarding HCV infection, in that HCV-specific T cells would be expected to home to the main site of infection. LIL include HCV-specific T cells, probably as a minor population. It seems reasonable to hypothesize that activated T cells become trapped in the liver, irrespective of their specificity. Furthermore, chronic inflammation in local tissues is capable of inducing additional T cell activation through the production of several cytokines, resulting in the expansion of activated T cells and switching of the TCR repertoire 33. This, indeed, may represent a mechanism that can enrich the liver tissue of expanded T cell clones, thus diversifying the intrahepatic T cell repertoire. Otherwise, as normal adult liver is devoid of constitutive lymphoid components, any intrahepatic T cells must have migrated into the liver 34. It can be argued that, during an immune response to hepatotrophic microorganisms, clonal expansions of antigen-specific lymphocytes occur in lymph nodes which drain the sites of infection. Then, activated lymphocytes enter the bloodstream and home to the liver. This implies that the pool of intrahepatic lymphocytes is maintained through the constant influx from extrahepatic sites. In this context, chemokines have been shown to orchestrate migration to and preferential sequestration in the liver of B and T cells 29,35. Intrahepatic T cell sequestration probably reflects deregulation of T cell traffic as the consequence of locally high production of chemokines. In this context, the formation of lymphoid follicle-like structures in the portal tracts of HCV-related MC patients is considered the morphological counterpart of ectopic lymphoid tissue which includes naive B cells in the central zone surrounded by mature B and T cells. These are structures that contribute to antigen presentation in situ and the clonal expansion of antigen-specific cells 36.
In the present series, 21·7% (five of 23) MC and 20% (three of 15) non-MC patients showed expanded T cell clones in the liver, but not in the circulation. This, indeed, may represent a biological condition potentially able to recruit a large number of inflammatory cells in the liver, suggesting that additional factors participate in the recruitment process. Similar mechanisms have been emphasized in an animal model of HBV chronic infection 37. Whether T regulatory CD4+CD25+ cells are defective in MC patients and influence the production of chemokines by stromal cells is a matter which deserves further investigation 38. Studies aimed at defining the role of in-situ production of factors involved in intrahepatic accumulation of inflammatory cells are warranted.
In 34·8% (eight of 23) and 40% (six of 15) MC and non-MC patients, respectively, clonal expansions of T cells in the peripheral blood reflected the intrahepatic population closely. Spectratype profiles between LIL and PBL appeared virtually identical, suggesting that T cell populations can circulate freely from one compartment to the other.
In a single instance, T cell clonalities were demonstrated in the peripheral blood, but not in the hepatic compartment. This possibly reflects an inability of the liver to entrap expanded T cells, or it could be the result of sampling error, in that T cell clonotypes occur in selected areas of liver sections, as demonstrated previously for expanded B cell clones 26.
T cell clonalities reflect down-regulation of the immune response in chronic HCV infection. It has been suggested that accumulated T cell clonal expansions represent a late differentiation stage of antigen-specific effector cells 39. However, viral clearance following IFN-α administration also suggests that this cytokine changes the quality of the cellular immune response 40.
Interestingly, the higher concentration of liver HCV RNA in patients with intraportal T cell clonal expansions supports the contention that HCV is involved directly in the pathogenesis of initial expansion and maintenance of T cell populations. Our data emphasize previous observations regarding clonally expanded B cell population in HCV-related MC patients 25. Overload of HCV-encoded proteins has been proposed to play a direct role in stimulating and maintaining intrahepatic B cell clones. CDR3 spectratyping determined the T cell repertoire regardless of antigen specificity, although clonal expansions of HCV-specific T cells could, potentially, be hidden by other T cell populations present in the liver 33. The existing data on TCR repertoires in HCV-infected patients do not clarify whether or not repertoire diversity is associated with HCV persistence. Public TCR were identified in response to two immunodominant HCV determinants (HSK and ATD) 41. The TCR repertoire selected against both antigens was comparable in patients with different disease outcome, suggesting that it is unlikely that resolution of HCV infection is controlled exclusively by the antigen-specific T cell repertoire 42. A formal differentiation between virus-specific and non-specific T cell populations must be defined at the site of infection and inflammation to assess the weight of the cellular components of the immune system 43.
Antigen-driven selection of T and B cell inflammatory populations has a direct impact on clinical and therapeutic outcomes of MC 21. In these patients, interaction between T and B cells may be of great relevance, in that intrahepatic production of RF molecules results in their reaction with human class I HLA molecules involved as part of the antigen-binding pocket, thus influencing peptide recognition by cell-mediated immune response 44,45.
It has been reported that clonally expanded intrahepatic T cells in HCV-infected patients without MC induce an immune regulatory environmental defect which predisposes to progressive inflammatory liver damage, thus establishing the basis for correlating clonal expansions of T cell populations to liver pathology 46,47. Restriction of the cellular immune response must be considered in the context of a limited host response to a pathogen capable of undergoing long-term spontaneous mutations that contribute heavily to the viral burden 44.
In conclusion, the present findings enhance the understanding of immune response in HCV-related MC patients in whom multi-faceted T cell response is comparable to that found in non-MC patients and is probably attributed to intrahepatic constant viral challenge. Further studies are needed to clarify the functional basis of TCR clonalities underlying the persistence/termination of chronic virus infection.
Acknowledgments
This study was supported in part by the Italian Ministry of University and Scientific and Technologic Research, National Project ‘Chronic liver damage induced by hepatitis C virus’ (DS); Agenzia Italiana del Farmaco (AIFA), funds for independent studies, 2007, contract no. FARM7SJX (DS); funds from University of Bari (D.S.); and funds from ‘Fondazione Cassa di Risparmio di Puglia’ (D.S.).
Disclosure
The authors declare no competing financial interests.
References
- 1.Whitcomb JM, Hughes SH. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 2.Butera ST, Roberts BD, Lam L, et al. Human immunodeficiency virus type 1 RNA expression by four chronically infected cell lines indicates multiple mechanisms of latency. J Virol. 1994;68:2726–2730. doi: 10.1128/jvi.68.4.2726-2730.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyashita EM, Yang B, Lam KM, et al. A novel form of Epstein–Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 4.McFadden G, Kane K. How DNA viruses perturb functional MHC expression to alter immune recognition. Adv Cancer Res. 1994;63:117–209. doi: 10.1016/s0065-230x(08)60400-5. [DOI] [PubMed] [Google Scholar]
- 5.Spriggs MK. Cytokine and cytokine receptor genes ‘captured’ by viruses. Curr Opin Immunol. 1994;6:526–529. doi: 10.1016/0952-7915(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 6.de Campos-Lima PO, Gavioli R, Zhang QJ, et al. HLA-A11 epitope loss isolates of Epstein–Barr virus from a highly A11+ population. Science. 1993;260:98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]
- 7.Phillips RE, Rowland-Jones S, Nixon DF, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 8.Cerny A, Chisari FV. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology. 1999;30:595–601. doi: 10.1002/hep.510300312. [DOI] [PubMed] [Google Scholar]
- 9.Wedemeyer H, He XS, Nascimbeni M, Davis AR, et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169:3447–3458. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 10.Ciuffreda D, Comte D, Cavassini M, et al. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. Eur J Immunol. 2008;38:2665–2677. doi: 10.1002/eji.200838336. [DOI] [PubMed] [Google Scholar]
- 11.Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol. 2004;58:391–424. doi: 10.1146/annurev.micro.58.030603.123836. [DOI] [PubMed] [Google Scholar]
- 12.Mehta SH, Lau B, Afdhal NH, et al. Exceeding the limits of liver histology markers. J Hepatol. 2009;50:36–41. doi: 10.1016/j.jhep.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonorino P, Leroy V, Dufeu-Duchesne T, et al. Features and distribution of CD8 T cells with human leukocyte antigen class I-specific receptor expression in chronic hepatitis C. Hepatology. 2007;46:1375–1386. doi: 10.1002/hep.21850. [DOI] [PubMed] [Google Scholar]
- 14.Garcia KC, Adams JJ, Feng D, et al. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubtsova K, Scott-Browne JP, Crawford F, et al. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci USA. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naylor K, Li G, Vallejo AN, et al. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174:7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 17.Giuggio VM, Bonkovsky HL, Rothman AL. Evolution of the intrahepatic T cell repertoire during chronic hepatitis C virus infection. Viral Immunol. 2005;18:179–189. doi: 10.1089/vim.2005.18.179. [DOI] [PubMed] [Google Scholar]
- 18.Wolfl M, Rutebemberwa A, Mosbruger T, et al. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J Immunol. 2008;181:6435–6446. doi: 10.4049/jimmunol.181.9.6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 20.Manfras BJ, Weidenbach H, Beckh KH, et al. Oligoclonal CD8+ T-cell expansion in patients with chronic hepatitis C is associated with liver pathology and poor response to interferon-alpha therapy. J Clin Immunol. 2004;24:258–271. doi: 10.1023/B:JOCI.0000025447.23473.ab. [DOI] [PubMed] [Google Scholar]
- 21.Sansonno D, Dammacco F. Hepatitis C virus, cryoglobulinaemia, and vasculitis: immune complex relations. Lancet Infect Dis. 2005;5:227–236. doi: 10.1016/S1473-3099(05)70053-0. [DOI] [PubMed] [Google Scholar]
- 22.Lauletta G, Russi S, Conteduca V, et al. Hepatitis C virus infection and mixed cryoglobulinemia. Clin Dev Immunol. 2012;2012:1–11. doi: 10.1155/2012/502156. 502156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sansonno D, Lauletta G, De Re V, et al. Intrahepatic B cell clonal expansions and extrahepatic manifestations of chronic HCV infection. Eur J Immunol. 2004;34:126–136. doi: 10.1002/eji.200324328. [DOI] [PubMed] [Google Scholar]
- 24.Dammacco F, Sansonno D, Piccoli C, et al. The lymphoid system in hepatitis C virus infection: autoimmunity, mixed cryoglobulinemia, and overt B-cell malignancy. Semin Liver Dis. 2000;20:143–157. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- 25.Sansonno D, De Vita S, Iacobelli AR, et al. Clonal analysis of intrahepatic B cells from HCV-infected patients with and without mixed cryoglobulinemia. J Immunol. 1998;160:3594–3601. [PubMed] [Google Scholar]
- 26.Sansonno D, Carbone A, De Re V, et al. Hepatitis C virus infection, cryoglobulinaemia, and beyond. Rheumatology (Oxf) 2007;46:572–578. doi: 10.1093/rheumatology/kel425. [DOI] [PubMed] [Google Scholar]
- 27.Sansonno D, Iacobelli AR, Cornacchiulo V, et al. Immunochemical and biomolecular studies of circulating immune complexes isolated from patients with acute and chronic hepatitis C virus infection. Eur J Clin Invest. 1996;26:465–475. doi: 10.1046/j.1365-2362.1996.162317.x. [DOI] [PubMed] [Google Scholar]
- 28.Ishak KG, Zimmerman HJ. Morphologic spectrum of drug-induced hepatic disease. Gastroenterol Clin North Am. 1995;24:759–786. [PubMed] [Google Scholar]
- 29.Sansonno D, Tucci FA, Troiani L, et al. Increased serum levels of the chemokine CXCL13 and up-regulation of its gene expression are distinctive features of HCV-related cryoglobulinemia and correlate with active cutaneous vasculitis. Blood. 2008;112:1620–1627. doi: 10.1182/blood-2008-02-137455. [DOI] [PubMed] [Google Scholar]
- 30.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 31.Gorochov G, Neumann AU, Kereveur A, et al. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4:215–221. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- 32.Shadrach B, Warshawsky I. A comparison of multiplex and monoplex T-cell receptor gamma PCR. Diagn Mol Pathol. 2004;13:127–134. doi: 10.1097/01.pdm.0000126419.92931.a3. [DOI] [PubMed] [Google Scholar]
- 33.Nascimbeni M, Rehermann B. Chronic HCV infection and the clonality of intrahepatic T cells. J Hepatol. 2003;38:677–680. doi: 10.1016/s0168-8278(03)00121-1. [DOI] [PubMed] [Google Scholar]
- 34.Jin Y, Fuller L, Carreno M, et al. The immune reactivity role of HCV-induced liver infiltrating lymphocytes in hepatocellular damage. J Clin Immunol. 1997;17:140–153. doi: 10.1023/a:1027326415164. [DOI] [PubMed] [Google Scholar]
- 35.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 36.Mosnier JF, Degott C, Marcellin P, et al. The intraportal lymphoid nodule and its environment in chronic active hepatitis C: an immunohistochemical study. Hepatology. 1993;17:366–371. [PubMed] [Google Scholar]
- 37.Sing GK, Li D, Chen X, et al. A molecular comparison of T lymphocyte populations infiltrating the liver and circulating in the blood of patients with chronic hepatitis B: evidence for antigen-driven selection of a public complementarity-determining region 3 (CDR3) motif. Hepatology. 2001;33:1288–1298. doi: 10.1053/jhep.2001.24026. [DOI] [PubMed] [Google Scholar]
- 38.Boyer O, Saadoun D, Abriol J, et al. CD4+CD25+ regulatory T-cell deficiency in patients with hepatitis C-mixed cryoglobulinemia vasculitis. Blood. 2004;103:3428–3430. doi: 10.1182/blood-2003-07-2598. [DOI] [PubMed] [Google Scholar]
- 39.Meyer-Olson D, Shoukry NH, Brady KW, et al. Limited T cell receptor diversity of HCV-specific T cell responses is associated with CTL escape. J Exp Med. 2004;200:307–319. doi: 10.1084/jem.20040638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pappas DJ, Coppola G, Gabatto PA, et al. Longitudinal system-based analysis of transcriptional responses to type I interferons. Physiol Genomics. 2009;38:362–371. doi: 10.1152/physiolgenomics.00058.2009. [DOI] [PubMed] [Google Scholar]
- 41.Tsai SL, Chen YM, Chen MH, et al. Hepatitis C virus variants circumventing cytotoxic T lymphocyte activity as a mechanism of chronicity. Gastroenterology. 1998;115:954–965. doi: 10.1016/s0016-5085(98)70268-9. [DOI] [PubMed] [Google Scholar]
- 42.Miles JJ, Thammanichanond D, Moneer S, et al. Antigen-driven patterns of TCR bias are shared across diverse outcomes of human hepatitis C virus infection. J Immunol. 2011;186:901–912. doi: 10.4049/jimmunol.1003167. [DOI] [PubMed] [Google Scholar]
- 43.Neumann-Haefelin C, Thimme R. Success and failure of virus-specific T cell responses in hepatitis C virus infection. Dig Dis. 2011;29:416–422. doi: 10.1159/000329807. [DOI] [PubMed] [Google Scholar]
- 44.Hahn YS. Subversion of immune responses by hepatitis C virus: immunomodulatory strategies beyond evasion? Curr Opin Immunol. 2003;15:443–449. doi: 10.1016/s0952-7915(03)00076-1. [DOI] [PubMed] [Google Scholar]
- 45.Williams RC, Jr, Malone CC, Kao KJ. IgM rheumatoid factors react with human class I HLA molecules. J Immunol. 1996;156:1684–1694. [PubMed] [Google Scholar]
- 46.Woitas RP, Sippel M, Althausen EM, et al. Differential expansion of T-cell receptor variable beta subsets after antigenic stimulation in patients with different outcomes of hepatitis C infection. Immunology. 2002;106:419–427. doi: 10.1046/j.1365-2567.2002.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pernollet M, Jouvin-Marche E, Leroy V, et al. Simultaneous evaluation of lymphocyte subpopulations in the liver and in peripheral blood mononuclear cells of HCV-infected patients: relationship with histological lesions. Clin Exp Immunol. 2002;130:518–525. doi: 10.1046/j.1365-2249.2002.01996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]