Abstract
Probiotics are live microorganisms which have beneficial effects on the host when ingested in adequate amounts. Probiotic bacteria may stimulate immune effector functions in a strain-specific manner. In this blind placebo-controlled trial, we investigated the effects on the immune system following daily intake of six different strains of lactobacilli or the Gram-negative bacterium Pseudomonas lundensis for 2 or 5 weeks. Blood lymphocyte subsets were quantified by fluorescence activated cell sorter and the expression of activation and memory markers was determined. The bacterial strains were also examined for their capacity to adhere to human intestinal cells and to be phagocytosed by human peripheral blood mononuclear cells. Intake of Lactobacillus plantarum strain 299v increased the expression of the activation marker CD25 (P = 0·01) on CD8+ T cells and the memory cell marker CD45RO on CD4+ T cells (P = 0·03), whereas intake of L. paracasei tended to expand the natural killer T (NK T) cell population (P = 0·06). The phagocytic activity of granulocytes was increased following intake of L. plantarum 299v, L. plantarum HEAL, L. paracasei or L. fermentum. In contrast, ingestion of L. rhamnosus decreased the expression of CD25 and CD45RO significantly within the CD4+ cell population. The observed immune effects after in-vivo administration of the probiotic bacteria could not be predicted by either their adherence capacity or the in-vitro-induced cytokine production. The stimulation of CD8+ T cells and NK T cells suggests that intake of probiotic bacteria may enhance the immune defence against, e.g. viral infections or tumours.
Keywords: lactobacilli, NK T cells, placebo-controlled, probiotics, T cell activation
Introduction
Probiotic bacteria are defined as live microorganisms which, when administered in adequate amounts, confer a health benefit on the host 1,2. Lactobacilli and bifidobacteria are the most frequently used bacteria in probiotic products as they are regarded as safe for human use, even for children and immunocompromised individuals 3. Intake of different probiotic bacteria has been shown to have clinical benefits in various physiological or pathological situations. The most clear-cut effects have been shown in diarrhoea caused by rotavirus infection 4,5 or antibiotic therapy 6–8. There are also studies showing positive clinical effects of probiotics on inflammatory bowel disease 9–12, irritable bowel syndrome 13–15, atopic dermatitis 16–18 and hypercholesterolaemia 19–21. The mechanisms by which probiotic bacteria contribute to clinical improvements in these conditions are not clear, and more human studies are needed to investigate basic mechanisms of probiotic function.
In theory, bacteria that are ingested in a sufficiently large dose are taken up into the Peyer's patches, where they are taken up by macrophages and/or dendritic cells and presented to the immune system. Studies in humans as well as in animals have shown that intake of probiotics stimulates cell-mediated innate immune functions, such as increased phagocytic activity of polymorphonuclear cells (PMN) and increased natural killer (NK) cell tumour killing activity 22–25.
Production of interleukin (IL)-12 by monocytes or dendritic cells in response to bacteria is of key importance for cell-mediated immunity, as IL-12 induces interferon (IFN)-γ production by T and NK cells which, in turn, activates phagocyte effector functions. Other cytokines, such as IL-6 and IL-10, may instead promote B cell activation, leading to antibody production (‘humoral immunity’). Ingestion of lactobacilli induces a humoral immune response, as demonstrated by increased circulation of antibody-producing B cell blasts 1–3 weeks after the start of probiotic intake 26–32. The antibodies produced are directed towards the ingested organism, but as a more broad activation of B cells occurs, antibodies also appear against other microorganisms and food proteins.
Lactobacilli belong to a very large and heterogeneous group of bacteria with widely differing properties, natural niches and metabolism. They have different adherence properties and may therefore differ in their ability to pass across the intestinal epithelium, a prerequisite for an interaction with antigen-presenting cells in the lamina propria. Furthermore, different species of lactobacilli may interact with antigen-presenting cells and induce different cytokine patterns leading to activation of different immune effector pathways. For example, Lactobacillus paracasei is a strong activator of IL-12 production, whereas L. rhamnosus induces more IL-10, a cytokine which counteracts cell-mediated immunity 33. In theory, it could be possible to tailor-make probiotics stimulating the desired immune reactions, but to date no systematic comparative studies exist. The immunomodulatory effects of six probiotic bacteria on human cells was studied recently after in-vitro stimulation of isolated peripheral blood mononuclear cells, cord blood cells and the monocyte/macrophage cell line CRL-9850 34.
In the present placebo-controlled study, we have investigated in vivo the effects on the innate and acquired immune system following daily intake of six different Lactobacillus strains, representing species predominant in the human intestinal or vaginal microbiota. The response was compared to placebo and to ingestion of the Gram-negative species Pseudomonas lundensis, a common dairy contaminant present in refrigerated milk. The strains were assessed in parallel for in-vitro adherence to intestinal epithelial cells and the cytokine pattern induced upon interaction with human blood leucocytes. The aim was to investigate, more systematically than performed previously, whether different species of lactobacilli differ in their capacity to stimulate cell-mediated immune functions and whether this pattern could be deduced by in-vitro behaviours of the strains. To our knowledge, this is the first systematic intervention study of the effects of different probiotic bacteria on selected immune functions in humans, as analysed in the peripheral blood.
Materials and methods
Study products
The study products were lyophilized bacteria belonging to the genera Lactobacillus or Pseudomonas, namely L. plantarum 299v (DSM9843) (n = 7), L. plantarum HEAL 19 (DSM15313) (n = 7), L. fermentum 35D (n = 7), L. paracasei 8700:2 (DSM13434) (n = 7), L. gasseri VPG44 (DSM16737) (n = 7) or L. rhamnosus 271 (DSM6594) (n = 7) or the Gram-negative bacterium P. lundensis (n = 7) or placebo (n = 10). The dose of bacteria was 1010 bacteria/day for lactobacilli and 109 bacteria/day for P. lundensis. The lower dose of the Gram-negative bacteria was based on calculation of the intake of 1000 ml milk having the maximal allowed amount of Gram-negative bacteria (106cfu/ml), according to food safety regulations. Bacteria were manufactured by Probi AB (Lund, Sweden) and packaged together with skimmed milk powder (Semper AB, Stockholm, Sweden) in 1-g lots in aluminium bags. The placebo product was skimmed milk powder in 1-g lots in aluminium bags. Randomization was performed by the manufacturer, and the code was broken after all analyses were made.
Subjects and trial criteria
Fifty-seven apparently healthy volunteers (37 female, 20 male) within the age range of 18–55 years (median 26 years) were recruited and randomized into eight groups, six groups receiving lactobacilli, one group the Gram-negative P. lundensis and one placebo group. The study participants were instructed to take the study product once daily, e.g. by mixing the content of one aluminium bag with a cold drink or sour milk. The intervention period was 2 weeks, and started after a 2-week-long wash-out period (Fig. 1a). However, for the groups receiving L. plantarum 299v or placebo there was a 5-week intervention after the initial wash-out period (Fig. 1b). Blood samples were taken at days 0, 14 and 35 (for only the 5-week treatment groups). The study was randomized and double-blind, with the limitation that the investigators were aware of whether subjects were in a 2- or 5-week treatment group. Each subject was supplied with a list of probiotic-containing products that should not be consumed during the time of the study. The subjects reported health status and adverse effects and confirmed their daily intake of study product in a diary kept during the trial. All study participants gave written consent. The study was approved by the ethics committee of the Medical Faculty, University of Gothenburg.
Fig. 1.
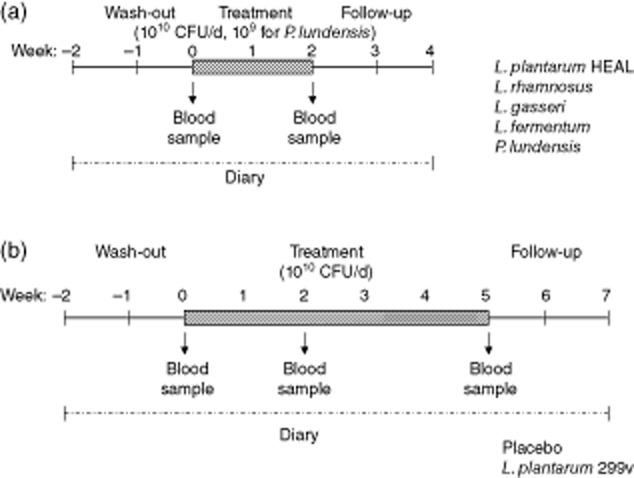
Subjects were assigned randomly to one of eight different study groups. The trial started with a wash-out period of 2 weeks. Thereafter, the treatment period followed. During this period, the subjects consumed one dose of study product per day for (a) 14 days (Lactobacillus plantarum HEAL, L. fermentum, L. paracasei, L. gasseri, L. rhamnosus, P. lundensis groups) or (b) 35 days (L. plantarum 299v and placebo group). Each dose contained 1010 (lactobacilli groups) or 109 bacteria (Pseudomonsas lundensis group). Blood samples were drawn on days 0, 14 and 35 (for only the 5-week treatment groups). Each subject was supplied with a list of probiotic-containing products that should not be consumed during the trial. Each participant kept a diary during the trial for reporting health status and adverse events, and for confirming the daily intake of the study product.
Flow cytometry
Phenotypic analysis of lymphocytes in whole blood was performed by flow cytometry. The following anti-human monoclonal antibodies purchased from Becton-Dickinson (Erembodegum, Belgium) were used as surface markers for different cell populations: CD3 fluorescein isothiocyanate (FITC) (SK7), CD4 allophycocyanin (APC) (SK3), CD8 peridinin chlorophyll (PerCP) (SK1), CD19 PerCP (SJ25C1), CD56 phycoerythrin (PE) (MY31), CD16 PE (B73·1) and CD5 FITC (L17F12). The following anti-human monoclonal antibodies were used for detection of different activation and memory markers: CD25 FITC (2A3), human leucocyte antigen D-related (HLA-DR) PE (L243), CD45RO PE (UCHL-1), CD38 PE (HB7), CD27 PE (L128) and CD11b PE (D12). Whole blood (100 μl) was incubated with antibodies (10 μl/antibody) for 30 min at 4°C in the dark. Thereafter, 2 ml of fluorescence activated cell sorter (FACS) lysing solution (Becton-Dickinson) was added and the mixtures were incubated for 15 min at 20°C in the dark. Cells were washed by adding 3 ml FACSFlow and centrifuged at 300 g for 5 min. Washed cells were resuspended in 200 μl FACSFlow and analysed on a FACSCalibur (Becton-Dickinson) with CellQuest software.
Phagocytosis assay
The phagocytic activity of granulocytes and monocytes was quantified with PHAGOTEST® (Orpegen Pharma, Heidelberg, Germany) according to the manufacturer's instructions, but with some modifications. Briefly, 20 × 106 FITC-labelled Escherichia coli or Staphylococcus aureus were added to precooled whole blood (100 μl). Blood cells and bacteria were incubated at 37°C for 10 min. Quenching solution was added and red blood cells were lysed. Washed cells were resuspended in 200 μl FACSFlow and analysed on a FacsCalibur (Becton-Dickinson) with CellQuest software. The granulocyte and monocyte cell populations were identified and gated in the forward-/side-scatter diagram. FITC-labelled phagocytosed bacteria were detected in the FL-1 channel and the mean fluorescence intensity was determined and used to measure the phagocytosed bacteria.
Preparation of bacteria for in-vitro stimulation of peripheral blood mononuclear cells (PBMC)
Lactobacillus strains were cultured aerobically on blood agar for 3 days and P. lundensis for 24 h. Bacteria were harvested in phosphate-buffered saline (PBS), washed and suspended to a concentration of 1 × 109 bacteria/ml, inactivated by exposure to ultraviolet (UV) light for 15 min and stored at–70°C. Inactivation was confirmed by negative viable counts.
Cell preparation and culture
PBMC cells were obtained from healthy blood donors. PBMC were isolated by density gradient centrifugation (Lymphoprep, Nyegaard, Norway) for 20 min at 1000 g. Cells were washed with RPMI-1640 medium (Gibco, Edinburgh, UK) and suspended in RPMI supplemented with 5% inactivated human serum from blood donors of blood group AB (Sigma, St Louis, MO, USA), 50 mM gentamycin (Sigma) and 2 mM L-glutamine (Sigma). Cell cultures containing 4 × 105 PBMCs in 200 μl were stimulated with 5 × 106 bacteria per well in round-bottomed 96-well microtitre plates (Nunc, Roskilde, Denmark). Cell cultures were incubated at 37°C in a humified atmosphere supplemented with 5% CO2. Supernatants were collected after 24 h of incubation for quantification of IL-10 and IL-12p70.
IL-10 and IL-12p70 determination
ELISA Eli-pair for human IL-10 and IL-12p70 (Diaclone, Besançon, France) were coated onto polystyrene microtitre plates (Costar Invitrogen, San Diego, CA, USA) overnight at 4°C. The plates were washed three times with PBS containing 0·05% Tween® 20 (Merck KgaA, Darmstadt, Germany) and blocked with 5% bovine serum albumin (BSA)–PBS (Sigma) for 2 h at room temperature. Thereafter supernatants were added in dilutions of 1:1, 1:5 and 1:25. Recombinant IL-10 and IL-12p70 (Diaclone) were used as standards. Standards and samples were incubated for 2 h at room temperature. After washing, biotinylated detecting antibody (Diaclone) was added and the plates were incubated for 3 h at room temperature. Plates were washed again and incubated for 20 min with streptavidin-horseradish peroxidase (HRP). After a final washing, tetramethylbenzidine (TMB) containing H2O2 was added. The reaction was blocked with 1 M H2SO4 after 20 min and the absorbance at 450 nm was measured.
Adherence to human intestinal epithelial cells
The ability of the lactobacillus strains and P. lundensis strain to adhere to cells of the human colonic carcinoma cell line HT-29 in the absence and presence of methyl-α-D-mannoside was tested as described previously 35. Briefly, the lactobacillus strains were cultured aerobically on Rogosa SL agar at 37°C for 3 days and P. lundensis on TSA agar plates for 24 h. Each well was also cultured in static Luria broth containing 0·1 % CaCl2 for 3 consecutive days with daily passages at 37°C. This represents the optimal culture conditions to promote expression of type 1 fimbriae, which mediate mannose-dependent adherence to a variety of cells, including human intestinal epithelial cells 36. Bacteria were harvested in PBS, washed and suspended to a concentration of 5 × 109 bacteria/ml. Cells of the human adenocarcinoma cell line HT-29 were cultured in Eagle's medium (PAA Laboratories GmbH, Pasching, Austria) supplemented with 5% fetal calf serum, 2 mM L-glutamine (Sigma) and 50 μg/ml gentamycin (Sigma). A few days after the cells had reached confluence they were detached with buffer containing 0·54 mM ethylenediamine tetraacetic acid (EDTA), washed and suspended in Hanks's balanced salt solution (HBSS; PAA Laboratories) at a concentration of 5 × 106 cells/ml. The HT-29 cells, bacteria and HBSS were mixed at a ratio of 1:1:3 and then incubated with end-over-end rotation for 30 min at 4°C. HT-29 cells and bacteria were incubated in the presence and absence of methyl-α-D-mannoside (Sigma), which blocks adherence to mannose-containing receptors, termed mannose-sensitive adherence. The cells were washed once with ice-cold PBS and fixed with neutral buffered formalin (Histofix; Histolab, Göteborg, Sweden). The number of bacteria attached to at least 40 cells was determined by using interference-contrast microscopy (magnification ×500; Nikon Optiphot microscope equipped with interference contrast equipment; Bergströms Instruments, Göteborg, Sweden), and the mean number of bacteria per cell was calculated. Mannose-resistant (MR) adherence was depicted as the number of bacteria adhering in the presence of mannose. Mannose-sensitive (MS) adherence was obtained by subtracting MR adherence from adherence in the absence of mannose (total adherence). Two transformed E. coli strains, which adhere to HT-29 cells by a mannose-sensitive or a mannose-resistant mechanism, were used as controls (506 MS and 506 MR, respectively).
Principal component analysis (PCA)
Principal component analysis was performed in order to find structures in the data set. This method transforms an original set of correlated variables into a smaller number of uncorrelated variables, termed principal components. The first principal component accounts for as much of the variability in the data as possible, and subsequent principal components as much of the remaining variability as possible. PCA was performed using Simca-p+ (Umetrics, Umeå, Sweden).
Statistics
Individual changes regarding different immune parameters were determined by calculating the ratio between the individual values obtained at days 14 and 0, or the values at days 35 and 0. These ratios were used for all descriptive data and statistics. Statistical analyses were performed with Statview software. The Mann–Whitney U-test was used to compare the two ratios (day 14/day 0 and day 35/day 0) between the different treatment groups.
Results
Clinical observations
Fifty-four of 57 volunteers completed the study. Two people were excluded due to infection leading to antibiotic treatment (one in the placebo group and one in the group receiving P. lundensis) and one was excluded on day 16 due to pregnancy (placebo group). Only mild adverse gastrointestinal side effects were reported following intake of study products and there was no apparent relation between symptoms and treatment group (Table 1).
Table 1.
Frequency of volunteers who reported any adverse gastrointestinal effects during the trial. Volunteers consumed 1010 lactobacilli or 109 Pseudomonas lundensis per day as freeze-dried product after a 2-week wash-out period. Symptoms were noted in a diary
| Wash-out period (weeks) | Treatment period (weeks) | Follow-up period (weeks) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | −2 | −1 | 1 | 2 | 3 | 4 | 5 | +1 | +2 |
| Lactobacillus plantarum 299v | 0/7 | 1/7 | 3/7 | 2/7 | 3/7 | 2/7 | 1/7 | 1/7 | 0/7 |
| L. plantarum HEAL | 0/7 | 1/7 | 1/7 | 2/7 | 2/7 | 1/7 | |||
| L. fermentum | 0/7 | 0/7 | 0/7 | 0/7 | 1/7 | 0/7 | |||
| L. paracasei | 0/7 | 0/7 | 1/7 | 0/7 | 0/7 | 0/7 | |||
| L. gasseri | 0/7 | 0/7 | 3/7 | 1/7 | 4/7 | 0/7 | |||
| L. rhamnosus | 1/7 | 1/7 | 0/7 | 0/7 | 1/7 | 0/7 | |||
| P. lundensis | 1/6 | 1/6 | 1/6 | 1/6 | 0/6 | 0/6 | |||
| Placebo | 0/9 | 0/9 | 2/9 | 3/9 | 1/8 | 1/8 | 0/8 | 0/8 | 0/8 |
Effect on different cell populations following intake of the study product
The baseline (day 0) numbers of different lymphocytes, percentages of CD4+ and CD8+ T cells expressing the activation markers CD25 and HLA-DR and geometric mean fluorescence intensity (GMFI) of the memory marker CD45RO on CD4+ and CD8+ T cells are shown in Table 2. No significant differences were observed between different groups at this time-point.
Table 2.
Baseline numbers (day 0) of different lymphocytes per ml blood, percentages of CD25 and human leucocyte antigen D-related (HLA-DR) on CD4+ and CD8+ T cells and geometric mean fluorescence intensity (GMFI) of CD45RO on CD4+ T cells and CD8+ T cells, respectively (mean ± standard error of the mean)
| No of cells (×103) | % of CD4+ expressing | % of CD8+ expressing | CD45RO on CD4+ (GMFI) | CD45RO on CD8+ (GMFI) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | CD4+ | CD8+ | NK T | CD25 | HLA-DR | CD25 | HLA-DR | ||
| Lactobacillus plantarum 299v | 650 (92) | 320 (37) | 64 (17) | 28 (3·1) | 12 (1·6) | 4·6 (1·2) | 23 (6·9) | 53 (10) | 27 (5·6) |
| L. plantarum HEAL | 820 (100) | 330 (43) | 56 (19) | 36 (3·0) | 11 (9·0) | 8·2 (1·8) | 31 (12) | 130 (39) | 61 (15) |
| L. fermentum | 910 (82) | 480 (51) | 87 (21) | 34 (1·9) | 10 (1·2) | 6·5 (1·2) | 32 (7·1) | 71 (13) | 36 (5·6) |
| L. paracasei | 790 (87) | 320 (64) | 98 (21) | 38 (2·1) | 19 (10) | 11 (5·9) | 35 (12) | 83 (13) | 50 (17) |
| L. gasseri | 770 (54) | 500 (110) | 110 (39) | 38 (4·6) | 9·0 (1·8) | 6·3 (3·3) | 23 (4·6) | 110 (96) | 45 (14) |
| L. rhamnosus | 780 (110) | 390 (50) | 110 (22) | 34 (2·3) | 7·0 (0·6) | 6·3 (1·8) | 23 (4·4) | 80 (24) | 40 (11) |
| Psudomonas lundensis | 730 (65) | 470 (84) | 87 (29) | 43 (8·2) | 24 (15) | 19 (12) | 40 (13) | 65 (9·4) | 38 (3·5) |
| Placebo | 650 (43) | 300 (34) | 110 (30) | 31 (2·5) | 10 (1·7) | 12 (13) | 28 (11) | 39 (8·2) | 23 (5·1) |
NK T: natural killer T cells.
The relative increase/decrease in population size of CD4+ T cells, CD8+ T cells, B cells including B-1 cells (CD19+CD5+), NK cells, granulocytes and monocytes were calculated as the ratio between the values at days 14 and 0 (the ratio between days 35 and 0 values was also calculated for the groups receiving L. plantarum 299v or placebo). Following intake of L. paracasei, there was a tendency towards increased proportion of lymphocytes being identified as NK T cells, i.e. cells expressing both the NK cell marker CD56 as well as the T cell marker CD3 (+40% on average, P = 0·06) (Fig. 2). No other cell populations were altered in any of the treatment groups (data not shown).
Fig. 2.
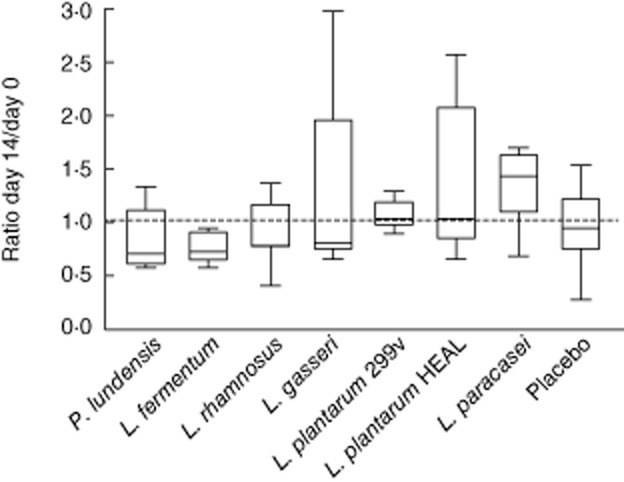
Percentages of lymphocytes positive for the natural killer T cell markers (CD56 CD16 CD3) were analysed by flow cytometry. Group means (± standard error of the mean) are shown based on individual ratios (day 14/day 0).
Expression of activation and memory markers on CD4+ and CD8+ T cells after intake of study product
The activation markers CD25 (the α-chain of the IL-2 receptor) and HLA-DR are expressed on the surface of different types of leucocytes such as activated T lymphocytes. Furthermore, T lymphocytes that have been activated may express the CD45RO memory marker. The expression of these markers on the surface of circulating CD4+ and CD8+ T cells after intake of bacteria is shown in Fig. 3. Regarding the CD4+ T cells (Fig. 3, left panel), treatment with L. plantarum 299v tended to increase the percentage of cells expressing CD25 within the CD4+ population (P = 0·13) after a 5-week intervention, while intake of either L. rhamnosus or L. fermentum led to decreased expression of CD25 (P = 0·03 and P = 0·04, respectively). Ingestion of the Gram-negative bacterium P. lundensis increased significantly the percentage of CD4+ T cells positive for the activation marker HLA-DR (P = 0·03). An increase for HLA-DR was also seen after ingestion of L. plantarum 299v, but the variation was large and the increase was not significant (P = 0·35) after 5 weeks. Furthermore, intake of L. plantarum 299v increased expression of CD45RO on CD4+ T cells, the difference being significant compared with the placebo group at day 35 (P = 0·03). Intake of L. rhamnosus and L. fermentum was associated with down-regulation of CD45RO expression on CD4+ T cells (P = 0·03 and P = 0·15, respectively).
Fig. 3.
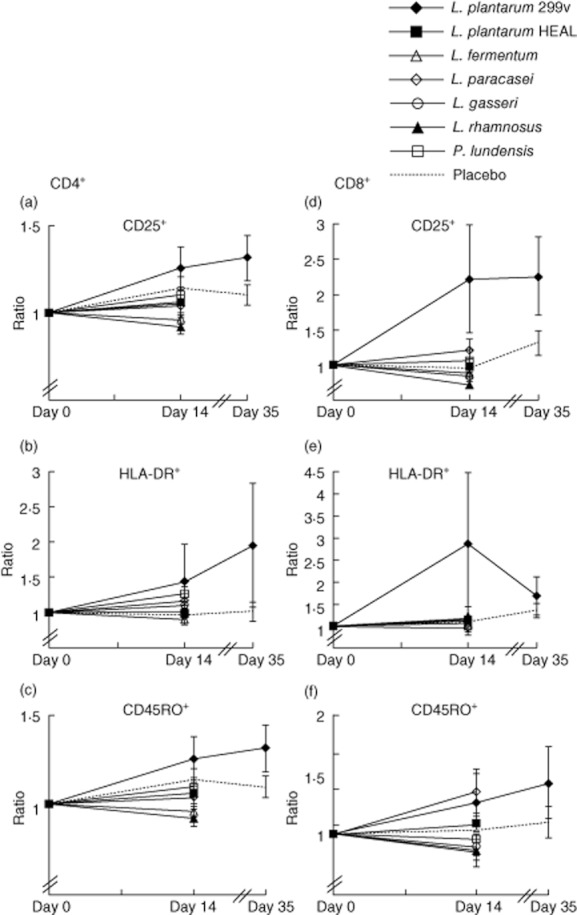
Percentages of CD4+CD3+ and CD8+ CD3+ cells positive for CD25 and human leucocyte antigen D-related were analysed by flow cytometry. Group means [± standard error of the mean (s.e.m.)] based on individual ratios, day 14/day 0 and day 35/day 0 (for Lactobacillus plantarum and placebo group only) are shown. Geometric means of the fluorescence intensity of the expression of the memory cell marker CD45RO on CD4+CD3+ and CD8+ CD3+ cells were analysed by flow cytometry. Group means (± s.e.m.) are shown based on individual ratios, day 14/day 0 and day 35/day 0 (for L. plantarum and placebo group only).
When examining circulating CD8+ T cells (Fig. 3, right panel), we observed a dramatic increase in the expression of the activation marker CD25 after intake of the study product containing L. plantarum 299v (P = 0·01). The difference was significant after 2 weeks, but not after 5 weeks, although CD25 expression remained highly elevated (Fig. 3). Intake of L. plantarum 299v also led to a strong up-regulation of HLA-DR on CD8+ T cells after 2 weeks, although the effect was not significant (P = 0·12). Treatment with L. rhamnosus was associated with a non-significant decrease in CD25 expression on CD8+ T cells (P = 0·30).
Intake of L. paracasei tended to increase the expression of the memory marker within the population of CD8+ T cells (P = 0·11). Ingestion of L. plantarum 299v for 5 weeks also seemed to increase the expression of the CD45RO memory marker, but due to the large variation in the group the increase did not reach significance (P = 0·56). In contrast, intake of L. rhamnosus and L. fermentum tended to decrease the expression of the CD45RO memory cell marker on CD8+ T cells (P = 0·37 and P = 0·27, respectively) (Fig. 3). No differences could be found among the different study groups regarding the expression of the memory marker CD27 on the surface B lymphocytes (data not shown).
In-vitro phagocytic activity following intake of study product
T cell activation enhances the phagocytic capacity of phagocytes. The ability of polymorphonuclear (PMN) leucocytes and monocytes to phagocytose FITC-labelled Gram-positive or Gram-negative bacteria in vitro was tested. As shown in Fig. 4, PMN cells from volunteers consuming L. paracasei were more efficient than PMN cells from placebo-treated volunteers in phagocytosis of the Gram-negative bacteria E. coli (P = 0·05). A similar tendency was seen for the two L. plantarum strains (P = 0·06 for both) and L. fermentum (P = 0·06). There were no significant differences between the treatment groups regarding phagocytosis of the Gram-positive bacteria S. aureus (data not shown). Furthermore, no differences were detected regarding the phagocytic activity of monocytes (data not shown).
Fig. 4.
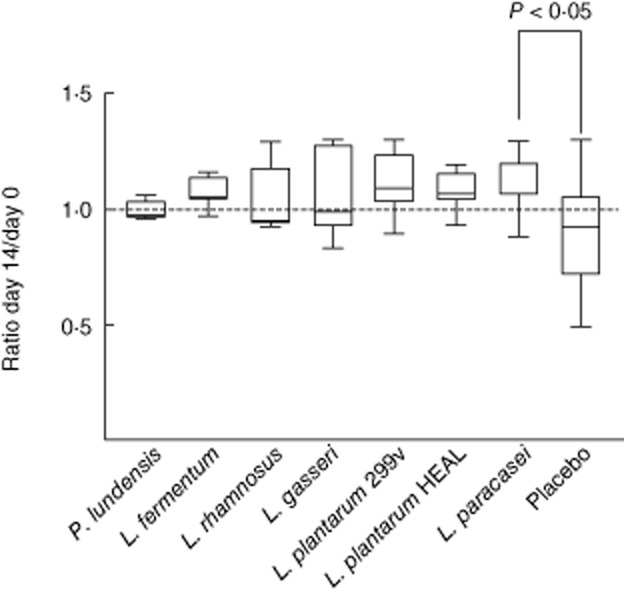
The in-vitro phagocytic activity of polymorphonuclear (PMN) cells was analysed by incubating whole blood cells with fluorescein isothiocyanate (FITC)-labelled Escherichia coli. Cells were analysed by flow cytometry. The PMN cell population was gated on the forward-/side-scatter diagram. Mean fluorescence intensity in the FITC-channel was determined. The ratio between mean fluorescence values obtained at days 14 and 0 was determined individually. Group means (± standard error of the mean) based on individual ratios (day 14/day 0) are shown.
Cytokine production after in-vitro stimulation of PBMC
We observed tendencies of T cell activation after ingestion of L. plantarum 299v and L. paracasei and T cell deactivation by L. rhamnosus and L. fermentum. Therefore, we sought to investigate whether this could be related to different patterns of mediator release from cells of the innate immune system after stimulation with the various bacteria. For example, IL-12 is a strong inducer of T cell activation, while IL-10 is an inhibitor of such activation. PBMC were isolated from healthy blood donors and stimulated in vitro with bacteria for 24 h, whereafter concentrations of IL-10 and IL-12p70 in the supernatants were quantified. L. fermentum, L. plantarum 299v, L. plantarum HEAL and L. paracasei induced high concentrations of the bioactive form of IL-12, while both L. rhamnosus and L. gasseri were weak inducers of IL-12p70 (Fig. 5). P. lundensis did not induce any detected levels of IL-12p70, which is compatible with the low IL-12-inducing capacity of Gram-negative bacteria 37. The Gram-negative species P. lundensis was a potent inducer of IL-10 (Fig. 6), which is in line with the fact that Gram-negative bacteria induce much more IL-10 than do Gram-positive bacteria 36. Interestingly, L. rhamnosus was also a quite efficient inducer of IL-10, as opposed to the other lactobacillus strains, which may fit with its T cell deactivating properties. However, L. fermentum, which behaved similarly to L. rhamnosus in vivo, induced no more IL-10 than L. paracasei, which was a T cell activator in vivo.
Fig. 5.
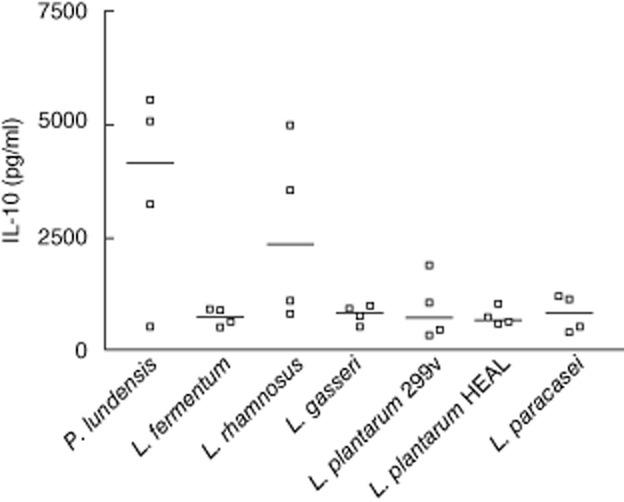
Peripheral blood mononuclear cells (PBMC, 4 × 105 cells) were stimulated for 24 h with 5 × 106 whole ultraviolet-inactivated lactobacilli or Pseudomonsas lundensis. Interleukin-10 levels in supernatants were quantified with enzyme-linked immunosorbent assay. Data are shown from four individual healthy blood donors and median.
Fig. 6.
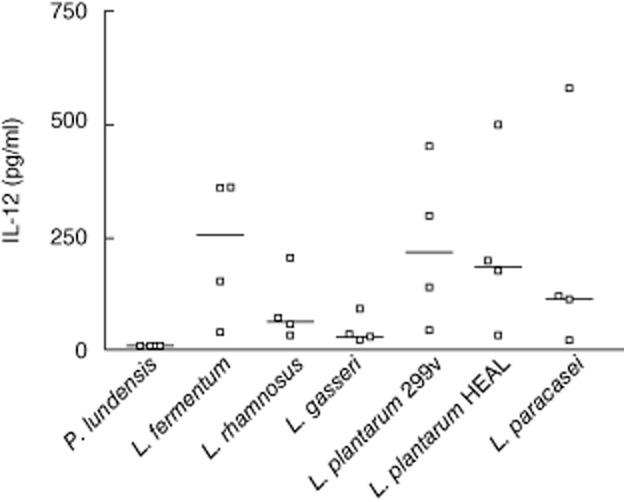
Peripheral blood mononuclear cells (PBMC, 4 × 105 cells) were stimulated for 24 h with 5 × 106 whole ultraviolet-inactivated lactobacilli or Pseudomonsas lundensis. Interleukin-12p70 levels in supernatants were quantified with enzyme-linked immunosorbent assay. Data are shown from four individual healthy blood donors and median.
Adherence to HT-29 cells
Another bacterial trait that might relate to a strain's capacity to stimulate the immune system when administered via the oral route is the capacity to adhere to intestinal epithelial cells. Thus, strongly adherent strains could be expected to colonize the small intestine more easily and be taken up by M cells covering the Peyer's patches, which are the inductive sites of mucosal immune responses. The six lactobacillus strains and the Gram-negative P. lundensis strain were tested for their ability to adhere to human colonic cell line HT-29 cells (Table 3). L. fermentum, L. paracasei and L. plantarum 299v adhered to HT-29 cells via mannose-sensitive mechanisms, while L. plantarum HEAL adhered via a mannose-resistant mechanism. L. gasseri and L. rhamnosus were non-adherent, as was P. lundensis.
Table 3.
Adherence of lactobacillus and Pseudomonas strains to the human colonic carcinoma cell line HT-29 in the presence or absence of methyl-α-D-mannoside
| Adherence (mean no. of bacteria/cell) | ||
|---|---|---|
| Strain | MS adherence = total adherence – MR adherence | MR adherence |
| Lactobacillus plantarum 299v | 4·0 | 2·6 |
| L. plantarum HEAL | 0 | 5·7 |
| L. fermentum | 6·6 | 7·5 |
| L. paracasei | 20 | 4·8 |
| L. gasseri | 0 | 1·0 |
| L. rhamnosus | 0·3 | 1·0 |
| P. lundensis (in Luria broth) | 0·1 | 1·6 |
| Escherichia coli 506 MS | 24 | |
| 506 MR | 12 | |
MR: mannose-resistant; MS: mannose-sensitive.
PCA
PCA was employed to investigate inherent structures in the variable data set. Differences in a number of immune parameters between days 14 and 0 for each individual consisted of the X variables, together with the treatment group. A significant model was generated (Fig. 7). As seen in the figure, all immune activation variables (increased expression of CD25 and HLA-DR on both CD4+ and CD8+ cells, increased NK T cell numbers and increased phagocytosis) were associated with one another, as the bars all point in the same direction (Fig. 7). Two of the lactobacilli, L. plantarum 299v and L. paracasei, were associated with such immune activation, while L. fermentum and L. rhamnosus pointed in the other direction, i.e. intake of these bacteria opposed this type of immune activation pattern. Treatment with placebo, L. plantarum HEAL and P. lundensis were all without effect on the measured parameters.
Fig. 7.
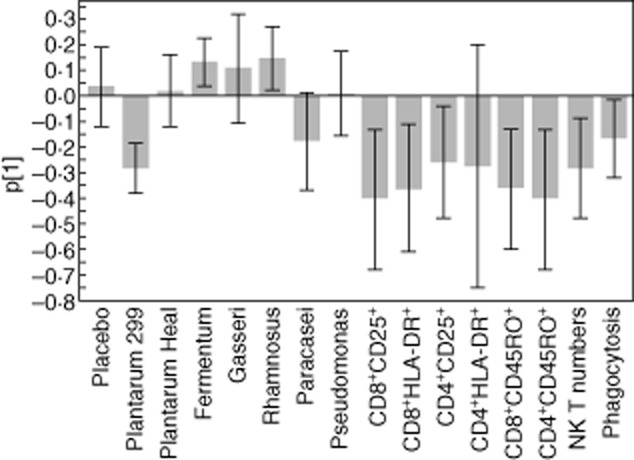
Principal component analysis. The immune response of each individual was calculated as the difference in the parameter in question between days 14 and 0, i.e. before ingestion of the product. The values on each parameter, as well as the treatment group, consisted of the X variables which were entered into the analysis. The y-axis shows p[1]. The X2X of the model was 0·20.
Discussion
This blind placebo-controlled study is, to our knowledge, the first study comparing in vivo the influence of different species of potentially probiotic bacteria on several immune parameters in humans. For comparison, the Gram-negative bacterium P. lundensis was included in the study. A ×10 lower dose was used for the Gram-negative bacterium due to safety reasons – the dose of 109 bacteria was chosen to reflect the intake of 1000 ml of milk containing the allowed limit of P. lundensis, a common contaminant of dairy milk. Even with this lower dose, we observed a 30% increase in HLA-DR expression on CD4+ T cells after intake of P. lundensis for 2 weeks.
Some of the lactobacilli induced strong immunomodulating effects when administered in a dose of 1010 colony-forming units (CFU)/day. The most pronounced finding of the present study was the unique ability of L. plantarum 299v to increase the expression of activation markers on cells participating in acquired cell-mediated immunity, i.e. CD4+ and CD8+ T cells. Ingestion of L. plantarum 299v was linked to a markedly increased expression of CD25 as well as HLA-DR on the surface of CD8+ cells and, to a lesser degree, also on the CD4+ cell population. Similar results were reported for L. casei Shirota after in-vitro stimulation of PBMC with different concentrations of the bacteria 38. There was a significant increase in the expression of CD25 on CD8+ but not on CD4+ cells.
We find it difficult to believe that the large proportion of CD4+ and CD8+ T cells that were activated by L. plantarum were specific for this bacterium. Antigen-specific T cells would make up a few per cent of all circulating T cells, while we noted a doubling of expression of CD25 and HLA-DR on all circulating CD8+ T cells and a 40% increase in the intensity of the CD45RO marker on T cells. We would therefore like to propose that intake of this bacterium has a broad activating function on antigen-presenting cells, leading to broadly enhanced T cell activity. Another striking observation was that the two strains of lactobacilli, namely L. fermentum and L. rhamnosus, seemed to down-regulate several parameters of cell-mediated immunity. Thus, consumption of either L. fermentum or L. rhamnosus for 2 weeks led to significantly reduced expression of the activation marker CD25 on CD4+ T cells and a non-significant decrease in the memory marker CD45RO on both CD4+ and CD8+ T cells.
It is often claimed that the in-vivo effect of probiotic strains could be deduced by their in-vitro behaviour. In the present study, we tested the capacity of the strains to elicit IL-12 and IL-10 from human PBMC and to adhere to a colonic cell line. Four of the six lactobacillus strains tested in this study were strong inducers of IL-12, i.e. L. paracasei, L. plantarum 299v, L. plantarum HEAL and L. fermentum, whereas L. rhamnosus and L. gasseri induced only moderate amounts. L. paracasei and L. plantarum 299v were strong stimulators of cell-mediated immune functions in vivo, whereas L. fermentum counteracted T cell activation and L. plantarum HEAL was apparently inactive. We also tested the capacity of the lactobacilli to induce IL-10 production. IL-10 opposes IL-12 functions, dampening T cell activation, reducing IFN-γ production by NK cells and decreasing antigen presentation by dendritic cells to T cells 39–41. As expected, the Gram-negative P. lundensis induced larger amounts of IL-10 than most of the lactobacilli, confirming that Gram-negative bacteria are more efficient IL-10 inducers than Gram-positive bacteria 37. Interestingly, however, we noted strong IL-10 production in response to the L. rhamnosus strain tested here, which is in accordance with our previous findings using a L. rhamnosus strain isolated from the human gastrointestinal tract 37. The tendency towards T cell deactivation in people consuming L. rhamnosus might, thus, be explained by induction of IL-10 in vivo. However, the fact that L. fermentum was an equally strong down-regulator of T cell responses, despite inducing large amounts of IL-12 and no more IL-10 than other lactobacilli in vitro, indicates that other mechanisms may operate. We have observed recently that whereas intact Gram-positive bacteria induce large amounts of IL-12, fragmented bacteria do not induce IL-12, but instead block IL-12 production induced by intact bacteria 42. To speculate, bacteria which tend to be fragmented in vivo before or upon contact with the gut immune system might be weak inducers of cell-mediated immunity and may even counteract such immune mechanisms. One additional factor that could affect in vivo effects on the immune system is the degree to which the bacterium in question reaches the gut-associated lymphoid tissue and systemic immune system, a necessary condition to modulate its function. For example, it has been described previously that the type of immune response after microbial invasion is related to the degree of bacterial uptake by dendritic cells in the lamina propria 43. Furthermore, dead bacteria are less efficient than live bacteria and invasive bacteria are more efficient than commensals in affecting the immune system.
In this study, L. paracasei adhered strongly to HT-29 cells and induced a high production of IL-12 by stimulated PBMC. Taken together, these qualities may be responsible for the observed effects on in-vivo immune functions. PMN cells from study participants having ingested L. paracasei for 2 weeks were better at phagocytosing E. coli compared to PMN cells from subjects consuming placebo. There was also a similar, but non-significant, tendency for the other lactobacilli.
Consumption of L. paracasei increased the phagocytic capacity of blood PMN. Previously, increased phagocytosis by blood cells has been reported after ingestion of different strains of lactobacilli and bifidobacteria 22–25. Furthermore, consumption of L. paracasei tended to increase the numbers of NK T cells, a lymphocyte subpopulation that co-expresses the NK cell marker CD56 and the T cell receptor–CD3 complex 44. These cells are strong inducers of IFN-γ and their expansion and activation may underlie the activation of blood phagocytes. NK T cells exert effector functions against tumour- 45,46 and virus-infected cells 47,48, but also appear to play a central role in the regulation of autoimmune diseases, such as multiple sclerosis 49, type 1 diabetes 50–54 and systemic lupus erythematosus 55. Other clinical studies evaluating the immunological effects of probiotic bacteria have shown that intake of L. casei Shirota or L. rhamnosus HN001 or Bifidobacterium lactis HN019 enhance the NK (including NK T) cell tumour killing activity of K562 cells 22,23,25,38. L. paracasei 8700:2 was used in combination with L. plantarum HEAL 9 in a recently conducted clinical study examining the effects of the probiotic bacteria on common cold infections 56. Interestingly, intake of the probiotic mixture during 12 weeks resulted in a reduced incidence of common cold and reduced severity of common cold symptoms, compared to a placebo group.
A puzzling observation in the present study was the relative inefficiency of the strain L. plantarum HEAL 19 to induce cell-mediated immunity, as opposed to strain L. plantarum 299v. We regard it unlikely that this strain of L. plantarum would have largely different effects on APC compared with L. plantarum 299v and, indeed, these two strains induced similar amounts of both IL-10 and IL-12 after in-vitro stimulation of PBMC. There is, however, one interesting difference between these two L. plantarum strains. L. plantarum 299v exhibits a mannose-specific adhesin, which enables it to bind to intestinal epithelial cells, a phenomenon that has been shown previously 35 and confirmed in the present study. This adhesin is shared by approximately 65% of all L. plantarum strains that are found at the intestinal mucosa 57. As shown in our study, L. plantarum HEAL 19 lacks this adhesin and may thus not share the property of L. plantarum 299v to interact with the intestinal mucosa 35. Both L. plantarum 299v and L. paracasei, which were strong inducers of cell-mediated immunity, shared the MS adherence capacity. However, L. fermentum, that also had MS adherence capacity, had T cell deactivating properties at the same time. Thus, the ability to adhere to mannose-containing receptors may enhance the bacterial uptake or target them to interaction with antigen-presenting cells, but the result of this interaction may depend upon other characteristics of the bacteria.
The limitations of the present study were the small size of the groups and the relatively short time of intervention, with the exception of L. plantarum 299v. We chose to test a relatively broad range of bacterial strains and species, rather than concentrating on a single strain, which is usually conducted in clinical studies. This precluded the use of large groups and long study periods, which would have been very costly. We used PCA to explore whether a pattern could be revealed between intake of the different species/strains and change in immune parameters. Indeed, we found that increased expression of CD25 and HLA-DR on both CD4+ and CD8+ cells, increased NK T cell numbers and increased phagocytosis were linked, suggesting that they constitute a coordinated set of reactions that occur in response to certain microbes. Interestingly, this type of response characterized individuals who had consumed L. plantarum 299v or L. paracasei, while consumption of L. rhamnosus and L. gasseri seemed to counteract this type of response. PCA is of value in exploring potential patterns of reactivity with limited numbers of observations. The effect patterns we observed in the present pilot study on innate and acquired immunity, as induced by different bacteria, can be exploited in more depth in future studies.
In addition to the clinical data, we investigated the in-vitro effect on cytokine production and adhesion to HT-29 cells of the selected strains. From this we conclude that in-vitro data might offer some tentative explanations for the observed effects on the immune system in vivo, but that most in-vivo results could not be deduced from in-vitro data. Therefore, it is extremely important to perform clinical studies in order to determine different effects on the immune system following intake of bacteria.
To summarize, intake of L. paracasei induced innate cell-mediated immune functions efficiently, presumably mediated via its strong adhesion capacity and capacity to induce IL-12 in human monocyte/macrophages. L. fermentum and L. rhamnosus seemed to down-regulate several aspects of cell-mediated immunity. In the case of L. rhamnosus, this could relate to its IL-10 inducing properties, while it remained unexplained for L. fermentum. Lastly, L. plantarum 299v seemed to be an activator of acquired T cell immunity. Clinical studies could reveal whether this ability may be used to promote, e.g. anti-viral or anti-tumour immunity.
Acknowledgments
The skilful technical assistance of Ingela Kinell is greatly appreciated. We also acknowledge Dr Anna Rudin for helpful discussions during the study. The study costs were supported by a grant of 50 000 euro from Probi AB (Lund, Sweden). The study was also funded by the Swedish Asthma and Allergy Associations Research Foundation and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning.
Disclosure
This study has been funded partly by Probi AB (Lund, Sweden). Anna Berggren and Irini Lazou Ahrén, who are co-authors in the present study, are employees at Probi AB and contributed to the analysis of the results and the writing of the manuscript, but were not involved in the conduction of the study.
References
- 1.Schrezenmeir J, de Vrese M. 2001 Probiotics, prebiotics, and synbiotics – approaching a definition. Am J Clin Nutr. 2001;73(Suppl 2):361S–364. doi: 10.1093/ajcn/73.2.361s. [DOI] [PubMed] [Google Scholar]
- 2.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. [PubMed] [Google Scholar]
- 3.Borriello SP, Hammes WP, Holzapfel W, et al. Safety of probiotics that contain lactobacilli or bifidobacteria. Clin Infect Dis. 2003;36:775–780. doi: 10.1086/368080. [DOI] [PubMed] [Google Scholar]
- 4.Muñoz JA, Chenoll E, Casinos B, et al. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl Environ Microbiol. 2011;77:8775–8783. doi: 10.1128/AEM.05548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–1049. doi: 10.1016/s0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- 6.Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2011;(11) doi: 10.1002/14651858.CD004827.pub3. CD004827. doi: 10.1002/14651858.CD004827.pub3. [DOI] [PubMed] [Google Scholar]
- 7.Cremonini F, Di Caro S, Nista EC, et al. Meta-analysis: the effect of probiotic administration on antibiotic-associated diarrhoea. Aliment Pharmacol Ther. 2002;16:1461–1467. doi: 10.1046/j.1365-2036.2002.01318.x. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza AL, Rajkumar C, Cooke J, Bulpitt CJ. Probiotics in prevention of antibiotic associated diarrhoea: meta-analysis. BMJ. 2002;324:1361. doi: 10.1136/bmj.324.7350.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelouhab K, Rafa H, Toumi R, Bouaziz S, Medjeber O, Touil-Boukoffa C. Mucosal intestinal alteration in experimental colitis correlates with nitric oxide production by peritoneal macrophages: effect of probiotics and prebiotics. Immunopharmacol Immunotoxicol. 2012;34:590–597. doi: 10.3109/08923973.2011.641971. [DOI] [PubMed] [Google Scholar]
- 10.Shanahan F. Probiotics in inflammatory bowel disease – therapeutic rationale and role. Adv Drug Deliv Rev. 2004;56:809–818. doi: 10.1016/j.addr.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Gionchetti P, Amadini C, Rizzello F, Venturi A, Campieri M. Review article: treatment of mild to moderate ulcerative colitis and pouchitis. Aliment Pharmacol Ther. 2002;16(Suppl 4):13–19. doi: 10.1046/j.1365-2036.16.s4.3.x. [DOI] [PubMed] [Google Scholar]
- 12.Gionchetti P, Rizzello F, Venturi A, Campieri M. Probiotics in infective diarrhoea and inflammatory bowel diseases. J Gastroenterol Hepatol. 2000;15:489–493. doi: 10.1046/j.1440-1746.2000.02162.x. [DOI] [PubMed] [Google Scholar]
- 13.Clarke G, Cryan JF, Dinan TG, Quigley EM. Review article: probiotics for the treatment of irritable bowel syndrome – focus on lactic acid bacteria. Aliment Pharmacol Therapy. 2012;35:403–413. doi: 10.1111/j.1365-2036.2011.04965.x. [DOI] [PubMed] [Google Scholar]
- 14.Ringel Y, Ringel-Kulka T. The rationale and clinical effectiveness of probiotics in irritable bowel syndrome. J Clin Gastroenterol. 2011;45(Suppl):S145–148. doi: 10.1097/MCG.0b013e31822d32d3. [DOI] [PubMed] [Google Scholar]
- 15.Nobaek S, Johansson ML, Molin G, Ahrné S, Jeppsson B. Alteration of intestinal microflora is associated with reduction in abdominal bloating and pain in patients with irritable bowel syndrome. Am J Gastroenterol. 2000;95:1231–1238. doi: 10.1111/j.1572-0241.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- 16.Drago L, Iemoli E, Rodighiero V, Nicola L, De Vecchi E, Piconi S. Effects of Lactobacillus salivarius LS01 (DSM 22775) treatment on adult atopic dermatitis: a randomized placebo-controlled study. Int J Immunopathol Pharmacol. 2011;24:1037–1048. doi: 10.1177/039463201102400421. [DOI] [PubMed] [Google Scholar]
- 17.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 18.Rosenfeldt V, Benfeldt E, Nielsen SD, et al. Effect of probiotic Lactobacillus strains in children with atopic dermatitis. J Allergy Clin Immunol. 2003;111:389–395. doi: 10.1067/mai.2003.389. [DOI] [PubMed] [Google Scholar]
- 19.Jones ML, Martoni CJ, Parent M, Prakash S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br J Nutr. 2012;107:1505–1513. doi: 10.1017/S0007114511004703. [DOI] [PubMed] [Google Scholar]
- 20.Anderson JW, Gilliland SE. Effect of fermented milk (yogurt) containing Lactobacillus acidophilus L1 on serum cholesterol in hypercholesterolemic humans. J Am Coll Nutr. 1999;18:43–50. doi: 10.1080/07315724.1999.10718826. [DOI] [PubMed] [Google Scholar]
- 21.Bertolami MC, Faludi AA, Batlouni M. Evaluation of the effects of a new fermented milk product (Gaio) on primary hypercholesterolemia. Eur J Clin Nutr. 1999;53:97–101. doi: 10.1038/sj.ejcn.1600683. [DOI] [PubMed] [Google Scholar]
- 22.Sheih YH, Chiang BL, Wang LH, Liao CK, Gill HS. Systemic immunity-enhancing effects in healthy subjects following dietary consumption of the lactic acid bacterium Lactobacillus rhamnosus HN001. J Am Coll Nutr. 2001;20(2 Suppl):149–156. doi: 10.1080/07315724.2001.10719027. [DOI] [PubMed] [Google Scholar]
- 23.Arunachalam K, Gill HS, Chandra RK. Enhancement of natural immune function by dietary consumption of Bifidobacterium lactis (HN019) Eur J Clin Nutr. 2000;54:263–267. doi: 10.1038/sj.ejcn.1600938. [DOI] [PubMed] [Google Scholar]
- 24.Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am J Clin Nutr. 2001;74:833–839. doi: 10.1093/ajcn/74.6.833. [DOI] [PubMed] [Google Scholar]
- 25.Chiang BL, Sheih YH, Wang LH, Liao CK, Gill HS. Enhancing immunity by dietary consumption of a probiotic lactic acid bacterium (Bifidobacterium lactis HN019): optimization and definition of cellular immune responses. Eur J Clin Nutr. 2000;54:849–855. doi: 10.1038/sj.ejcn.1601093. [DOI] [PubMed] [Google Scholar]
- 26.Paineau D, Carcano D, Leyer G, et al. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol Med Microbiol. 2008;53:107–113. doi: 10.1111/j.1574-695X.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 27.Fukushima Y, Kawata Y, Hara H, Terada A, Mitsuoka T. Effect of a probiotic formula on intestinal immunoglobulin A production in healthy children. Int J Food Microbiol. 1998;42:39–44. doi: 10.1016/s0168-1605(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 28.de Vrese M, Rautenberg P, Laue C, Koopmans M, Herremans T, Schrezenmeir J. Probiotic bacteria stimulate virus-specific neutralizing antibodies following a booster polio vaccination. Eur J Nutr. 2005;44:406–413. doi: 10.1007/s00394-004-0541-8. [DOI] [PubMed] [Google Scholar]
- 29.Link-Amster H, Rochat F, Saudan KY, Mignot O, Aeschlimann JM. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol Med Microbiol. 1994;10:55–63. doi: 10.1111/j.1574-695X.1994.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 30.Kaila M, Isolauri E, Virtanen E, Arvilommi H. Preponderance of IgM from blood lymphocytes in response to infantile rotavirus gastroenteritis. Gut. 1992;33:639–642. doi: 10.1136/gut.33.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaila M, Isolauri E, Soppi E, Virtanen E, Laine S, Arvilommi H. Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus strain. Pediatr Res. 1992;32:141–144. doi: 10.1203/00006450-199208000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Majamaa H, Isolauri E, Saxelin M, Vesikari T. Lactic acid bacteria in the treatment of acute rotavirus gastroenteritis. J Pediatr Gastroenterol Nutr. 1995;20:333–338. doi: 10.1097/00005176-199504000-00012. [DOI] [PubMed] [Google Scholar]
- 33.Hessle C, Hanson LA, Wold AE. Lactobacilli from human gastrointestinal mucosa are strong stimulators of IL-12 production. Clin Exp Immunol. 1999;116:276–282. doi: 10.1046/j.1365-2249.1999.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donkor ON, Ravikumar M, Proudfoot O, et al. Cytokine profile and induction of T helper type 17 and regulatory T cells by human peripheral mononuclear cells after microbial exposure. Clin Exp Immunol. 2012;167:282–295. doi: 10.1111/j.1365-2249.2011.04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adlerberth I, Ahrne S, Johansson ML, Molin G, Hanson LA, Wold AE. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol. 1996;62:2244–2251. doi: 10.1128/aem.62.7.2244-2251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duguid JB, Robertson WB. Mechanical factors in atherosclerosis. Lancet. 1957;272:1205–1209. doi: 10.1016/s0140-6736(57)91786-5. [DOI] [PubMed] [Google Scholar]
- 37.Hessle C, Andersson B, Wold AE. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while Gram-negative bacteria preferentially stimulate IL-10 production. Infect Immun. 2000;68:3581–3586. doi: 10.1128/iai.68.6.3581-3586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong H, Rowland I, Tuohy KM, Thomas LV, Yaqoob P. Selective effects of Lactobacillus casei Shirota on T cell activation, natural killer cell activity and cytokine production. Clin Exp Immunol. 2010;161:378–388. doi: 10.1111/j.1365-2249.2010.04173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J Exp Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiorentino DF, Zlotnik A, Vieira P, et al. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J Immunol. 1991;146:3444–3451. [PubMed] [Google Scholar]
- 41.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barkman C, Martner A, Hessle C, Wold AE. Soluble bacterial constituents down-regulate secretion of IL-12 in response to intact Gram-positive bacteria. Microbes Infect. 2008;10:1484–1493. doi: 10.1016/j.micinf.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Rescigno M, Rotta G, Valzasina B, Ricciardi-Castagnoli P. Dendritic cells shuttle microbes across gut epithelial monolayers. Immunobiology. 2001;204:572–581. doi: 10.1078/0171-2985-00094. [DOI] [PubMed] [Google Scholar]
- 44.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 45.Kawano T, Nakayama T, Kamada N, et al. Antitumor cytotoxicity mediated by ligand-activated human V alpha24 NKT cells. Cancer Res. 1999;59:5102–5105. [PubMed] [Google Scholar]
- 46.Smyth MJ, Thia KY, Street SE, et al. Differential tumor surveillance by natural killer (NK) and NKT cells. J Exp Med. 2000;191:661–668. doi: 10.1084/jem.191.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakimi K, Guidotti LG, Koezuka Y, Chisari FV. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J Exp Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Exley MA, Bigley NJ, Cheng O, et al. CD1d-reactive T-cell activation leads to amelioration of disease caused by diabetogenic encephalomyocarditis virus. J Leukoc Biol. 2001;69:713–718. Erratum in: J Leukoc Biol 2001; 70(2):340. [PubMed] [Google Scholar]
- 49.Illés Z, Kondo T, Newcombe J, Oka N, Tabira T, Yamamura T. Differential expression of NK T cell V alpha 24J alpha Q invariant TCR chain in the lesions of multiple sclerosis and chronic inflammatory demyelinating polyneuropathy. J Immunol. 2000;164:4375–4381. doi: 10.4049/jimmunol.164.8.4375. [DOI] [PubMed] [Google Scholar]
- 50.Hong S, Scherer DC, Singh N, et al. Lipid antigen presentation in the immune system: lessons learned from CD1d knockout mice. Immunol Rev. 1999;169:31–44. doi: 10.1111/j.1600-065x.1999.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 51.Hammond KJ, Poulton LD, Palmisano LJ, Silveira PA, Godfrey DI, Baxter AG. Alpha/beta-T cell receptor (TCR)+ CD4– CD8– (NKT) thymocytes prevent insulin-dependent diabetes mellitus in non-obese diabetic (NOD)/Lt mice by the influence of interleukin (IL)-4 and/or IL-10. J Exp Med. 1998;187:1047–1056. doi: 10.1084/jem.187.7.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehuen A, Lantz O, Beaudoin L, et al. Overexpression of natural killer T cells protects Valpha14–Jalpha281 transgenic non-obese diabetic mice against diabetes. J Exp Med. 1998;188:1831–1839. doi: 10.1084/jem.188.10.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gombert JM, Tancrède-Bohin E, Hameg A, et al. IL-7 reverses NK1+ T cell-defective IL-4 production in the non-obese diabetic mouse. Int Immunol. 1996;8:1751–1758. doi: 10.1093/intimm/8.11.1751. [DOI] [PubMed] [Google Scholar]
- 54.Godfrey DI, Kinder SJ, Silvera P, Baxter AG. Flow cytometric study of T cell development in NOD mice reveals a deficiency in alphabetaTCR+CDR–CD8– thymocytes. J Autoimmun. 1997;10:279–285. doi: 10.1006/jaut.1997.0129. [DOI] [PubMed] [Google Scholar]
- 55.Mieza MA, Itoh T, Cui JQ, et al. Selective reduction of V alpha 14+ NK T cells associated with disease development in autoimmune-prone mice. J Immunol. 1996;156:4035–4040. [PubMed] [Google Scholar]
- 56.Berggren A, Lazou Ahrén I, Larsson N, Önning G. Randomised, double-blind and placebo-controlled study using new probiotic lactobacilli for strengthening the body immune defence against viral infections. Eur J Nutr. 2011;50:203–210. doi: 10.1007/s00394-010-0127-6. [DOI] [PubMed] [Google Scholar]
- 57.Ahrné S, Nobaek S, Jeppsson B, Adlerberth I, Wold AE, Molin G. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol. 1998;85:88–94. doi: 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]


