Abstract
There are now 7 nucleoside/tide analogues, along with interferon-α, that are approved by the FDA for the management of chronic hepatitis B virus (HBV) infection, a disease affecting hundreds of millions of people worldwide. These medications, however, are limited in usefulness, and significant side effects and the emergence of viral escape mutants make the development of novel and updated therapeutics a pressing need in the treatment of HBV. With this in mind, a library containing 2,000 compounds already known to be safe in both humans and mice with known mechanisms of action in mammalian cells were tested for the possibility of either antiviral activity against HBV or selective toxicity in HBV producing cell lines. A modified real-time immune-absorbance-polymerase chain reaction (IA-PCR) assay was developed for this screen, utilizing cells that produce and secrete intact HBV virions. In this procedure, viral particles are first captured by an anti-HBs antibody immobilized on a plate. The viral load is subsequently assessed by real-time PCR directly on captured particles. Using this assay, eight compounds were shown to consistently reduce the amount of secreted HBV viral particles in the culture medium under conditions that had no detectable impact on cell viability. Two compounds, proparacaine and chlorophyllide, were shown to reduce HBV levels 4- to 6-fold with an IC50 of 1 and 1.5μM respectively, and were selected for further study. The identification of these compounds as promising antiviral drug candidates against HBV, despite a lack of previous recognition of HBV antiviral activity, supports the validity and utility of testing known compounds for “off- pathogen target” activity against HBV, and also validates this IA-PCR assay as an important tool for the detection of anti-viral activity against enveloped viruses.
Keywords: Hepatitis B virus, Polymerase Chain Reaction, Immune-absorbance Polymerase Chain Reaction, Proparacaine, Chlorophyllide, anti-viral drug
1. Introduction
Although interferon-α and seven different nucleoside/tide analogues (NAs) have been approved for use in the management of chronic hepatitis B, these treatments are not “curative” and remain unsatisfactory (Yuen and Lai, 2011). The interferons are effective in reaching clinical endpoints in fewer than 50% (possibly even less than 20%) of those in whom they are used, and are associated with a number of undesirable side effects (Jafri and Lok, 2010; Janssen et al., 1992; Kartal et al., 2007; Lok, 1994). The NAs are HBV DNA polymerase inhibitors and are recommended for use in less than 50% of the infected population. Although, they are highly effective at reducing viremia, the development of escape mutants and the need for long-term, possibly life long, use limits enthusiasm. Clearly, additional anti-HBV therapeutics are needed (Stein and Loomba, 2009; Wu et al., 2012; Zoulim, 2004).
Despite the need, the cost of bringing a new drug to the market can be staggering. It has been estimated that the cost of bringing a new molecular entity from discovery to approval is on average more than $750 million and takes at least 8 years (Weisman et al., 2006). Strategies to reduce the time between discovery of a compound’s antiviral activity and its introduction to the market as a drug would be appealing, especially where commercial interest is lacking. Although there are more than 15,000 compounds that have been tested for therapeutic use in humans, many have not reached the market place (Weisman et al., 2006). There has been, however, a growing interest in “repurposing” compounds with known activity and safety profiles for new indications (Weisman et al., 2006). This strategy follows the logic that a molecule that has already been tested and shown to be safe in humans will require less study, time, and money to reach the market. Therefore, screening compounds with known safety profiles for uses other than their original indication could uncover candidate molecules that can more easily be developed as a therapeutic than developing a completely new drug. This strategy has already successfully been used to discover some potential therapeutics for hepatitis C virus (HCV) (Gastaminza et al., 2010). The effectiveness of this approach, of course, depends on the potency and on the illness being treated. The other advantage of this approach is that even if candidate molecules cannot be optimized and developed into therapeutics, new strategies for treating the off-target disease could be developed based on the known activity of candidate compounds.
We have developed a program to screen libraries of compounds known to be safe in people. The intent of this screening program is to identify candidates that can be used, alone or in combination, for activity against various steps in the HBV life cycle. Our initial screen, described here, measures the ability of a particular compound to reduce the amount of secreted intact viral particles in tissue culture systems known to produce and secrete infectious virus (Guo et al., 2007a; Ladner et al., 1997; Sells et al., 1988). To do this, a real-time “immune-absorbance-polymerase chain reaction” (real-time IA-PCR) assay was developed. Because this technology is specifically designed to analyze the end-point of the viral life cycle, it can identify compounds that inhibit HBV replication, nucleocapsid formation, virus assembly, and/or cellular virus transportation and secretion. This approach utilizes robotic high throughput screening in a 96-well format coupled with antibody-based capture of HBV envelope proteins and quantitative PCR to measure minimal amounts of viral DNA. By combining specific antibody-capture and qPCR we are able to confirm that we are specifically analyzing intact viral particles secreted into the culture medium of HBV producing cells and exclude subviral particles and viral intermediates.
Using this technology, a MicroSource (Gaylordsville, CT) compound library containing 2000 compounds with known safety profiles was screened. All compounds in this library are approved for animal or human use and their pharmacological and toxicological profiles have been defined and published. Furthermore, most of the compounds in this library are off-patent, so patent rights have been extinguished and the compounds can be sold without restriction. This creates the optimal situation for quickly developing an anti-HBV drug for clinical use (Kocisko et al., 2003; Weisman et al., 2006; Weissmann and Aguzzi, 2005). The strength and validity of this assay as well as the potential of the compounds identified are discussed.
2. Materials and Methods
2.1. Chemicals and library screened
The chemical library Spectrum from MicroSource (Gaylordsville, CT) contains 2000 compounds and is a collection intended to survey a wide range of biological activities and structural diversity for screening programs. It is further described elsewhere (Kocisko et al., 2003), and published online at http://www.msdiscovery.com. Each compound has a minimum of 95% purity. All compounds are dissolved in DMSO and used at a final concentration of 0.5% DMSO.
2.2. Cell culture and drug treatment
The HepG2.2.15 (G2.215) cell line, which expresses HBV from two integrated head to tail dimers of the HBV genome, was originally kindly provided by George Acs (Mt. Sinai Medical College, New York, N.Y.) (Sells et al., 1988). HepG2 cells were purchased from the American Type Culture Collection (Rockville, MD). The generation of the HepAD38 and HepDE19 cell line, in which HBV expression is tetracycline regulated, has been described previously (Guo et al., 2007b). HepDE19 and HepAD38 cells were cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS), 1mg/ml Penicillin, 100untis/ml Streptomycin (PS) and 50ug/mL tetracycline (tet) to suppress HBV expression. HepG2 and G2.215 cells were maintained in RPMI 1640 medium (GIBCO-BRL, Gaithersburg, Md.) supplemented with 10% FBS, with 200μg/ml of G418 included in the medium for G2.215 cells. For drug screening, G2.215 cells were seeded in 96-well plates and grown until 60% confluent. Cultured cells were then incubated with medium containing 10μM compounds or control medium (no drug) containing 0.5% Dimethyl sulfoxide (DMSO) the same concentration as in the medium containing drugs. After 6 days of treatment, the medium was collected for measurement of secreted enveloped HBV DNA (virion), and the cells were tested for viability as described.
2.3. Real-time Immune-absorbance-PCR to detect enveloped HBV DNA
To quantifiably detect enveloped HBV DNA (virion) in the culture medium, a method called real-time immune-absorbance-PCR (IA-PCR) was developed. Briefly, 96-well plates (Applied Biosystems, Foster City, CA) were first coated at 4°C overnight with 250ng per well monoclonal anti-HBs antibody (Meridian Life Science, Saco ME) diluted in 50μl 0.01M sodium bicarbonate coating buffer (pH 9.6). After washing with phosphate buffered saline containing 0.5% tween 20 (PBS-T), the plates were blocked with 75μl 2% bovine serum albumin (BSA) in PBS at 37°C for 1.5 hours. Wells were again washed with PBS-T after blocking. The samples were diluted 4 fold in normal RPMI medium and 50μl of this dilution was added to the wells and incubated overnight at 4°C with shaking. The wells were again washed with PBS-T and all liquid was removed from the wells. PCR mastermix containing forward primer, reverse primer, FAM/TAMRA probe and 2X Universal Fast Real-Time PCR Mastermix (as per manufacturer’s instructions, Applied Biosystems) was added to each well and real time PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Two standard curves were included on each plate to assure consistency between assays.
The forward and reverse primers, with sequences 5′-ACTCGTGGTGGACTTCTCTCA-3′ and 5′ –AAGATGAGGCATAGCAGCAGG 3′ respectively, amplify a fragment encoding a portion of the HBV small surface protein from genomic position 252nt to 432nt. The probe, located between the two primers, was labeled with the reporter dye FAM at the 5′ terminus and the quencher dye TAMRA at the 3′ terminus (5′ FAM-TGGATGTGTCTGCGGCGTTTTATC–TAMRA3′). The reaction was performed with a 10 minute incubation at 95° followed by 40 cycles of 95° for 15 seconds and 52° for 30 seconds. The CT (Threshold cycle) value for each sample was calculated and compared with controls and standards. The reported CT value of each compound represents the average of duplicate wells, and in some instances the CT value is presented as HBV copy number using the standard curve of HBV DNA. Compounds causing >4 fold reduction in HBV DNA signal compared to untreated samples were considered positive anti-HBV candidates and selected for further testing.
2.4. Particle gel examination of enveloped HBV DNA
The secreted HBV virions in the culture medium were assayed as previously described (Guo et al., 2007a). Briefly, virions were precipitated from 1ml of clarified culture medium by adding PEG8000 to a final concentration of 10% followed by incubation at 4°C for 1 h. The precipitates were collected by centrifugation at 10,000 rpm for 10 min and dissolved in 40μl TNE buffer (10mM Tris-HCl, pH7.4; 150mM NaCl and 1mM EDTA). The virions were directly fractionated by electrophoresis through non- denaturing 1% agarose gels and transferred to a nitrocellulose filter by blotting in TNE buffer. The DNA-containing particles on the filter were then denatured, neutralized, and the HBV DNA was detected by hybridization with a α-32P-UTP (800Ci/mmol, Perkin Elmer) labeled minus strand specific full-length HBV riboprobe.
2.5. Southern blot analysis of HBV replication
Intracellular HBV core DNA was extracted as described previously (Guo et al., 2007a). Briefly, cells from one well of a 6-well plate were lysed with 0.5ml of lysis buffer containing 10mM Tris-HCl (pH 8.0), 1mM EDTA, 1% NP40 and 2% sucrose at 37°C for 10 minutes. Cell debris and nuclei were removed by centrifugation and the supernatant was mixed with 130μl of 35% polyethylene glycol (PEG) 8000 containing 1.5M NaCl. After 2 hour incubation at 4°C, viral nucleocapsids were pelleted by centrifugation at 10,000 rpm for 5 min at 4°C, followed by 1 hour digestion at 37°C in 400μl of digestion buffer containing 0.5mg/ml pronase (Calbiochem), 0.5% SDS, 150mM NaCl, 25mM Tris- HCl (pH 8.0) and 10mM EDTA. The digestion mixture was extracted with phenol and DNA was precipitated with ethanol and dissolved in TE (10mM Tris, pH8.0, 1mM EDTA) buffer. Half of the DNA sample was resolved by electrophoresis into a 1.5% agarose gel. The gel was then soaked in 0.2N HCl for 10 min and then subjected to denaturation in a solution containing 0.5M NaOH and 1.5M NaCl for 1 h, followed by 1 h neutralization in a buffer containing 1M Tris-HCl (pH 7.4) and 1.5M NaCl. DNA was then blotted onto Hybond-XL membrane (GE Healthcare) in 20X SSC buffer. HBV DNA was examined by hybridization with α-32P-UTP labeled probe specific for HBV.
3. Results
3.1. Specificity and sensitivity of real time immune-absorbance-PCR
The goal of this work was to screen compound libraries for activity against hepatitis B virus (HBV), however, infectivity assays for HBV in tissue culture are technically difficult and not very robust. Because of this an efficient method to measure decreases in the amount of enveloped particles containing viral DNA secreted from HBV producing cells, such as G2.215 cells, was developed. It is reasoned that since production and release of enveloped viral DNA, or virions, by G2.215 cells requires that all synthetic steps in the viral life cycle be functional except entry, drugs suppressing any post-entry stage of the HBV life cycle can be identified by measuring levels of secreted virion in culture medium. By utilizing an antibody to first capture virions based on their envelope protein and then specifically amplifying HBV DNA, we are excluding analysis of subviral particles and viral intermediates and ensuring specific analysis of infectious virions. To take advantage of this, a high-throughput screening system was developed, allowing us to robotically administer our compound library in 96-well microtiter plates. The sensitivity of real time PCR combined with the specificity of immune-absorbance allows for detecting the small amounts of secreted virion and differences in virion levels that would be expected from the low number of cells within each well of a 96-well plate.
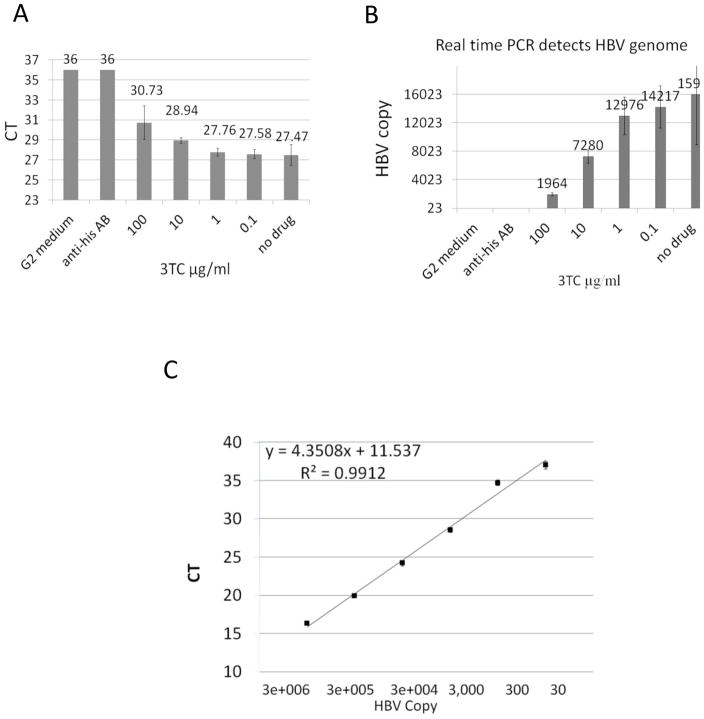
The linearity, reproducibility and dynamic range of this assay were first evaluated to determine the strength and validity of the assay (Fig. 1). Successful PCR, resulting from the amplification of DNA associated with HBs captured by the immune-absorbance step, was shown to be specific for HBV virions and did not result in non-specific amplification (Fig. 1A). Because a lower CT value in a real time PCR assay indicates that the level of DNA amplification reached the set threshold value earlier in the reaction, a lower CT value can be interpreted as a greater amount of starting HBV DNA present in the sample (Materials and Methods). With this in mind, the results show the lowest value, 27.47, CT was amplified in medium from untreated G2.215 cells (Fig. 1A, no drug). Replacing the anti-HBs antibody in the immune-absorbance portion of the assay with an irrelevant anti-His tag antibody resulted in a loss of PCR signal, with a CT value of 36 meaning it was below the threshold of detection as determined by the standard curve (Fig. 1A, anti-his AB). The specificity of detection was further supported by the fact that no HBV DNA was amplified from medium for HepG2 cells (CT of 36), which are the parental cells of the G2.215 cells and do not produce HBV (Fig. 1A HepG2). Suppression of HBV replication by treatment of cells with the known HBV inhibitor lamivudine (3TC) resulted in a dose-dependent decrease of HBV DNA signal in the assay (Fig. 1A, 3TC treated). Incubation of G2.215 cells with varying concentrations of 3TC for 6 days, the time point optimized for drug incubation in this assay, resulted in a dose dependent suppression of the captured HBV DNA with a maximum reduction in CT value of ~3.3 (Fig. 1A), or ~8 fold when converted to viral DNA copies (Fig. 1B). Together these results demonstrate the specificity of the assay to successfully pull down and amplify HBV DNA without isolating or amplifying nonspecific DNA products.
Figure 1. Real-time IA-PCR specifically detects enveloped HBV DNA in G2.215 culture medium.
G2.215 cells were seeded in 96-well plates and incubated with lamivudine at different doses for 6 days. Cell culture medium was incubated with anti-HBs pre-immobilized on 96-well PCR plates at 4°C overnight. HBV DNA in captured virions was then amplified by real-time PCR. Medium from HepG2 cells and G2.215 cell medium incubated in wells coated with an anti-His antibody instead of anti-HBs were used as negative controls. A.) A dose-dependent increase in CT value was seen in medium from cells incubated with lamivudine. B.) The effect on CT levels was converted to reduction of HBV DNA by correlating to virus copy number using a standard curve of HBV genomic DNA. C). Serial CT dilution of HBV genomic DNA served as the standard curve for the real-time PCR assay.
In addition to the specificity, this assay is also sensitive enough to detect as few as 30 copies of HBV genome (Fig. 1C). Using a standard curve of HBV DNA, the linear range of the assay was determined to be from 30 copies to 3×106 copies of HBV genome (Fig. 1C). Thus, at the standard conditions used, the CT differences are large enough to distinguish between viral levels in the medium of the cells producing HBV from that in the “negative” non-producing control (G2.215 versus HepG2 cells). Similar results were obtained using the HepAD38 cell line, which produces intact HBV virions under the control of a tetracycline-responsive promoter (Ladner et al., 1997; Zhu et al., 2009). HepAD38 cells produce HBV virus derived from transcripts from an integrated transgene after removal of tetracycline. HBV DNA is amplified with a CT of 35.1, which is near the lower limit of detection, when the HBV transgene is repressed, while transgene activated cells have a CT of 26.7 (Suppl 1). For ease of use, and because consistent results were seen in both systems, G2.215 cells were used as the positive control in these studies.
3.2. The Z′ value for robotic high-throughput screening
To determine the extent to which the assay lends itself to the high-throughput screening platform, the Z′ value of the assay was determined, as defined previously (Muller et al., 2008; Zhang et al., 1999). In this assay, the Z′ value reflects the well-to-well variations in assay results with regard to the presence or absence of HBV DNA. To determine the Z′ value of our assay, 48 wells of a 96 well plate were utilized to measure HBV DNA levels from G2.215 cell culture medium and the other half of the plate (48 wells) was used to measure HBV DNA from HepG2 cell culture medium (Fig. 2). The average CT values and standard deviation (SD) of positive wells (with HBV) and negative wells (without HBV) were determined and the Z′ value was calculated. The Z′ value of this assay is calculated as 0.801 (Fig. 2), and since Z′ values of greater than 0.7 are generally considered adequate for robotic high-throughput screens, the real-time IA-PCR assay described here meets this threshold and was advanced for further use (Muller et al., 2008; Zhang et al., 1999).
Figure 2. The Z′ value of the real-time IA-PCR as a high-throughput screening platform was determined.
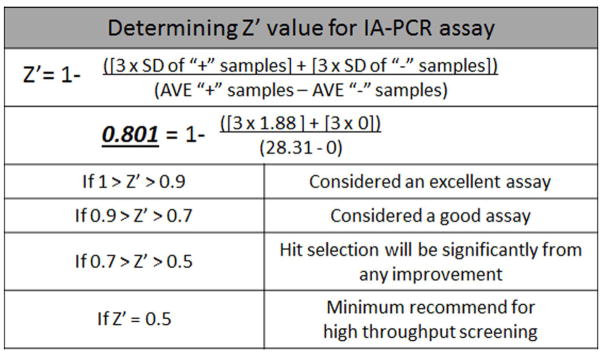
The equation for calculation of Z′ value is shown. The Z′ value of this assay was determined by the average (AVE) and standard deviation (SD) of positive controls (+) and negative controls (−). Averages represent data from half of the wells (48) of a 96 well plate for both positive and negative samples. Medium from G2.215 cells was used as the positive control, and medium from HepG2 cells, which do not produce HBV, was used as the negative control.
3.3. Screening results overview
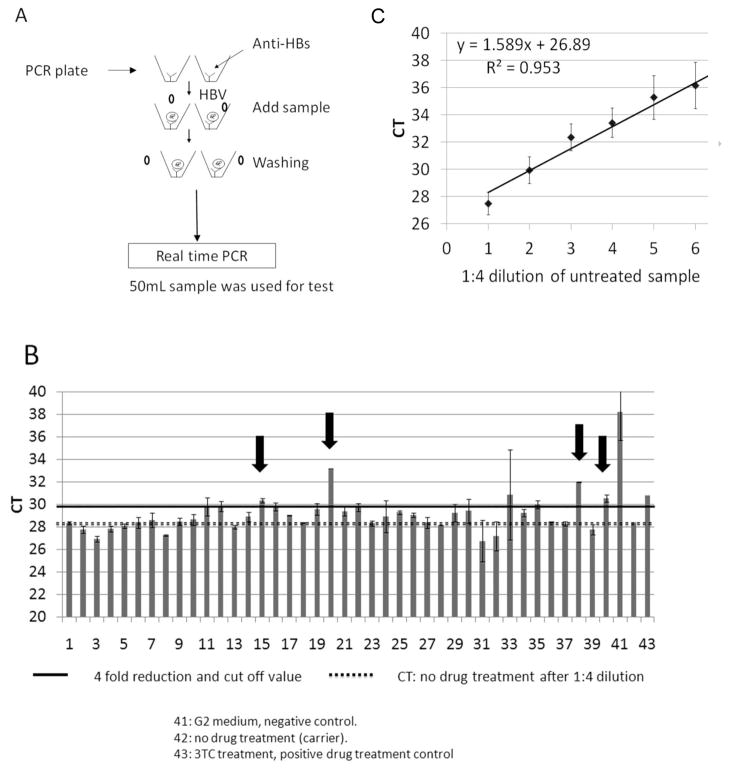
After verification, the assay was used to screen a commercial compound library with ~2,000 compounds with known safety profiles and mechanisms of action (Microsource Inc. described in Materials and Methods) for drugs that lowered the amount of HBV DNA detected in the culture medium. Briefly, G2.215 cells were seeded in 96-well plates and incubated with either “carrier” (the 0.5% DMSO medium in which the compounds were dissolved), 30μM lamivudine (as a positive control for inhibition of HBV) or 10μM of each of the test compounds from the library. After 6 days of treatment, cell culture medium was collected for use in the real-time PCR assay as described in the Materials and Methods, and illustrated in Fig. 3A. Duplicate wells were run for each sample and the average CT value corresponding to each compound was calculated. Two standard curves, one with a 1:4 serial dilution of medium from G2.215 cells treated with carrier alone (Fig. 3C) and another with a 1:10 serial dilution of HBV genome copies from plasmid P13 (Lamontagne et al., 2010), were included on each IA-PCR plate for calculation of level of reduction of HBV signal and copy number of HBV DNA respectively. While it would be ideal to run each sample in triplicate, the cost of supplies and reagents made this prohibitive.
Figure 3. IA-PCR assay setup and typical results.
A.) Work-flow describing the procedure for the real-time IA-PCR assay. B.) Representative data shows the typical results from an assay plate. Samples were diluted 1:4 from culture medium before being added to the plate in duplicate. After immune- absorbance, enveloped HBV DNA was amplified by real-time PCR. The solid black line across the data represents a 4-fold reduction compared to the untreated control. The dotted line represents the CT value of the untreated control, diluted 1:4 along with the treated samples. Arrows indicate compounds that reduced HBV DNA >4 fold, and are examples of samples that were selected for retesting. C.) The standard curve for the complete drug screening assay, derived cumulatively from the 1:4 serial dilutions of untreated, positive control medium run on the 24 independent assays. This was used as a reference for reduction of secreted HBV DNA in each assay.
As expected from the assay development stages (Fig. 1), undiluted medium from cells treated with carrier alone amplified to a CT of ~26, and this signal was reduced, proportionally, with 1:4 serial dilutions (Fig. 3C), indicating the linearity and dynamic range of the assay. Also as expected, levels of HBV DNA in medium from HepG2 cells were below the level of detection, confirming the specificity seen during assay design and setup (Fig. 3C and Fig. 1). In addition, medium from G2.215 cells incubated with lamivudine had a CT of ~30–31 (Fig. 3B, Sample 43), indicating a significant reduction in HBV DNA compared to cells treated with carrier alone. This is important because this not only confirms the ability of the assay to detect compounds which successfully reduce HBV DNA levels, but also defines the level of reduction that could be expected from candidate drugs during the screen of this compound library. A CT of 30, representing a 4-fold reduction in HBV DNA, was therefore used as a cut-off to define primary hits in the assay. The viability of cells was determined by MTT assay of cell culture plates used for drug treatment after media collection (data not shown).
Figure 3B shows the results of a typical IA-PCR plate, where 3–4 of the 40 compounds run per plate usually met our criteria of a “hit”. Compounds with high variation (SD) were not counted as hits regardless of their average CT value. An example of such a sample would be Sample 36 in Fig. 3B. The names of the screened compounds are listed in Supp.2.
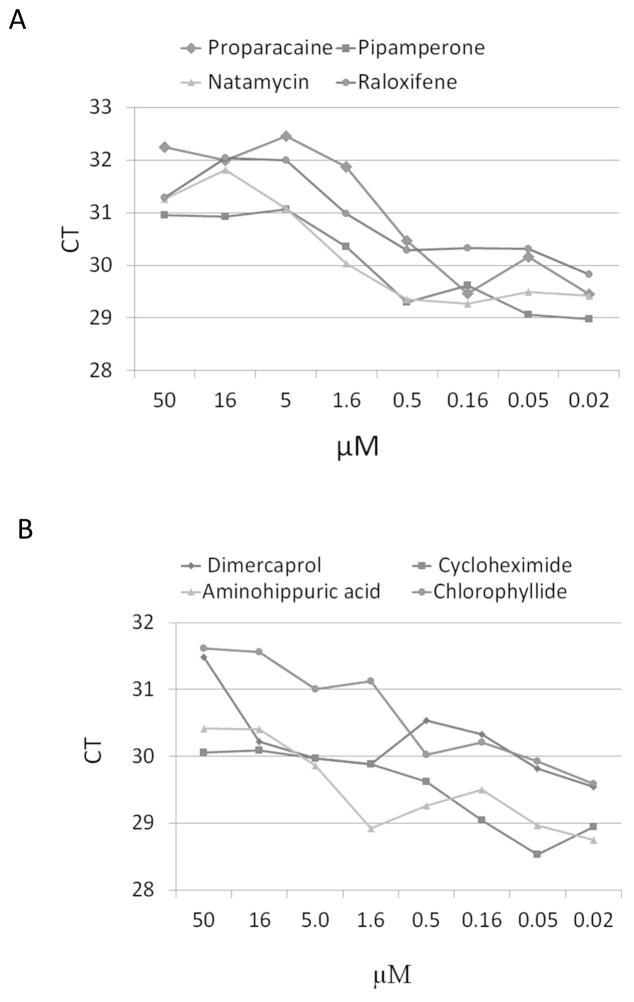
From the screen of 2,000 compounds, 153 were scored as hits in the primary screen. Each primary hit that was found by MTT test to be non-cytotoxic was retested in a second run. During retesting eight compounds were confirmed as effective in reducing HBV DNA and their names as well as reported functions and mechanisms are shown in the Table 1. One of these identified hits, cycloheximide, is a known inhibitor of protein synthesis and inducer of cell stress (Paoletti et al., 2010). Because of its known mechanism, the reduction of HBV DNA in cycloheximide treated cells most likely was caused by non-selective cellular toxicity and thus was not pursued further. Therefore, seven compounds were ordered for re-synthesis and their activity was confirmed in dose response assays using the same real-time IA-PCR assay (Fig. 4A, B). Proparacaine and chlorophyllide showed the most significant and consistent activity in this system. At a concentration of 5μM, proparacaine reduced HBV virion levels in culture medium by ~3 CT (P<0.01), whereas chlorophyllide reduced the amount of HBV enveloped virus by ~2 CT at the same concentration (P<0.05) (Fig. 4A, B). The IC50 for proparacaine against HBV was 1.0μM, and was 1.5μM for chlorophyllide (Fig. 4A, B). Therefore, these two candidates were evaluated further.
Table 1.
Candidate compounds that showed a dose-dependent effect on HBV DNA in treated G2.215 cells upon retesting.
| Compound Name1 | Fold HBV reduction2 | Approved or intended use3 | Mechanism4 | Reference |
|---|---|---|---|---|
| Proparacaine | 6 | Eye drop, Anti-HSV; | Prevent viral envelopment; Reduce pain and discomfort | Romanowski et. al Storey et. al |
| Chlorophyllide | 4 | Anti- cancer including liver cancer, anti-HSV | interaction with a carcinogen or removal of toxic aflatoxins | Breinholt et. Al; Egner et. al |
| Natamycin | 4 | Anti-mycotic agents | Suppress microorganism growth | Maher et. al |
| Raloxifene | 3.5 | Anticonvulsant | Estrogen receptor modulators | Rey et. al |
| Aminohippuric Acid | 3 | Renal function diagnosis | Organic ion transporter | Morisaki et. al |
| Pipamperone | 3.8 | Commercial name: Dipiperon. Antipsychotic | Inhibitor of G protein- receptor | Van Craenenbroeck et. al |
| Cycloheximide | 2 | Protein synthesis inhibitor | Suppression of protein synthesis, cell stress | Greenberger et. al |
| Dimercaprol | 3 | Chelating agent (As, Au, Hg antidote) | Interfere protein interaction | Morris et. al |
G2.215 cells were incubated with 10 μM of indicated compound for 6 days. No cytotoxicity was observed at this concentration.
The amount of HBV DNA in the culture medium was determined by IA-PCR, using HBV specific primers, immune-absorbance of an aliquot of medium with mAb to HBV envelope protein. Methods are described in text.
Approved or intended use is the use for which the compound has received US FDA approval (a) or for which it has been used in animal or human trials.
Mechanism is the action attributed to the compound as stated on either its “label” or literature.
Figure 4. Eight candidate compounds were confirmed by retesting using a dosage gradient.
G2.215 cells were incubated with a serial dilution of candidate compound and the CT value was determined using the IA-PCR assay. A.) IA-PCR results for the first sample set including the candidate drug, proparacaine, one of two compounds selected for further testing. B.) IA-PCR results for the second sample set, including chlorophyllide, the second compound selected for further testing.
3.4. Secondary assays to confirm antiviral activity of compounds identified for the ability to reduce enveloped HBV DNA in a cell culture system
Further study suggested that proparacaine reduced enveloped virus in culture medium after 6 days of treatment, but did not reduce intra-cellular HBV DNA levels. Unlike empty spherical particles, intact HBV virions contain the large surface protein (LHBs). Therefore, the presence and amount of LHBs was examined by western blot using the anti-LHBs monoclonal antibody MA18/7 (Lu et al., 1996). The reduction of LHBs usually suggests the reduction of viral particles, though filamentous particles also have LHBs. Figure 5A shows that 6 day treatment with 10 or 20μM proparacaine inhibited secretion of HBV particles containing LHBs in culture medium ~3- to 4 fold, which agrees with the results seen in the primary screens.
Figure 5. Western and Southern blot analysis of the role of proparacaine in reducing HBV DNA.
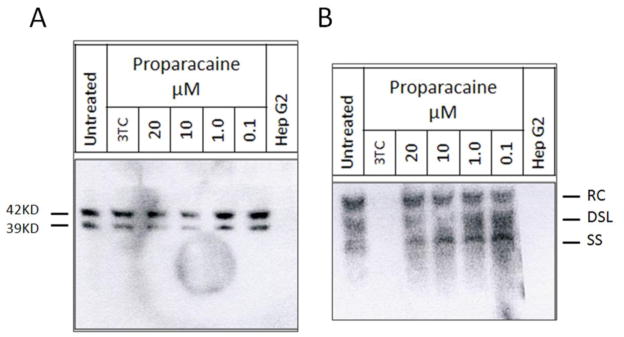
G2.215 cells were treated with different doses of proparacaine. A.) Western blot detecting LHBs. B.) Southern blot detecting cellular HBV replication intermediates. RC, relaxed circular; DSL, double- stranded linear; SS, single-stranded DNA.
To determine whether proparacaine inhibits HBV replication, cellular HBV DNA was isolated from G2.215 cells after treatment as described previously (Lu et al., 1996). HBV replication intermediates were analyzed by Southern blot with a labeled HBV probe. Compared to the untreated control, no reduction in the level of relaxed circular HBV DNA (RC-DNA, Fig. 5B) was seen at any concentration tested. There was some evidence of a reduction in double stranded linear (DSL) and single stranded DNA in the 10μM sample, although this was not consistently seen in other experiments (Fig. 5B). These potential reductions were modest, and more work is needed to determine if this is indeed a definitive effect. These results, however, support the idea that proparacaine may work on post-replication stages perhaps by preventing HBV envelopment and/or secretion.
To analyze the impact of chlorophyllide on HBV we used non-denaturing particle gel analysis. Because G2.215 cells do not produce enough HBV virions for this assay, we used HepDE19 cells. HepDE19 is a stable cell line expressing HBV under the control of tetracycline, allowing the secretion of high levels of HBV viral particles after removal of tetracycline. Similar to G2.2.15 cells, HepDE19 cells also secrete naked nucleocapsids into the medium for unknown reasons (Guo et al., 2011; Guo et al., 2007b). Under non-denaturing conditions the naked nucleocapsids can be separated from the virions based on size. HBV DNA both in virions and nucleocapsids can then be examined by Southern blot using a labeled HBV-specific probe. After a 5 day treatment with chlorophyllide, HBV virions and nucleocapsids were resolved on a 1% non-denaturing agarose gel, transferred to a nylon membrane and hybridized with a labeled HBV-specific probe (Guo et al., 2007b). Figure 6A suggests that treatment with chlorophyllide reduces viral particles in the cell culture medium in a dose-dependent manner, however, the formation and secretion of nucleocapsids were not affected. Analysis of cellular HBV DNA by Southern blot suggested that chlorophyllide did not inhibit HBV replication. Treatment with up to 10 μM chlorophyllide did not repress either RC or SS DNA levels (Fig. 6B). The anti-viral activity of chlorophyllide and its analogies have been extensively studied and published elsewhere (Guo et al., 2011). These results support the idea that, like proparacaine, chlorophyllide may also prevent HBV virion formation and/or secretion.
Figure 6. Particle gel and Southern blot analysis of the role of chlorophyllide in reducing HBV DNA.
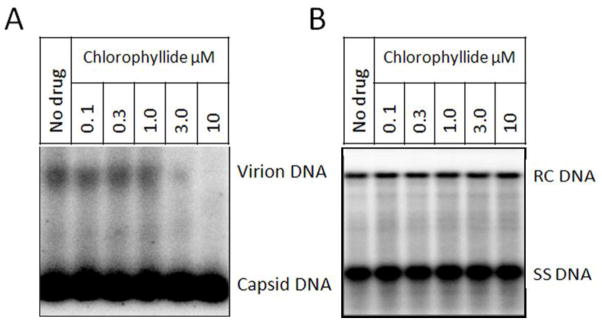
HepDE19 cells were cultured with Tet- medium for 5 days to initiate HBV expression. Cells were then treated with chlorophyllide at different doses. A.) HBV virions and nucleocapsids in culture medium were resolved in a 1% non-denaturing gel and transferred to membrane. The membrane was then hybridized with a HBV-specific probe. B.) Southern blot to detect cellular HBV DNA.
4. Discussion
There are a growing number of examples where libraries of compounds known to be safe in humans are being tested for the possibility of activity for indications other than those for which they were intended. This phenomenon of “repurposing” has, in addition to the library described here, been applied to other screens of safe compound libraries (Bader and Korba, 2010; Gastaminza et al., 2010; O’Connor and Roth, 2005; Thomas et al., 2009). This approach is particularly appealing in the field of HBV where current therapies are lacking, and it was with this in mind that the current screen was performed.
The high throughput screening system, utilizing the real time IA-PCR assay described, is both highly sensitive and highly specific. Because of the PCR platform for this assay, the sensitivity is high enough to detect as few as 30 copies of HBV DNA. In addition, the 96-well plate format allows large numbers of compounds to be quickly screened and lends itself to robotic handling. Moreover, this assay has the ability to identify not only drugs which inhibit HBV replication such as the nucleoside analog lamivudine, but also can identify drugs such as proparacaine and chlorophyllide which likely work on post-replication stages such as viral assembly, transport and/or secretion. The principle used in this technology, first capturing protein associated virus by use of an immobilized antibody, followed by amplification and detection of viral DNA, can easily be applied to screen for inhibitors of other enveloped viruses or pathogens.
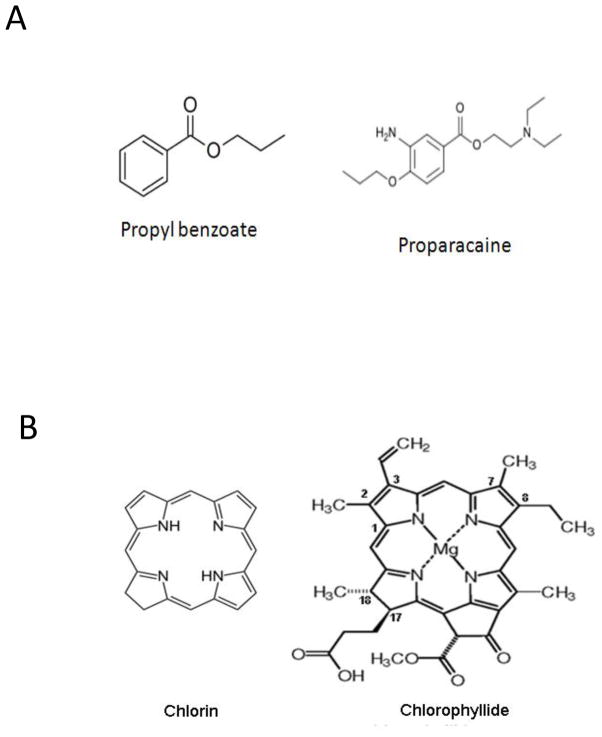
Two compounds proparacaine and chlorophyllide were identified to have activity against HBV in this screen. Although their mechanisms still require additional study, the data suggests they may work on post-replication stages of the HBV life cycle. The proparacaine family members all have a common propylbenzoate core structure (Fig. 7A). Chlorophyllide is a chlorophyll derived from a compound named chlorin, the core structure of which is a large heterocyclic aromatic ring consisting of three pyrroles and one pyrroline coupled through four methine linkages (Fig. 7B). Magnesium- containing chlorins are called chlorophylls (Godnev et al., 1963). Both of these compounds as well as their analogs have previously been used in humans or animals. Proparacaine, known commercially as Alcaine, is an approved drug used primarily as an ophthalmic anesthetic (as eye drops) to reduce pain and discomfort. Chlorophyllide is an alkylated porphoryn, structurally related to chlorophyll. The most studied chlorophyll is chloropyllin, which has anticancer activity that has been shown to be effective against liver cancer (Reinbothe et al., 2006; Sarkar et al., 1994). Although the mechanism is still unclear, the interaction of chlorophyllin with a carcinogen or the removal of toxic aflatoxins may play an important role in its anticancer activity (Breinholt et al., 1995; Egner et al., 2001; Guo et al., 2011).
Figure 7. The structure of selected candidate compounds.
A.) The structure of proparacaine, along with the core structure for the family of compounds is presented. B.) The structure of chlorophyllide, along with the core structure for its analogs is presented.
The antiviral activity of these two compounds has previously been reported. For example, chlorophyllide has been reported to inactivate Varicella Zoster Virus and influenza virus (Mekler et al., 1969), and proparacaine can inactivate equine herpesvirus-4 and feline herpesvirus-1 (Romanowski et al., 1999; Storey et al., 2002). Neither of these compounds, however, can inactivate nonenveloped viruses such as feline calicivirus (Storey et al., 2002), suggesting that the antiviral activity of these two compounds is associated with viral envelopment. This is consistent with our observations that the antiviral mechanism of these two compounds is dependent on preventing the envelopment or secretion of the virus. In vitro, the infectivity of HSV and influenza virus is lost following treatment with chlorophyllide and proparacaine, which implies that these two drugs might also prevent viral entry (Mekler et al., 1969; Romanowski et al., 1999; Storey et al., 2002). Based on these findings, further research is currently underway to investigate the effect of these two compounds on HBV infectivity using an in vitro infection system.
5. Conclusion
We have successfully established a novel high throughput screening technology, real-time IA-PCR, based on the capture of viral particles by an antibody immobilized on a PCR plate and then subsequent assessment of viral load by real-time PCR. Using this technology to screen 2000 compounds we were able to identify eight compounds that were shown to consistently reduce the amount of secreted HBV viral particles in the culture medium under conditions that had no detectable impact on cell viability. The anti-HBV activity of two of these, proparacaine and chlorophyllide, which were shown to reduce HBV levels 4- to 6-fold with an IC50 of 1 and 1.5μM respectively, have been further confirmed by other technologies. Additionally, the basis for this assay makes it easily adapted to the identification of compounds against other enveloped viruses by simply using different antibodies specific for the envelope of the virus being studied.
Supplementary Material
Highlights.
A novel high throughput screening system Immune-absorbance PCR was established.
Using this system, 2000 compounds from safety MicroSource library were screened.
8 compounds have been proved having anti-Hepatitis B virus (HBV) activity.
The anti-HBV mechanism of Proparacaine and chlorophyllide has been further studied.
Acknowledgments
This work is supported by an Appropriation from the Commonwealth of Pennsylvania, Hepatitis B Foundation USA, National Cancer Institute grant CA165314-01A1 (to X.L.) and ImCare Biotech.
Glossary
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- IA-PCR
Immune-absorbance PCR
- LHBs
HBV large surface protein
- NAs
Nucleotide analogues
- PCR
Polymerase chain reaction
Footnotes
Disclosure
X.L receives research support from ImCare Biotech.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bader T, Korba B. Simvastatin potentiates the anti-hepatitis B virus activity of FDA-approved nucleoside analogue inhibitors in vitro. Antiviral research. 2010;86:241–245. doi: 10.1016/j.antiviral.2010.02.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinholt V, Schimerlik M, Dashwood R, Bailey G. Mechanisms of Chlorophyllin Anticarcinogenesis against Aflatoxin B1: Complex Formation with the Carcinogen. Chemical Research in Toxicology. 1995;8:506–514. doi: 10.1021/tx00046a004. [DOI] [PubMed] [Google Scholar]
- Egner PA, Wang JB, Zhu YR, Zhang BC, Wu Y, Zhang QN, Qian GS, Kuang SY, Gange SJ, Jacobson LP, Helzlsouer KJ, Bailey GS, Groopman JD, Kensler TW. Chlorophyllin Intervention Reduces Aflatoxin-DNA Adducts in Individuals at High Risk for Liver Cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14601–14606. doi: 10.1073/pnas.251536898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Whitten-Bauer C, Chisari FV. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc Natl Acad Sci U S A. 2010;107:291–296. doi: 10.1073/pnas.0912966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godnev TN, Akulovich NK, Rotfarb RM. Total synthesis of chlorophyll and its biosynthesis. Usp Sovrem Biol. 1963;55:204–218. [PubMed] [Google Scholar]
- Guo H, Jiang D, Zhou T, Cuconati A, Block TM, Guo JT. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol. 2007a;81:12472–12484. doi: 10.1128/JVI.01123-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Pan X, Mao R, Zhang X, Wang L, Lu X, Chang J, Guo JT, Passic S, Krebs FC, Wigdahl B, Warren TK, Retterer CJ, Bavari S, Xu X, Cuconati A, Block TM. Alkylated porphyrins have broad antiviral activity against hepadnaviruses, flaviviruses, filoviruses, and arenaviruses. Antimicrobial agents and chemotherapy. 2011;55:478–486. doi: 10.1128/AAC.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhou T, Jiang D, Cuconati A, Xiao GH, Block TM, Guo JT. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J Virol. 2007b;81:10072–10080. doi: 10.1128/JVI.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri SM, Lok AS. Antiviral Therapy for Chronic Hepatitis B. Clin Liver Dis. 2010;14:425–438. doi: 10.1016/j.cld.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Janssen HL, Berk L, Schalm SW, Heijtink RA, Hess G, Rossol S, Meyer zum Buschenfelde KH, Chamuleau RA, Jansen PL, Reesink HW, et al. Antiviral effect of prolonged intermittent lymphoblastoid alpha interferon treatment in chronic hepatitis B. Gut. 1992;33:1094–1098. doi: 10.1136/gut.33.8.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartal ED, Alpat SN, Ozgunes I, Usluer G. Adverse effects of high-dose interferon-alpha-2a treatment for chronic hepatitis B. Advances in therapy. 2007;24:963–971. doi: 10.1007/BF02877700. [DOI] [PubMed] [Google Scholar]
- Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J Virol. 2003;77:10288–10294. doi: 10.1128/JVI.77.19.10288-10294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrobial agents and chemotherapy. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamontagne J, Pinkerton M, Block TM, Lu X. Hepatitis B and hepatitis C virus replication upregulates serine protease inhibitor Kazal, resulting in cellular resistance to serine protease-dependent apoptosis. J Virol. 2010;84:907–917. doi: 10.1128/JVI.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok AS. Treatment of chronic hepatitis B. J Viral Hepat. 1994;1:105–124. doi: 10.1111/j.1365-2893.1994.tb00110.x. [DOI] [PubMed] [Google Scholar]
- Lu X, Block TM, Gerlich WH. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. Journal of Virology. 1996;70:2277–2285. doi: 10.1128/jvi.70.4.2277-2285.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler LB, Bychovsky AF, Krikun BL. Electron Microscope Study of the Viricidal Properties of Sodium Magnesium-chlorophyllin. Nature. 1969;222:574–575. doi: 10.1038/222574b0. [DOI] [PubMed] [Google Scholar]
- Muller J, Schust J, Berg T. A high-throughput assay for signal transducer and activator of transcription 5b based on fluorescence polarization. Analytical biochemistry. 2008;375:249–254. doi: 10.1016/j.ab.2008.01.017. [DOI] [PubMed] [Google Scholar]
- O’Connor KA, Roth BL. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat Rev Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- Paoletti F, Ainger K, Donati I, Scardigli R, Vetere A, Cattaneo A, Campa C. Novel fluorescent cycloheximide derivatives for the imaging of protein synthesis. Biochemical and biophysical research communications. 2010 doi: 10.1016/j.bbrc.2010.04.075. [DOI] [PubMed] [Google Scholar]
- Reinbothe C, Bartsch S, Eggink LL, Hoober JK, Brusslan J, Andrade-Paz R, Monnet J, Reinbothe S. A role for chlorophyllide a oxygenase in the regulated import and stabilization of light-harvesting chlorophyll a/b proteins. Proc Natl Acad Sci U S A. 2006;103:4777–4782. doi: 10.1073/pnas.0511066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski EG, Barnhorst DA, Kowalski RP, Gordon YJ. The survival of herpes simplex virus in multidose office ophthalmic solutions. American journal of ophthalmology. 1999;128:239–240. doi: 10.1016/s0002-9394(99)00114-2. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Sharma A, Talukder G. Chlorophyll and chlorophyllin as modifiers of genotoxic effects. Mutation Research/Reviews in Genetic Toxicology. 1994;318:239–247. doi: 10.1016/0165-1110(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Sells MA, Zelent AZ, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LL, Loomba R. Drug targets in hepatitis B virus infection. Infectious disorders drug targets. 2009;9:105–116. doi: 10.2174/187152609787847677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey ES, Gerding PA, Scherba G, Schaeffer DJ. Survival of equine herpesvirus-4, feline herpesvirus-1, and feline calicivirus in multidose ophthalmic solutions. Vet Ophthalmol. 2002;5:263–267. doi: 10.1046/j.1463-5224.2002.00234.x. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Moloughney JG, Toney JH. Old drugs resurrected? Curr Opin Investig Drugs. 2009;10:119–120. [PubMed] [Google Scholar]
- Weisman JL, Liou AP, Shelat AA, Cohen FE, Guy RK, DeRisi JL. Searching for new antimalarial therapeutics amongst known drugs. Chemical biology & drug design. 2006;67:409–416. doi: 10.1111/j.1747-0285.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C, Aguzzi A. Approaches to therapy of prion diseases. Annu Rev Med. 2005;56:321–344. doi: 10.1146/annurev.med.56.062404.172936. [DOI] [PubMed] [Google Scholar]
- Wu F, Wu MJ, Zhuge XL, Zhu SM, Zhu B. Correlation of the occurrence of YMDD mutations with HBV genotypes, HBV-DNA levels, and HBeAg status in Chinese patients with chronic hepatitis B during lamivudine treatment. Hepatobiliary & pancreatic diseases international: HBPD INT. 2012;11:172–176. doi: 10.1016/s1499-3872(12)60144-1. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. Journal of gastroenterology and hepatology. 2011;26(Suppl 1):138–143. doi: 10.1111/j.1440-1746.2010.06545.x. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Curtis M, Qi X, Miller MD, Borroto-Esoda K. Anti-hepatitis B virus activity in vitro of combinations of tenofovir with nucleoside/nucleotide analogues. Antivir Chem Chemother. 2009;19:165–176. doi: 10.1177/095632020901900404. [DOI] [PubMed] [Google Scholar]
- Zoulim F. Mechanism of viral persistence and resistance to nucleoside and nucleotide analogs in chronic hepatitis B virus infection. Antiviral research. 2004;64:1–15. doi: 10.1016/j.antiviral.2004.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.