Abstract
The isolation and identification of unknown membrane proteins offers the prospect of discovering new pharmaceutical targets and identifying key biochemical receptors. However, interactions between membrane protein targets and soluble ligands are difficult to study in vitro due to the insolubility of membrane proteins in non-detergent systems. Nanodiscs, nanoscale discoidal lipid bilayers encircled by a membrane scaffold protein belt, have proven to be an effective platform to solubilize membrane proteins and have been used to study a wide variety of purified membrane proteins. This report details the incorporation of an unbiased population of membrane proteins from Escherichia coli membranes into Nanodiscs. This solubilized membrane protein library (SMPL) forms a soluble in vitro model of the membrane proteome. Since Nanodiscs contain isolated proteins or small complexes, the SMPL is an ideal platform for interactomics studies and pull-down assays of membrane proteins. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the protein population before and after formation of the Nanodisc library indicates that a large percentage of the proteins are incorporated into the library. Proteomic identification of several prominent bands demonstrates the successful incorporation of outer and inner membrane proteins into the Nanodisc library.
Keywords: Nanodisc, Membrane Proteins, Proteomics, Solubilized Membrane Protein Library, Bioanalytical methods, Genomics / Proteomics, Nanoparticles / Nanotechnology
Introduction
Identification of membrane proteins receptors for soluble ligands has the potential to reveal new drug targets and to elucidate important biochemical interactions. However, isolating membrane protein targets is challenging due to the difficulty of solubilizing membrane proteins outside of the lipid bilayer without disrupting native interactions [1,2]. In vivo studies, including yeast two-hybrid [3] and fluorescence resonance energy transfer, have been applied to mapping membrane protein interactions. However, these whole-cell measurements suffer from high levels of false positives and false negatives, and they require genomic modification of proteins to introduce appropriate labels [4]. In vitro measurements are complicated by the need to solubilize membrane proteins using detergents, which can disrupt key protein-protein interactions and interfere with down-stream analysis methods [1].
Nanodiscs have proven to be an effective technology for solubilizing membrane proteins in detergent-free buffers [5]. Nanodiscs are nanoscale lipid bilayers encircled by two amphipathic membrane scaffold proteins (MSP) [6]. Previous studies have incorporated a range of proteins and complexes into Nanodiscs [7–11]. Typically, the protein of interest is purified and isolated prior to incorporation. Controlling the ratios of membrane proteins and MSP allows the assembly of membrane protein-Nanodisc complexes with defined stoichiometry [12,13]. Civjan et al. demonstrated that a functional cytochrome P450 may also be incorporated directly from a solubilized membrane and can be purified post-incorporation [14].
Nanodiscs have been used in previous proteomics applications as “bait” for isolating and identifying glycolipid and membrane protein interaction partners. Borch et al. assembled Nanodiscs with ganglioside GM1 [15]. Co-immunoprecipitation of the GM1 Nanodiscs with culture media from Escherichia coli isolated heat labile enterotoxin B. Another study assembled Nanodiscs with membrane transporters SecYEG and MalFGK, and each membrane protein-Nanodisc system was incubated with stable isotope-labeled cell culture extracts [16]. Soluble proteins interacting with the membrane protein bait in Nanodiscs were isolated, separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and identified with liquid chromatography-tandem mass spectrometry (LC-MS/MS). Both studies demonstrated the utility of Nanodiscs as a platform for interactomics.
Although previous studies have measured the interaction of a heterogeneous soluble protein pool with homogenous Nanodiscs containing a single membrane protein species, it is possible to reverse the experiment to measure the interaction of a single homogenous soluble ligand with a heterogeneous Nanodisc-solubilized membrane protein library (SMPL). In such an experiment, Nanodiscs serve as the “prey” towards immobilized ligand bait. This inversion allows isolation and identification of unknown membrane protein targets in vitro without the need for detergent buffer. However, the utility of this approach depends on the formation of SMPL Nanodiscs that faithfully reflects the membrane proteome. This report details a general protocol for optimization of SMPL formation and examines the degree to which the SMPL reflects the membrane proteome of pooled inner and outer membranes from E. coli.
Experimental Methods
Materials
Lysozyme, octyl-beta-glucoside, imidazole, ampicillin, ethylenediaminetetraacetic acid (EDTA), and Amberlite XAD-2 beads were purchased from Sigma-Aldrich (St. Louis, MO). Phenylmethanesulfonyl fluoride (PMSF) was obtained from Fisher Scientific (Pittsburgh, PA). Sodium Cholate as obtained from Affymetrix (Maumee, OH). Sodium dodecyl sulfate was purchased from Bio Rad (Hercules, CA). 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was obtained from Avanti Polar Lipids (Alabaster, AL).
Escerichia coli Growth and Membrane Isolation
Library competent DH5α E. coli cells were purchased from Invitrogen and transformed with pUC 19 control DNA plasmid. Cells were cultured in terrific broth media with 100 μg/mL ampicillin and were grown for 24 hours. Cell pellets were harvested by centrifugation and frozen at −80 °C. Cell pellets were resuspended in cold lysozyme buffer (75 mM Tris HCl, 0.25 M sucrose, 0.25 mM EDTA, 0.02 mg/mL lysozyme, pH 8) and pelleted again by centrifugation. Pelleted cells were resuspended in cold lysis buffer (50 mM Tris HCl, 1 mM PMSF, pH 8) and were lysed by sonicating on ice. Lysed cells were centrifuged at 2000×g for 10 minutes at 4 °C to remove any unbroken cells or debris. The supernatant was removed and centrifuged at 90000×g (maximum radial centripetal force) for 1 hour to harvest the cell membrane.[17] The membrane pellet was resuspended in 0.1 M phosphate buffer, pH 7.7, and flash frozen. The concentration of membrane proteins in the resuspension was typically 2 mg/mL, as measured by a bicinchoninic acid (BCA) assay (Thermo Scientific, Rockford, IL).
Detergent Extraction of Membrane Proteins
Assembly of membrane proteins into Nanodiscs relies on the initial solubilization of the membrane protein in detergent. The membrane solution was centrifuged at 12000×g for 30 minutes. Membranes were resuspended in an equal volume of either octyl-glucoside (OG) buffer (1% w/v OG, 0.1 M phosphate, pH 7.7) or sodium dodecyl sulfate (SDS) buffer (0.1% w/v SDS, 20 mM Tris, 0.1 M NaCl, 0.1% w/v sodium azide). Membranes were thoroughly mixed and incubated at room temperature for 15 minutes to extract membrane proteins. Insoluble membranes were removed by centrifugation at 12000×g for 15 minutes. The concentration of membrane proteins in the detergent extract was measured by a BCA assay. Typically, the membrane protein concentration was 1–2 mg/mL in the detergent extract.
Nanodisc-Solubilized Membrane Protein Library Preparation
Expression and purification of membrane scaffold protein (MSP) was described previously [6,18]. MSP1E3D1, a variant of MSP with three extended helices and a poly-histidine tag, was used because this scaffold makes Nanodiscs with a 12.1 nm Stokes diameter that will accommodate large membrane proteins. This represents an upper limit on the size of membrane proteins or complexes that will be incorporated in the Nanodisc, as anything that extends more than 12 nm along the membrane will not fit inside the Nanodisc. Membrane proteins that extend less than 12 nm along the membrane may be incorporated in Nanodiscs and may extend any length perpendicular to the membrane surface, which will increase the overall Stokes diameter of the Nanodisc complex. MSP from a stock concentration of around 175 μM was added in a ratio ranging from 2 to 20 μg membrane proteins in detergent per nmol MSP. POPC in chloroform was dried under nitrogen and solvated in 0.1 M sodium cholate to 50 mM. The molar ratio of POPC:MSP was tested within the range of 40 to 130 as described below. Detergent-solubilized membrane proteins, MSP, and cholate-solubilized POPC were combined, and extra cholate was added to bring the final cholate concentration to 20 mM. This reconstitution mixture was incubated for two hours at 4 °C. Self assembly of the Nanodisc library was initiated by adding 0.5–0.8 g Amberlite XAD-2 hydrophobic beads per mL of reconstitution mixture, and the mixture was incubated overnight on an orbital shaker at 4 °C. The Amberlite XAD-2 hydrophobic beads serve to remove the detergents and drive Nanodiscs assembly.
Nanodiscs were purified by immobilized metal affinity chromatography. The reconstitution mixture was removed from the hydrophobic beads and filtered through a 0.22 μm filter from Millipore (Marlborough, MA). Filtered reconstitution mixture was loaded on nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Qiagen, Valencia, CA), washed with buffer containing 15 mM imidazole, and eluted in buffer containing 500 mM imidazole. The Ni-NTA column captured Nanodiscs by the poly-histidine tag on MSP1E3D1, washing away proteins or lipids not incorporated into Nanodiscs.
Size exclusion chromatography (SEC) was performed with a calibrated Superdex HR 200 10/30 size exclusion column (Amersham-Pharmacia Biotech, Piscataway, NJ). Samples were filtered prior to analysis and injected using a 500 μL sample loop and a flow rate of 0.5 mL/min. To evaluate the optimal protein to lipid ratio, a chromatogram from lipid-only POPC Nanodiscs was aligned with chromatograms from SMPL Nanodiscs of various lipid ratios with a custom program in Mathematica 8.0.4. The sum of squared errors was calculated between the SMPL Nanodisc chromatograms and the aligned control Nanodisc chromatogram.
SDS-PAGE Analysis
SDS-PAGE analysis of membrane protein libraries in Nanodiscs is complicated by the large excess of MSP in the sample. To observe the proteins in the Nanodisc library, MSP was removed by a Ni-NTA column. Samples were loaded on the column, washed with a 15 mM imidazole buffer, and eluted by adding buffer with 50 mM sodium cholate. The cholate disassembled the Nanodiscs allowing the membrane proteins to elute from the column while MSP remained bound. Membrane proteins released from the Nanodiscs, proteins from the raw membranes, and proteins from the detergent extract were prepared for SDS-PAGE using a methanol-chloroform-water precipitation to remove lipids, salts, and detergents [19]. Around 10 μg total protein was used for each precipitation. Precipitated samples were dissolved in Laemmli buffer, incubated at 70 °C for 20 min, and run on Bio Rad Mini-PROTEAN TGX 12% PAGE gels. Gels were stained with Imperial Protein Stain (Thermo Scientific) and imaged with a Molecular Dynamics Typhoon 9400 Multilaser Scanner. Multiple gels were analyzed with similar results, but data is shown for a single gel.
Image analysis of gels was performed using a custom program in Mathematica 8.0.4. Median value of the pixel intensity was calculated for each row of pixels in a lane for the region ranging from 20 kDa to 100 kDa. Peaks in the line trace were identified using a custom continuous wavelet transform algorithm [20]. Peaks were considered overlapping if they were within 16 pixels; the total region of consideration was around 1600 pixels.
Proteomic Identification
Gel slices were excised, destained, and digested in 25 μL sequencing-grade trypsin (G-Biosciences, St. Louis, MO) at a concentration of 12.5 ng/μL with a CEM Discover microwave digester (Mathews, NC) for 15 minutes at 55 °C. Peptides were extracted using 50% acetonitrile with 5% formic acid, dried, and resuspended in 13 μL of 5% acetonitrile with 0.1% formic acid. Ten μL were injected for LC-MS/MS.
Mass spectrometry (MS) was performed on a Waters quadrupole time-of-flight connected to a Waters nano Acquity Ultra Performance Liquid Chromatography system. A Waters Atlantis C-18 column (0.003 mm particle, 0.075 mm by 150 mm) was used at a flow rate of 250 nL/min. Peptide elution was performed using a linear gradient of water with acetonitrile containing 0.1% formic acid with the gradient ramping from 0–60% acetonitrile over one hour. Mass spectrometry utilized data dependent acquisition with MS/MS scans being performed on the four most abundant peaks at a given time. Data analysis was performed using the Waters Lynx Global Server 2.2.5 and Mascot (Matrix Sciences). Mascot searches were performed with the NCBI NR database and the SwissProt database specifying E. coli as the organism. Both databases gave similar results. The peptide tolerance was set to 0.5 Da for both MS and MS/MS measurements. One missed trypsin site as well as variable oxidation of methionine were allowed.
Results and Discussion
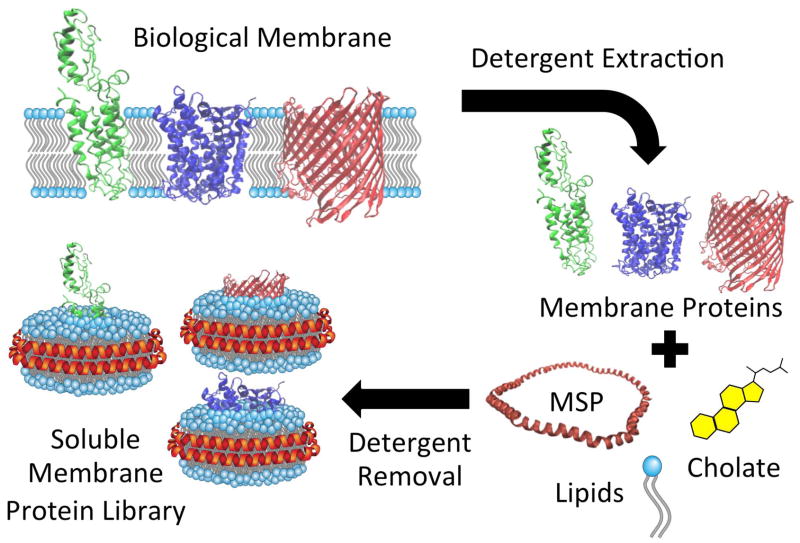
There are several important steps and variables to consider when preparing a Nanodisc library (see Figure 1 for schematic). The best approach to each step will depend on the specific biological system of interest. This report outlines a generalized protocol for the formation of Nanodisc-solubilized membrane protein libraries and characterizes the method as applied to a prototype E. coli membrane system.
Fig. 1.
Schematic of SMPL formation. Membrane proteins are extracted from the membranes with detergent and added to a mixture of MSP, lipids, and cholate. As the detergents are removed, the components self-assemble into a Nanodisc library
Isolation and Detergent Extraction of Membrane Proteins
SMPL formation begins with isolated membranes. As a prototype system, we used inner and outer membranes from E. coli, which provide a well-characterized assortment of membrane proteins for establishing benchmarks for SMPL formation. The membrane protein pool is solubilized in detergent to extract proteins. The type, concentration, and molar amount of detergent will all be potentially important parameters in solubilization because any unextracted proteins will not be incorporated into Nanodiscs. A detailed investigation of detergent extraction is outside the scope of this report but may be found in several reviews [17,21–24]. In general, the best detergent system will efficiently extract the membrane proteins without denaturing them.
Two detergent systems were tested for SMPL formation, 1% octyl-glucoside (OG) and 0.1% sodium dodecyl sulfate (SDS). Each was used at a ratio of around 1–2 mg of membrane protein per mL detergent solution. Both detergent systems efficiently extracted membrane proteins and were compatible with Nanodisc formation as seen by SEC and SDS-PAGE analysis (data shown for OG only). Because it is impossible to analyze whether every extracted protein retains its native fold and activity, it is advisable to screen several detergents to find the best system to maintain the specific biological interaction of interest. Due to its widespread use to solubilize functional membrane proteins [24,25] and its compatibility with SMPL formation, 1% OG was used for the remainder of the studies.
Nanodisc Formation
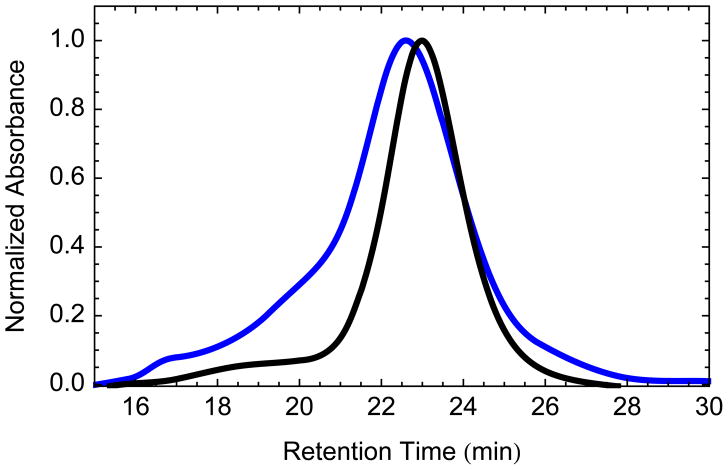
There are two key ratios to consider for SMPL formation. The first is the ratio of membrane protein to MSP. In general, an excess of MSP is used to drive the system towards a single protein or complex in each Nanodisc [26,12,27]. At higher loading ratios, the probability increases of multiple proteins randomly incorporating into a single Nanodisc. This is undesirable for most applications as it could lead to nonspecific interaction or co-localization of proteins that randomly become incorporated into the same Nanodisc complex. However, at lower ratios, the excess of empty Nanodiscs dilutes the membrane protein concentration leading to higher levels of background MSP and lipids. We evaluated a range of loadings from 2 to 20 μg membrane protein (MP) per nmol MSP. Increasing the membrane protein loading showed a shift in the SEC peak as the incorporation of membrane proteins into the Nanodiscs increased the average Stokes diameter of the particles. A ratio of 10 μg MP/nmol MSP was used for optimization of lipid:MSP ratios. At this ratio, the average Stokes diameter is 12.7 nm, 0.6 nm larger than empty Nanodiscs (Figure 2). Assuming an average membrane protein (MP) molecular weight of 50 kDa, the ratio, 10 μg MP/nmol MSP, will be 0.2 nmol MP/nmol MSP or 0.4 nmol MP/nmol Nanodiscs, since each Nanodisc contains two MSP molecules. Therefore, a maximum of 2 out of every 5 Nanodiscs will contain a membrane protein assuming a perfect efficiency in incorporation.
Fig. 2.
Size exclusion chromatogram for control Nanodiscs (black) and purified SMPL Nanodiscs (blue) made with a loading ratio of 10 μg MP/nmol MSP and a lipid ratio of 100 POPC:MSP
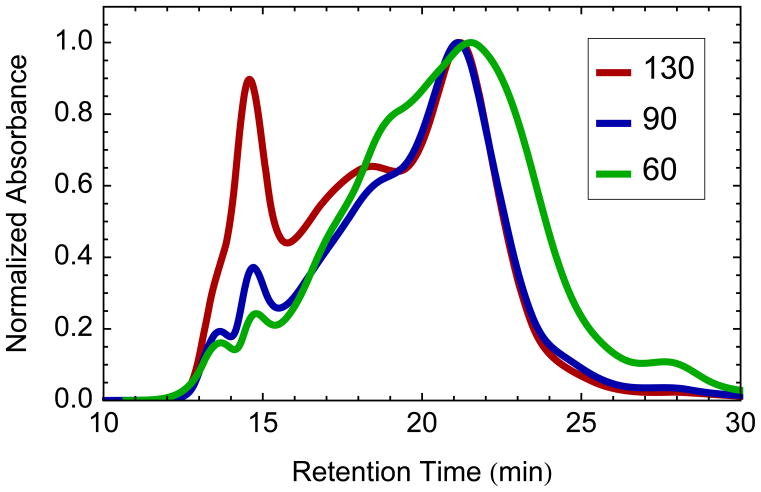
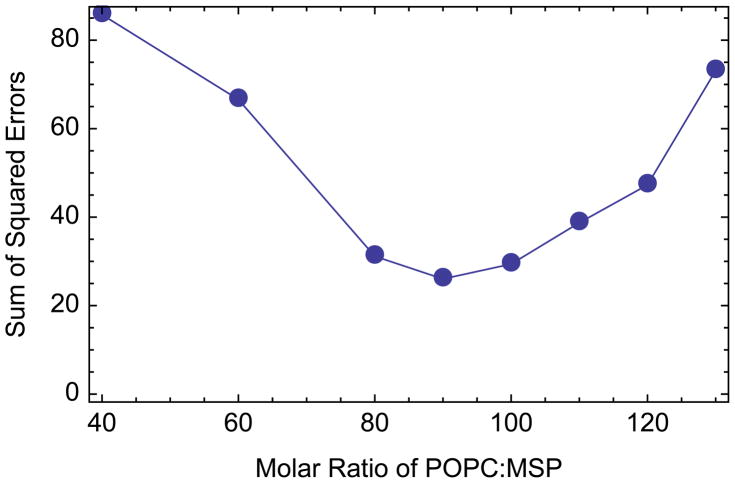
After determining the proper amount of MP and MSP, the second key ratio is the proportion of supplemental synthetic lipid to MSP. Too many lipids lead to the formation of large aggregate species. Too few lipids causes poor formation of Nanodiscs, which leads to a polydisperse size exclusion chromatogram (Figure 3). For POPC, the optimal molar ratio for MSP1E3D1 Nanodiscs has been established previously as 130 POPC:MSP for Nanodiscs without membrane proteins [18]. However, the addition of detergent-solubilized lipids from the starting membrane and the displacement of lipids by the incorporated proteins lower the ratio of lipids that must be added. Thus, the optimal ratio of POPC to MSP will depend on the protein to lipid ratio in the starting membrane, the detergent extraction [28], and the membrane protein to MSP loading ratio. Libraries were formed with a range of lipid ratios to determine the optimal value. Samples were prepared from the same detergent extract (1% OG) with a fixed amount of membrane proteins, 10 μg MP/nmol MSP. Each sample was analyzed by SEC and compared with a control sample of lipid-only POPC Nanodiscs. To account for the shift caused by the increasing Stokes diameter, the control Nanodisc chromatogram was shifted to align the maxima. The sum of squared errors between the aligned control sample and each chromatogram showed the minimum difference and hence optimal lipid ratio at 90 POPC:MSP, about 70% of what is required for POPC-only Nanodiscs (Figure 4). Even at optimal lipid loading, a small shoulder is observed on a shifted Nanodisc peak (see Figure 2). This may be attributed to incorporation of protein complexes with large extracellular or cytosolic domains, which significantly increase the Stokes diameter of the Nanodisc [29,30]. In general, performing a pilot study similar to this for each new system to find the optimal ratio of lipid to MSP is advisable.
Fig. 3.
Size exclusion chromatogram of unpurified Nanodiscs following SMPL formation at 130 (red), 90 (blue), and 60 (green) POPC:MSP molar ratio. Large aggregate species are observed for at 130 POPC:MSP while poorly-formed polydisperse Nanodiscs are formed at 60 POPC:MSP
Fig. 4.
Optimization of POPC:MSP ratio for SMPL Nanodiscs made from 1% OG-solubilized membrane proteins at a loading ratio of 10 μg MP/nmol MSP with different amounts of lipid added. Unpurified reconstitution mixtures were analyzed by SEC. Chromatograms were aligned with a chromatogram of control Nanodiscs, and the sum of squared error was calculated. The optimal ratio is the minimum difference, 90 POPC:MSP
Analysis of the SMPL Proteome
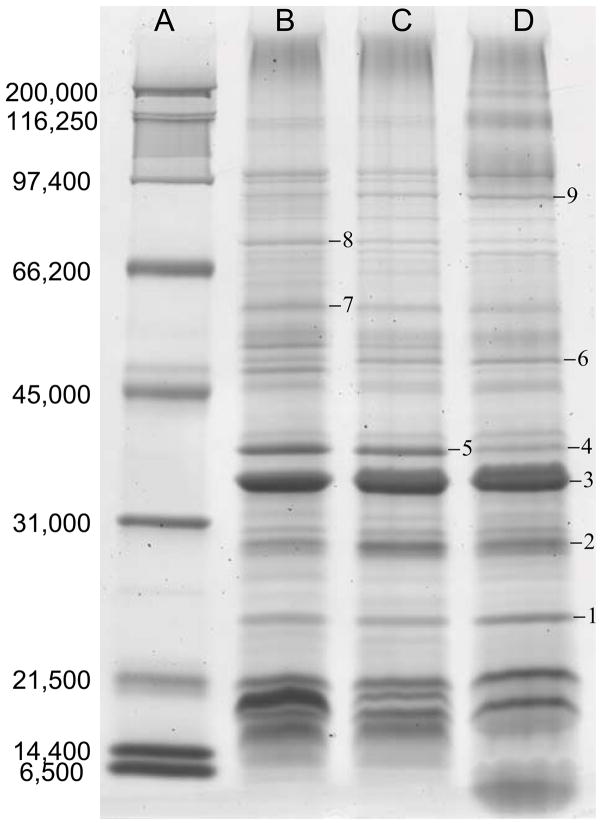
To evaluate the degree to which the Nanodisc library mirrored the protein content of the starting membrane proteome, delipidated protein extracts were analyzed with SDS-PAGE from the starting membranes (lane B), detergent extracts (lane C), and SMPL (lane D) as shown in Figure 5. To avoid interference from MSP in the SMPL sample, Nanodiscs were captured on a Ni-NTA column. Membrane proteins were eluted by disassembling the Nanodiscs with cholate, leaving MSP bound to the column. Image analysis was performed to count and compare bands between lanes. The SMPL bands overlapped with 89% of bands in the detergent extract (25 of 28 bands) and 86% of bands in the raw membranes (25 of 29 bands). These data demonstrate that the SMPL contains a large percentage of the proteins present in the membrane proteome. It was also observed that 93% of bands in the detergent extract overlapped with bands in raw membranes (27 of 29 bands). This suggests that some of the proteins absent in the SMPL are lost due to incomplete detergent extraction, while others are efficiently solubilized but not efficiently inserted into Nanodiscs.
Fig. 5.
SDS-PAGE gel of protein extracts of the Nanodisc library (lane D) compared to the raw membranes (lane B) and detergent extracted proteins (lane C). Standards are annotated (in Da) in lane A. The SMPL was formed with a loading ratio of 10 μg MP solubilized in 1% OG per nmol MSP and a lipid ratio of 100 POPC:MSP. Bands 1–9 were excised and identified with mass spectrometry as given in Table 1
Although it is difficult to ascertain precise quantitative data from Figure 5, the intensities for many of the bands are similar from lane to lane. Comparing the peak intensities of matching bands reveals that on average they differ by around 15% from lane to lane. This suggests that the incorporation efficiency is roughly similar for many of the proteins. However, there are several bands, most notably bands 4 and 5, where the incorporation efficiency is significantly lower. Future studies will seek to determine the mechanism behind the quantitative differences in incorporation efficiency.
Prominent bands in the gel were excised and digested for mass spectrometric identification. Several bands were selected from the detergent and membrane lanes due to slightly higher levels of protein in those bands, which improved confidence in identification as seen in bands 4 and 5. The bands are annotated in Figure 5 and proteomic results are given in Table 1. Several outer membrane proteins were observed, including OmpA, OmpW, OmpC, Maltoporin, and BamA. OmpA and OmpW are both monomeric proteins while OmpC and Maltoporin form trimers in native membranes [31]. BamA is a larger outer membrane assembly factor with a significant extra-membrane domain [32]. These proteins demonstrate the successful incorporation of integral outer membrane proteins of a range of sizes.
Table 1.
Proteomic identification of select bands from Figure 5 with proteomic statistics
| Band no.a | Protein Name | Accession no.c | Mascot Score | Mass (Da) | Sequence Coverage |
|---|---|---|---|---|---|
| 1 | Outer Membrane Protein W | OMPW_ECOLI | 1958 | 22913 | 48% |
| 2b | Outer Membrane Protein A (fragment) | gi|195940407 | 1578 | 28716 | 80% |
| 3 | Outer Membrane Protein A | OMPA_ECOLI | 2901 | 37178 | 61% |
| 4 | Outer Membrane Protein C | OMPC_ECOLI | 554 | 40343 | 53% |
| 5 | Outer Membrane Protein C | OMPC_ECOLI | 3471 | 40343 | 91% |
| 6 | Maltoporin | LAMB_ECOBW | 1402 | 49881 | 37% |
| 7 | ATP synthase subunit alpha | ATPA_ECOLI | 1014 | 55188 | 40% |
| 8 | Chaperone protein DnaK | DNAK_ECOLI | 975 | 69072 | 24% |
| 9 | Outer membrane protein assembly factor BamA | BAMA_ECOLI | 970 | 90496 | 26% |
According to Figure 5.
This OmpA fragment was found in the NCBI database but was not in the SwissProt database. Results are given for the NCBI database search.
Accession numbers are given for SwissProt database except for band 2 which was determined using the NCBI NR database.
In addition to outer membrane proteins, two inner membrane proteins were identified, the alpha subunit of ATP synthase and molecular chaperone DnaK [33]. The detection of ATP synthase is to be expected as purified ATP synthase was previously studied in Nanodiscs under similar sample preparation conditions [34]. Detection of these proteins demonstrates both the incorporation of inner membrane proteins and the incorporation of peripheral membrane proteins.
These proteomic data suggest that SMPL Nanodiscs provide a faithful model of the membrane proteome in a soluble form useful for a range of novel biochemical analyses. In addition, the utility of SMPL Nanodiscs for probing the functional characteristics of membrane proteins in heterogeneous populations is demonstrated by their recent use in incorporating membrane proteins from synapses, used for mechanistic studies of Aβ oligomer receptors and drug discovery ([35], manuscript in preparation).
It is important to note that the SMPL is best considered a model of the starting membrane proteome rather than an exact duplicate. We do not expect that 100% of the proteins in the starting membrane will be functionally incorporated in the SMPL. As such, this approach could lead to false negatives and should not be used to exclude a particular protein. Other approaches to membrane protein solubilization, including detergent-based techniques, face the same challenge. However, the above results show that the SMPL contains a large percentage of the protein pool and serves as a useful in vitro model of the membrane proteome.
Conclusion
Nanodisc libraries offer a novel technique for solubilizing membrane proteomes. Although the specifics for SMPL formation will depend on the system of interest, the general protocol, optimization, and analysis techniques described in this report will guide future studies. We have explored the formation of SMPL Nanodiscs from E. coli inner and outer membranes. Membrane proteins were extracted and incorporated into Nanodiscs with different loading ratios. At a relatively high loading of membrane proteins, displacement of lipids by the incorporated membrane proteins and the presence of natural lipids from the membrane in the detergent extract caused the optimal lipid to MSP molar ratio to be significantly less than is required for empty Nanodiscs. SDS-PAGE and proteomic analysis of the library suggest that the Nanodisc library faithfully reflects the starting membrane proteome.
Acknowledgments
We thank Dr. Peter Yau and the University of Illinois Urbana-Champaign Protein Science Facility for assistance in proteomic identification of bands. We thank Michelle Yoo for her assistance in preparing samples. This work was funded by the National Institutes of Health (R01-GM31756 and R01-GM33775 to S.G.S). M.T.M. was supported by the Robert C. and Carolyn J. Springborn Endowment.
Abbreviations
- BCA
bicinchoninic acid
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MP
membrane protein
- MS
mass spectrometry
- MSP
membrane scaffold protein
- Ni-NTA
nickel-nitrilotriacetic acid
- OG
octyl-glucoside
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SEC
size exclusion chromatography
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SMPL
soluble membrane protein library
References
- 1.Hooker BS, Bigelow DJ, Lin C-T. Methods for mapping of interaction networks involving membrane proteins. Biochem Biophys Res Commun. 2007;363 (3):457–461. doi: 10.1016/j.bbrc.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 2.Piehler J. New methodologies for measuring protein interactions in vivo and in vitro. Curr Opin Struct Biol. 2005;15 (1):4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BDM, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, Fong V, Tam YYC, Davey M, Hnatshak O, Bajaj N, Chandran S, Punna T, Christopolous C, Wong V, Yu A, Zhong G, Li J, Stagljar I, Conibear E, Wodak SJ, Emili A, Greenblatt JF. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012 doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 4.Stagljar I, Fields S. Analysis of membrane protein interactions using yeast-based technologies. Trends Biochem Sci. 2002;27 (11):559–563. doi: 10.1016/s0968-0004(02)02197-7. [DOI] [PubMed] [Google Scholar]
- 5.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584 (9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayburt TH, Grinkova YV, Sligar SG. Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2002;2 (8):853–856. [Google Scholar]
- 7.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. Functional reconstitution of β2-adrenergic receptors utilizing self-assembling nanodisc technology. BioTechniques. 2006;40(5):601–602. 604, 606, 608, 610, 612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 8.Pandit A, Shirzad-Wasei N, Wlodarczyk LM, van Roon H, Boekema EJ, Dekker JP, de Grip WJ. Assembly of the Major Light-Harvesting Complex II in Lipid Nanodiscs. Biophys J. 2011;101 (10):2507–2515. doi: 10.1016/j.bpj.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ham M-H, Choi JH, Boghossian AA, Jeng ES, Graff RA, Heller DA, Chang AC, Mattis A, Bayburt TH, Grinkova YV, Zeiger AS, Van Vliet KJ, Hobbie EK, Sligar SG, Wraight CA, Strano MS. Photoelectrochemical complexes for solar energy conversion that chemically and autonomously regenerate. Nat Chem. 2010;2 (11):929–936. doi: 10.1038/nchem.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12 (11):2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisov IG, Sligar SG. Cytochromes P 450 in nanodiscs. Biochim Biophys Acta, Proteins Proteomics. 2011;1814 (1):223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayburt TH, Leitz AJ, Xie GF, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282 (20):14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 13.Grinkova YV, Denisov IG, Sligar SG. Functional reconstitution of monomeric CYP3A4 with multiple cytochrome P450 reductase molecules in Nanodiscs. Biochem Biophys Res Commun. 2010;398 (2):194–198. doi: 10.1016/j.bbrc.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. BioTechniques. 2003;35 (3):556–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 15.Borch J, Roepstorff P, Moeller-Jensen J. Nanodisc-based co-immunoprecipitation for mass spectrometric identification of membrane-interacting proteins. Mol Cell Proteomics. 2011;10(7):O110 006775, 006779. doi: 10.1074/mcp.O110.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XX, Chan CS, Bao H, Fang Y, Foster LJ, Duong F. Nanodiscs and SILAC-Based Mass Spectrometry to Identify a Membrane Protein Interactome. J Proteome Res. 2012;11 (2):1454–1459. doi: 10.1021/pr200846y. [DOI] [PubMed] [Google Scholar]
- 17.Arnold T, Linke D. Current Protocols in Protein Science. John Wiley & Sons, Inc; 2001. The Use of Detergents to Purify Membrane Proteins. [DOI] [PubMed] [Google Scholar]
- 18.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126 (11):3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 19.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138 (1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 20.Du P, Kibbe WA, Lin SM. Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics. 2006;22 (17):2059–2065. doi: 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
- 21.Duquesne K, Sturgis JN. Membrane Protein Solubilization. In: Mus-Veteau I, editor. Heterologous Expression of Membrane Proteins, vol 601. Methods in Molecular Biology. Humana Press; 2010. pp. 205–217. [DOI] [PubMed] [Google Scholar]
- 22.Garavito RM, Ferguson-Miller S. Detergents as Tools in Membrane Biochemistry. J Biol Chem. 2001;276 (35):32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 23.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta, Biomembr. 2004;1666 (1–2):105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Jones MN. Surfactants in membrane solubilisation. Int J Pharm. 1999;177 (2):137–159. doi: 10.1016/s0378-5173(98)00345-7. [DOI] [PubMed] [Google Scholar]
- 25.Speers AE, Wu CC. Proteomics of Integral Membrane Proteins: Theory and Application. Chem Rev. 2007;107 (8):3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- 26.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450 (2):215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464 (Part F):211–231. doi: 10.1016/S0076-6879(09)64011-8. Liposomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee P, Joo JB, Buse JT, Dawson G. Differential solubilization of lipids along with membrane proteins by different classes of detergents. Chem Phys Lipids. 1995;77 (1):65–78. doi: 10.1016/0009-3084(95)02455-r. [DOI] [PubMed] [Google Scholar]
- 29.Boldog T, Grimme S, Li MS, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103 (31):11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in Cytochrome P450 3A4: Linkages in Substrate Binding, Spin State, Uncoupling, and Product Formation. J Biol Chem. 2007;282 (10):7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 31.Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta, Biomembr. 2002;1565 (2):308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- 32.Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156 (9):2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- 33.Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, Daley DO. Protein Complexes of the Escherichia coli Cell Envelope. J Biol Chem. 2005;280 (41):34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 34.Ishmukhametov R, Hornung T, Spetzler D, Frasch WD. Direct observation of stepped proteolipid ring rotation in E. coli FoF1-ATP synthase. EMBO J. 2010;29 (23):3911–3923. doi: 10.1038/emboj.2010.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilcox KC, Das A, Velasco P, Marty MT, Kuhns B, Luan CH, Sligar SG, Klein WL. Alzheimer’s drug discovery using Nanodisc-stabilized receptors for oligomeric Aβ. New Orleans, LA: Society for Neuroscience; 2012. Program No. 748.014. [Google Scholar]