Abstract
Recently, Brachypodium distachyon has emerged as a model plant for studying monocot grasses and cereal crops. Using assembled expressed transcript sequences and subsequent mapping to the corresponding genome, we identified 1219 alternative splicing (AS) events spanning across 2021 putatively assembled transcripts generated from 941 genes. Approximately, 6.3% of expressed genes are alternatively spliced in B. distachyon. We observed that a majority of the identified AS events were related to retained introns (55.5%), followed by alternative acceptor sites (16.7%). We also observed a low percentage of exon skipping (5.0%) and alternative donor site events (8.8%). The ‘complex event’ that consists of a combination of two or more basic splicing events accounted for ∼14.0%. Comparative AS transcript analysis revealed 163 and 39 homologous pairs between B. distachyon and Oryza sativa and between B. distachyon and Arabidopsis thaliana, respectively. In all, we found 16 AS transcripts to be conserved in all 3 species. AS events and related putative assembled transcripts annotation can be systematically browsed at Plant Alternative Splicing Database (http://proteomics.ysu.edu/altsplice/plant/).
Keywords: alternative splicing, Brachypodium distachyon, expressed sequence tags, functional ontology, intron retention
1. Introduction
Recent advances in the next generation sequencing technologies presented us with a wider computational challenge to analyze and correlate the abundance of the protein diversity with the gene content. In eukaryotes particularly, the presence of introns within the protein-coding genes can generate more than one functional mRNA isoforms from one pre-mRNA transcript, through a post-transcriptional event called as alternative splicing (AS), which in turn increases the protein complexity and mRNA abundance without increasing the gene content. It has been proposed that the glycine-rich RNA binding protein AtGRP7 has been suggested as a potential factor to regulate AS in Arabidopsis thaliana.1 In principle, four basic and major type AS types have been reported, including exon skipping (ExonS), alternative donor (AltD) or alternative acceptor (AltA) site, and intron retention (IntronR).2 However, other types such as alternate terminal exon, retained exon (RE) and skipped exon (SE), initiation within an intron, termination within an intron, and spliced intron have also been reported in Oryza sativa.3 Some other basic types also have been classified as AS events such as alternative transcription initiation, alternative transcription termination, and mutually exclusive exons.4 However, complex events can be formed by combinations of the basic AS types. AS may generate a functional transcript that encodes distinct functional proteins, or a nonfunctional transcript that harbors a premature termination codon (PTC). These nonfunctional isoforms are degraded through a process called ‘regulated unproductive splicing and translation.’5 AS transcripts also play an important role in protein–protein interactions (PPIs) networks in addition to the differential gene expression in describing the tissue-specific cellular functions.6 Conditional splicing using ExonS of a suicide exon has been shown to regulate transgene and gene activation in Nicotiana benthamiana.7
AS transcripts are generally generated through three pathways: (i) IntronR in the mature mRNA, accounting for 30–50% of AS in A. thaliana and O. sativa, (ii) alternative exon usage, resulting in ExonS, and (iii) the use of cryptic splice sites that may elongate or shorten an exon.2 Approximately 60–75% of AS events occur within the protein-coding regions of mRNAs, resulting in changes in binding properties, intracellular localization, protein stability, enzymatic, and signaling activities.8 Expressed sequence tags (ESTs) have been used to discover and delineate these alternative-splicing variants through homology mapping-based approach.2,3,9 In plants, the functional relevance of certain AS-derived isoforms in responses to biotic and abiotic stresses has been reviewed.10,11 RNA-seq has played an important role in revealing the transcript diversity through AS events and revealed the abundance of the alternative isoforms with PTC.12 Recently, using RNA-seq AS events have also been revealed in plant circadian clock-associated genes in A. thaliana.13 The roles of these genes involved in the processes of photosynthesis, plant disease resistance, flowering, and grain quality in O. sativa have been well illustrated (see Reddy for review).10 Complex networks of regulation of gene expression and variation in AS have played a major role in the adaptation of plants to their corresponding habitat and environment.14 A recent review describes the role of the splicing regulatory elements and emerging experimental and computational approaches to identify cis-elements involved in regulating of AS in plants.15
Brachypodium distachyon is considered to be the model temperate grass for studying the evolutionary genomics and for the elucidation of the biologic pathways in grass genomics.16–18 It has also been suggested to be a model species for studying the evolutionary insights into biofuel crops such as genus Miscanthus and Panicum virgatum.19 The genome of B. distachyon was recently sequenced, and a total of 25 532 protein-coding genes were predicted as a part of genome analysis pipeline.20 B. distachyon genome exhibits a much higher level of co-linearity and synteny to the genomes of temperate cereal crops and also forage grasses such as Lolium and Festuca,17,20,21 and, thus, it can serve as a model genome to understand the functional protein diversity generated through alternative splice events in cereal crops. ESTs generated from B. distachyon were used for gene finding and phylogenetic analysis.22 In an earlier study, through mapping of 3818 tentative consensus sequences on the assembled genome scaffolds, we have demonstrated the presence of 128 AS events involving 120 genes in B. distachyon.9 In recent years, transcript abundance has been observed in the identification of the ESTs from this model organism, which presents an opportunity to study the AS at a wider level because of the availability of the larger number of transcripts with a higher genomic coverage. RIKEN has recently established a full-length cDNA (FLcDNA) project of B. distachyon consisting of the 79 872 ESTs generated using the biotinylated CAP trapper methodology. BrachyFLcDNA DB has been integrated with other available resources to make it is an integrated repository. The availability of the FLcDNA in B. distachyon will also aid in redefining the earlier gene models and will also aid in the functional analysis of the gene patterns.23 In the present study, we have reinvestigated the landscape of the AS events and demonstrated the presence of 1219 AS events involving 2204 putatively assembled transcripts generated from 941 genes. We have also classified the AS events according to functional ontology. In addition, we have created an online portal (http://proteomics.ysu.edu/altsplice/plant/) to access the annotation of all ESTs and AS events identified in this study.
2. Materials and methods
2.1. Sequence datasets
A total of 30 991 assembled transcript sequences also known as PlantGDB-assembled unique transcripts (PUTs) for B. distachyon were downloaded from PlantGDB (http://www.plantgdb.org/).24 The PUTs sets were generated by assembling 128 098 ESTs and mRNA sequences (version 175a) using the methods described at the site (http://www.plantgdb.org/prj/ESTCluster/PUT_procedure.php). Our previous evaluation confirmed that assemblies generated by CAP3 (parameters –p 95, –o 49, and –t 10 000), which was used by PlantGDB, were suitable for identification of AS isoforms.25,26 The option t with value 10 000 improves the quality of the assembly using the maximum available memory, and, thus, it avoids the misassembling of the ESTs and formation of counterfeit longer assemblies. The genome sequence of B. distachyon (diploid inbred line Bd21) was downloaded from Phytozome (v7.0).20,27 The assembled genome of B. distachyon measures approximately 272 Mb and spans across 5 chromosomes and 78 unmapped scaffolds.
2.2. Homology mapping, identification, and functional ontology of AS isoforms
Homology mapping was achieved by mapping the PUTs (hereafter referred as ESTs) of B. distachyon to the corresponding genome sequence, and identification of AS isoforms was done using ASFinder (http://proteomics.ysu.edu/tools/ASFinder.html/).28 ASFinder uses SIM4 program to map ESTs to the corresponding genome.29 It subsequently identifies those ESTs that are mapped to the same genomic location, but have variable exon–intron boundaries as AS isoforms. For our mapping, we used the following thresholds: a minimum of 97% identity of aligned ESTs with genomic sequences, a minimum of 80 bp of aligned length, and >85% of EST sequence aligned to the genome. The output of ASFinder was subsequently analyzed, and AS events were identified using AStalavista server (http://genome.crg.es/astalavista/).30 Additional AS isoforms from ESTs that were not mapped to the genome were identified using ASFinder BLASTN method to detect those EST pairs that have two or more segmented alignments. This similar method of using BLAT tool, rather than BLAST, has been previously used to identify AS from ESTs in plants.31 The functionalities of genes having AS events were identified using rpsBLAST against Pfam and the Conserved Domain Database.32 Gene ontology (GO) information was retrieved by mapping Pfam to GO (http://www.geneontology.org/external2go/pfam2go) and analyzed using GO SlimViewer with plant-specific GO terms.33 ESTs were also compared with predicted protein-coding DNA sequences (CDS) in Phytozome using BLASTN, with a cutoff E-value of 1e-10, and a total of 22 591 ESTs had one or more CDS match with ≥97% identity. The information was available in the database for comparison.
2.3. Conserved alternatively spliced genes in B. distachyon, O. sativa, and Arabidopsis
To identify potentially conserved alternatively spliced genes between B. distachyon, O. sativa, and A. thaliana, we downloaded previously detected mRNA sequences of AS genes in O. sativa and A. thaliana from Plant GDB ASIP portal (http://www.plantgdb.org/ASIP/Download/).2 Conserved AS genes were identified using O. sativa and A. thaliana AS mRNAs to search against B. distachyon AS ESTs using BLASTN with E-value of 1e-5. The best BLASTN hit with an identity of >70% and a minimum aligned length of >100 bp were identified as conserved AS pairs.
2.4. Data access and visualization of AS
We annotated all ESTs according to function and structure, which involves the prediction of the protein-coding regions in the ESTs using OrfPredictor (http://proteomics.ysu.edu/tools/OrfPredictor.html),34 mapping to genomic sequences, BLASTN matching ESTs to predicted protein CDS (≥97% identity used) of this species downloaded from Phytozome, and Pfam and conserved domain search. We further developed an online web portal for the systematic display of the mapped ESTs and associated AS events (http://proteomics.ysu.edu/altsplice/plant/). The database can be queried using EST ID, keywords, or AS event types. The AS events can be systematically browsed using GBrowse, and the CDS annotations were added using the GFF file of the annotated genome downloaded from Phytozome. We provide a functionality to query for all ESTs and AS events using BLASTN.
3. Results and discussion
3.1. Detection and classification of AS events
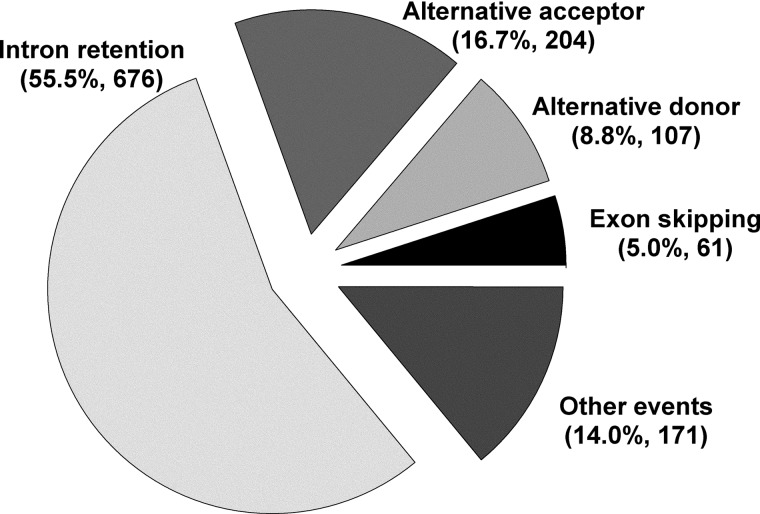
In our study, a total of 23 693 ESTs were successively mapped to 14 841 genomic loci of B. distachyon genome sequence. We identified a total of 1219 pairs of ESTs (2021 unique ESTs), which were mapped to 941 genomic loci having AS, suggesting that ∼6.3% of expressed genes in B. distachyon are under the potential influence of AS (Supplementary Table S1). The observed AS percentage is relatively low in comparison to the percentage of the genes observed in A. thaliana (21.8%) and O. sativa (21.2%).2 This potential difference in the distribution of the AS genes versus non-AS genes may be due to the fact that relatively more transcript resources are available for A. thaliana and O. sativa in comparison to B. distachyon. Classification of the AS events showed that the majority of the observed events (676) corresponded to IntronR (55.5%), followed by AltA (204, 16.7%), AltD (107, 8.8%), and ExonS (61, 5.0%). We observed 171 ‘complex events’ that correspond to AS events having more than 1 basic event (14.0%) (Fig. 1). Inspite of the difference in the transcript abundance, the observed AS events, particularly IntronR, showed conservation across the plant lineage, which may be an indication of dominance of the splice site recognition by intron definition as previously suggested in A. thaliana and O. sativa.2
Figure 1.
Landscape of AS events in B. distachyon.
The abundance of IntronR is consistent with previous several reports in organism such as Medicago truncatula (39%), Populus trichocarpa (34%), A. thaliana (56%), O. sativa (54%), and Chlamydomonas reinhardtii (50%).2,35,36 Recently, using a RNA-seq study, IntronR was also observed as a major event in mRNAs of the CCA1/LHY-like subfamily of MYB transcription factors in A. thaliana.13 The prevalence of IntronR type in B. distachyon and in other plants supports the intron definition model, i.e. introns are identified by the splicing machinery splicesomes, for pre-mRNA splicing in plants, that was in contrast to the exon definition model in animals. Supportive views for the plant introns retention suggest that they are not derived just as a cause of incomplete splicing, but they are maintained as potential cytoplasmic translatable transcripts.31 The results observed in the present study expanded the landscape of the AS events in B. distachyon by incorporating a higher number of ESTs when compared with the previous published reports of AS events in B. distachyon.9 The events of AltA were approximately twice frequent as the events of AltD, which are also consistent with previous observations in other plants.2,3,36
Recently, it has been observed that alternative 3′ splice site has been observed as the most dominant AS event in contrast to IntronR in A. thaliana using annotated genomic data.37 Such a discrepancy might be caused by insufficient coverage of AS types in the study of Koralewski and Krutovsky as EST data were not used and only about one-quarter of AS events were identified when compared with the previous study.2,37 Recently, using the data generated from ultrahigh-throughput RNA sequencing (RNA-seq) in A. thaliana revealed that at least 42% of intron-containing genes were alternatively spliced.12 However, despite the difference in the percentage of AS genes using EST mapping and deep transcriptome sequencing, these results were consistent in identifying that IntronR was the most prevalent form of AS in plants.2,12
There was a whole genome duplication event that occurred before the diversification of the grasses, and also grass-specific tandem gene expansion in B. distachyon was observed.20 In an earlier study, the relation between gene family size and AS was studied by comparing the distribution of AS frequency in single-copy genes versus gene families, and, interestingly, a higher percentage of AS was found in gene families than in single-copy genes in A. thaliana and O. sativa.38 In our analysis, we clustered paralogs using the blastclust tool (–S 50 and –L 0.7) in BLAST package and identified 3407 gene families and 15 169 single genes. Then, mapping AS transcripts to them revealed 7.7 and 5.9% of AS in gene families and in single-copy genes, respectively. Recent analysis showed that gene duplication and AS evolved independently, which is further exemplified with the confirmation of the independent evolution of these two processes using isochorismate synthase in Arabidopsis and Populus.39 AS divergence between duplicated genes may have contributed to gene functional evolution and led to preservation of some duplicated genes.40 Our results are in accordance with the recent finding which suggests that the moderately duplicated gene families have potentially higher AS isoforms when compared with the singleton genes. AS, along with gene duplication, represents an evolutional strategy that rapidly increases genome plasticity to deal with various plant stresses.11
In addition to the AS genes and events identified by mapping ESTs to the genome, using the BLASTN method implemented in ASFinder, we also identified 56 genes (involving 129 ESTs) from 7298 ESTs, which were not mapped to the genome. However, specific category of AS events in this subset of genes cannot be identified because of the lack of the genomic information. All ESTs and spliced variants can be queried, visualized, explored, and browsed at the Plant Alternative Splicing Database (http://proteomics.ysu.edu/altsplice/plant/) using several query patterns such as EST IDs, gene, and AS event type. One similar resource for the visualization of AS events, which has been developed for A. thaliana, is available at ArabiTag; http://transvar.org/arabitag/.41
3.2. Features of exons and introns in protein-coding genes
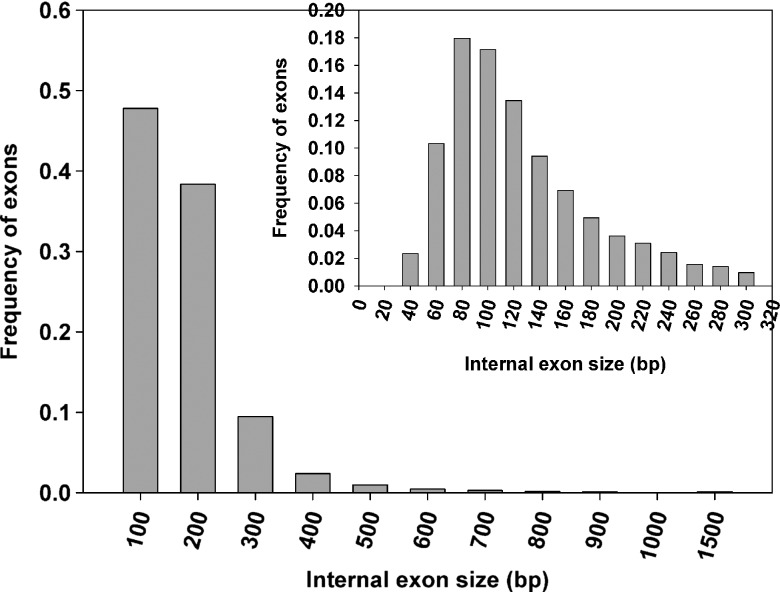
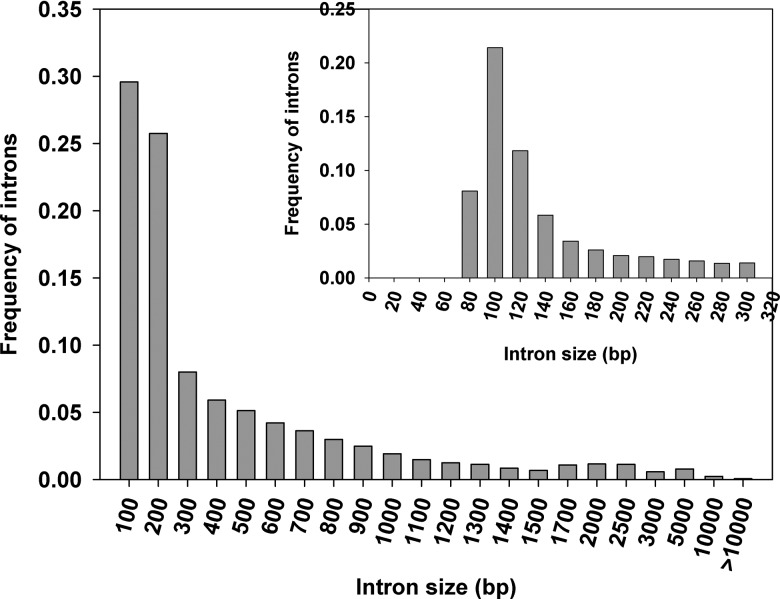
Following the mapping of the ESTs to the corresponding genomic sequence, we extracted the lengths of 34 087 internal exons and 48 000 introns. We observed size variation with internal exons that had a size ranging from 15 to 1461 bp with an average size of 130 ± 87 bp. We further observed that approximately 96% of exons had a size below 300 bp and 58% lied between 60 and 140 bp (Fig. 2). The observed exon length is in line with the earlier observed mean internal exon length in A. thaliana (172 bp) and O. sativa (193 bp).2 It has been previously shown that the shorter exons have high potential for AS events by exon shuffling events coupled with exon duplication and may promote the genome and transcriptomic complexity.37 It has been demonstrated previously that organisms having the shorter exons have higher gene content ratio.37 However, when compared with exon size, the distributions of intron size were more variable, ranging from 10 bp to >10 kb. There were 32 introns (0.07%) having a size >10 kb. However, in considering possible errors in EST assembly and genome assembly, these long introns identified in this study need to be further examined experimentally. Excluding those long introns with >10 kb in size, the remaining introns had an average size of 420 bp with a standard deviation of 622 bp, ∼63% had a size below 300 bp, and ∼47% of introns lied in 60–140 bp (Fig. 3). The average intron size in B. distachyon was very close to rice intron size (433 bp), and both of them were much longer than the average intron size (173 bp) in Arabidopsis, calculated using a similar method.2
Figure 2.
Distribution of internal exon size: The x-axis indicates the size of internal exons. Bin sizes are right inclusive (e.g. bin 100 comprises sequences of lengths 1–100 bp). The y-axis indicates the frequency of internal exons. The inset shows a detailed distribution of smaller internal exons.
Figure 3.
Distribution of intron size: The x-axis indicates the size of introns. Bin sizes are right inclusive (e.g. bin 100 comprises sequences of lengths 1–100 bp). The y-axis indicates the frequency of introns. The inset shows a detailed distribution of smaller introns.
3.3. Features of retained introns, SEs, and other DNA fragments involved in AS events
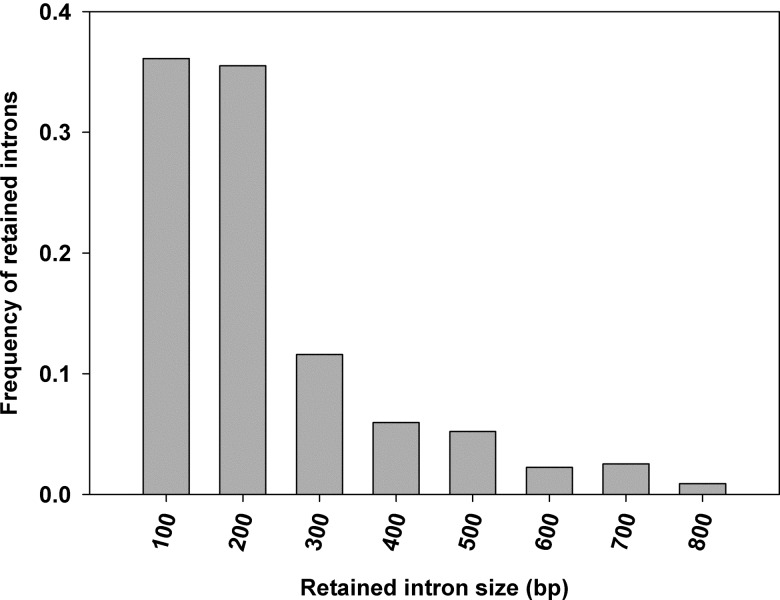
We examined the size distribution of DNA fragments involved in AS events, including retained introns, SEs, and fragments in AltD or AltA sites (Table 1). The retained introns had a range from 8 to 1142 bp with an average size of 184 bp, which is quite small in comparison to the average size of the introns in the species. Although the overall frequency distribution of retained intron size was similar to the distribution of all introns, there were no introns longer than 1150 bp in the retained intron set (Fig. 4). The SEs (ExonS) had a range of 23–376 bp with an average size of 111 bp. The fragments involved in AltD or AltA sites ranged from 3 to 468 bp (average size 49 bp) in AltA and from 3 to 553 bp (average size 67 bp) in AltD. Overall, the average size of the fragments involved in AS was relatively short. It has been previously suggested that the presence of the retained intron within the coding sequence may give rise to the nonsense-mediated decay (NMD).42 However, recently it has been proposed in Arabidospis that splice isoform with the retained introns is not sensitive to NMD.43 Although the high proportion of retained introns in AS events in plants supports the intron definition model in splicing, the evidence that the size of the retained introns was more similar to typical exons, lack of long introns, makes the exon definition model an alternative reasonable explanation.
Table 1.
Summary of DNA fragment sizes involved in AS events
| IntronR | AltA | AltD | ExonS | |
|---|---|---|---|---|
| Size range (bp) | 8–1142 | 3–468 | 3–553 | 23–376 |
| Mean | 184 | 49 | 67 | 111 |
| Standard deviation | 152 | 82 | 103 | 68 |
Figure 4.
Distribution of retained intron size: The x-axis indicates the size of retained introns with bin sizes that are right inclusive (e.g. bin 100 comprises sequences of lengths 1–100 bp). The y-axis indicates the frequency of retained intron size.
3.4. Functional ontology of AS genes
Functional annotation of the alternative spliced transcripts and their association with certain domains can give the mechanistic overview of the functional impact of the AS on the domain and also on domain-mediated regulation of AS. Recently, it has been predicted that AS events have potential effects on the protein domain function. In a recent study, it was demonstrated that MIKC MADS-box genes are under the strong influence of the AS events, and conserved AS events were observed across the borders or with in the K-box domain.44 In a recent study, it has been demonstrated that two spliceosomal proteins, U1-70K and U2AFb35 are known to function in 5′ and 3′ splice site selection and regulate the AS in SR45 that is a serine/arginine-rich (SR)-like protein with two arginine/serine-rich (RS) domains.45
To annotate the AS transcripts and to identify their possible association with functional domains, we performed a BLASTX search of all ESTs of B. distachyon against UniProt database. The protein-coding sequences [open reading frames (ORFs)] of ESTs were identified using OrfPredictor webserver.34 The functional domains of the AS genes, using the longest ORF of each AS gene, were predicted using rpsBLAST searching Pfam and CDD database. Among a total of 941 proteins encoded by AS genes, 651 were predicted having functional domains (Supplementary Table S1). We have further classified the genes according to the gene functional ontology (GO) categories, i.e. cellular component, molecular functions, and biological processes (Table 2, Supplementary Table S2). It was observed that the overall distribution of molecular functions in genes having AS and non-AS genes was similar (Table 2). Most of the functionally annotated transcripts were found to be associated with binding activity or catalytic activity, and some of which were found to be associated with DNA and RNA binding, respectively (Table 2). These transcripts with functional annotation may be of functional importance in revealing the genes in various biological pathways. It has been suggested that the binding of the proteins to cis-regulatory sequences in exons and introns and associated splicing regulators may regulate the loading of the splicing machinery to splice site.10
Table 2.
GO and classification of associated molecular functions of AS genes and non-AS genes
| Molecular function | AS genes |
Non-AS genes |
||
|---|---|---|---|---|
| GO term count | % | GO term count | % | |
| Binding | 100 | 21.0 | 1936 | 22.3 |
| Catalytic activity | 76 | 16.0 | 1253 | 14.4 |
| Nucleotide binding | 54 | 11.3 | 1059 | 12.2 |
| Transferase activity | 52 | 10.9 | 1049 | 12.1 |
| Hydrolase activity | 44 | 9.2 | 843 | 9.7 |
| Protein binding | 32 | 6.7 | 539 | 6.2 |
| Kinase activity | 31 | 6.5 | 546 | 6.3 |
| DNA binding | 25 | 5.3 | 396 | 4.6 |
| Nucleic acid binding | 13 | 2.7 | 220 | 2.5 |
| Structural molecule activity | 13 | 2.7 | 251 | 2.9 |
| Transporter activity | 13 | 2.7 | 308 | 3.5 |
| RNA binding | 11 | 2.3 | 141 | 1.6 |
| Others | 12 | 2.5 | 145 | 1.7 |
| Total | 476 | 8686 | ||
Earlier, it has been shown that the AS also plays an important role in promotion of the floral transitions, which gives a layout of the involvement of the AS events in the biological pathways. In a study, it has been shown that the AS of the FCA transcript, which encodes a RNA-binding domain and a putative PPI domain, promotes floral transition in A. thaliana fca mutants.46 Recently, it has been demonstrated that a splice variant (IDD14β) of Arabidopsis INDETERMINATE DOMAIN14 (IDD14) transcription factor regulates the function of the IDD14α in starch metabolism by acting as an inhibitor forming heterodimers, which potentially explains the role of the AS in regulation of the transcription factor activity.47 Recent reports suggest that AS also plays a potential role in the regulation of the freezing tolerance through the splicing of the clock compounds (CIRCADIAN CLOCK-ASSOCIATED1; CCA1).48 We observed GO categories enriched in carbohydrate metabolic process (GO:0005975) and in response to stress (GO:0006950) in the AS transcripts (Supplementary Table S2), which gives a strong indication of the modulation of the biosynthetic pathway and increases the protein diversity for a subset of genes using AS events. Recently, using a systematic investigation of an intronic miR400 under heat stress in A. thaliana, it has been shown that the AS acts as a regulatory link and possibly regulates the interactions of miRNAs and environmental stress.49 Several recent reports postulate the association of the AS events with biosynthesis in a couple of genes involved in auxin biosynthesis shows the patterns of tissue-specific splicing.50,51 However, all these functions still remain uncovered in B. distachyon that is a model organism for grass monocot lineage. Further experimental verification of the AS events with functional annotation, identified in the present study, may highlight some important biological phenomenon in detail, which are specific to grass lineage.
3.5. Conserved alternatively spliced genes
Identification of the conserved AS events can also assist us in understanding the evolution of the functional genes and their regulation at the transcriptional and at the translational level, which may shed light on the understanding of the trade-off going on between the plant machinery and the environmental adaptation. The identification of the AS isoforms in plants may explain and postulate plausible answers to adaptation to environmental stress, domestication, and also may likely explain the trait selection.51 It has been stressed that the detection of the conserved AS events may potentially help in the identification of the AS isoforms containing PTC.52 Recent analysis further revealed that AS underlies the exonization of the 5S rRNA, which regulates the TFIIA.53 The serine/arginine-rich (SR) protein family plays an important role in constitutive AS and regulates AS in a tissue-specific and stress-responsive manner.54 Three sets of AS events were conserved between A. thaliana and rice in the plant-novel-SR protein, SC35-like and two Zn-knuckles-type 9G8 subfamilies.54 It was reported that 41.7% of AS genes in A. thaliana were found to have close homologs in O. sativa that were also identified to be alternatively spliced, and 30% of the intron-retained transcripts also showed conservation between A. thaliana and O. sativa.2
In our study, we identified 163 homologous pairs of AS genes between B. distachyon and O. sativa, ∼17.3% of all AS genes identified in B. distachyon and 39 homologous pairs of AS genes between B. distachyon and A. thaliana (Supplementary Table S3). Among the functionally annotated AS events, 16 AS genes were found be conserved among all the species (Table 3). Our study postulates the potential list of 16 conserved genes that may act as potential marker genes/transcripts for further, detailed experimental examination of their biological significance of AS in plant growth, development, and tracing the evolution of certain biological pathways.
Table 3.
Conserved alternatively spliced genes in B. distachyon, O. sativa, and A. thaliana
| BD_PUT_ID | Rice_gi | Arabidopsis_gi | Functional domain |
|---|---|---|---|
| PUT_Bd26530 | 7212633 | 315633 | pfam13 893, RRM_5, RNA recognition motif |
| PUT_Bd2017 | 4878380 | 14532699 | pfam04928, PAP_central, poly(A) polymerase central domain |
| PUT_Bd87 | 428653 | 935690 | pfam03949, Malic_M, malic enzyme, NAD-binding domain |
| PUT_Bd26130 | 702742, 2280709 | 2763737 | pfam02991, Atg8, autophagy protein Atg8 ubiquitin like |
| PUT_Bd28467 | 428194 | 937533 | pfam01717, Meth_synt_2, cobalamin-independent synthase, catalytic |
| PUT_Bd27829 | 702009 | 496827 | pfam01370, Epimerase, NAD-dependent epimerase/ dehydratase family |
| PUT_Bd1551 | 571572 | 17474 | pfam01287, eIF-5a, eukaryotic elongation factor 5A hypusine |
| PUT_Bd31189 | 701361 | 166645 | pfam00504, Chloroa_b-bind, chlorophyll A–B-binding protein |
| PUT_Bd940 | 431832, 3762355 | 16406 | pfam00230, MIP, major intrinsic protein |
| PUT_Bd3432 | 572134, 2310784 | 16869 | pfam00179, UQ_con, ubiquitin-conjugating enzyme |
| PUT_Bd4338 | 701921 | 935078 | pfam00085, thioredoxin, thioredoxin |
| PUT_Bd2952 | 427824, 2310078, 2434385 | 937253 | pfam00071, Ras, Ras family |
| PUT_Bd2364 | 428011, 2311034 | 398603 | Function unknown |
| PUT_Bd7431 | 2312070 | 1045041 | Function unknown |
| PUT_Bd4686 | 287194, 571706, 1631641 | 1053979 | Function unknown |
| PUT_Bd10494 | 286390 | 1217256 | Function unknown |
In summary, B. distachyon is currently a model plant for studying grass system and evolutionary biology. Identification of alternatively spliced genes and AS events is the first step for understanding which categories of genes are regulated post-transcriptionally and to identify the potential mechanisms for transcriptomic complexity. Our systematically analyzed data provide a visualization portal to B. distachyon community researchers who aim to identify potential mechanisms of transcriptomic diversity and adaption to stress and to expand the functional relevance of the identified AS events through experimental approaches.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Authors' contribution
B.W. and G.L. contributed to the database construction, G.S. and X.J.M. contributed to the experiment design, data analysis, and preparation of the manuscript. All authors have read and approved the final version of the manuscript.
Funding
The work was funded by Youngstown State University (YSU) Research Council (Grant 12-11) and was also supported by YSU Research Professorship and the College of Science, Technology, Engineering, and Mathematics Dean's reassigned time for research to X.J.M. Open Access publication fee was provided by Ohio Plant Biotechnology Consortium (Grant 2011-001) through Ohio State University, Ohio Agricultural Research and Development Center to X.J.M.
Supplementary Material
Footnotes
Edited by Dr Masahiro Yano
References
- 1.Streitner C., Köster T., Simpson C.G., et al. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 2012;40:11240–55. doi: 10.1093/nar/gks873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang B., Brendel V. Genome wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA. 2006;103:7175–80. doi: 10.1073/pnas.0602039103. doi:10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell M.A., Haas B.J., Hamilton J.P., Mount S.M., Buell C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. doi:10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts G.C., Smith C.W. Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol. 2002;6:375–83. doi: 10.1016/s1367-5931(02)00320-4. doi:10.1016/S1367-5931(02)00320-4. [DOI] [PubMed] [Google Scholar]
- 5.Lareau L.F., Brooks A.N., Soergel D.A.W., Meng Q., Brenner S.E. The coupling of alternative splicing and nonsense mediated mRNA decay. In: Blencowe B.J., Graveley B.R., editors. Alternative Splicing in the Postgenomic Era. Austin, TX: Landes Biosciences; 2007. pp. 191–212. [Google Scholar]
- 6.Burgess D.J. Alternative splicing: proteomic rewiring through transcriptomic diversity. Nat. Rev. Genet. 2012;13:518–9. doi: 10.1038/nrg3288. doi:10.1038/nrg3288. [DOI] [PubMed] [Google Scholar]
- 7.Hickey S.F., Sridhar M., Westermann A.J., et al. Transgene regulation in plants by alternative splicing of a suicide exon. Nucleic Acids Res. 2012;40:4701–10. doi: 10.1093/nar/gks032. doi:10.1093/nar/gks032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamm S., Ben-Ari S., Rafalska I., et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. doi:10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Sablok G., Gupta P.K., Baek J.M., Vazquez F., Min X.J. Genome-wide survey of alternative splicing in the grass Brachypodium distachyon: an emerging model biosystem for plant functional genomics. Biotechnol. Lett. 2011;33:629–36. doi: 10.1007/s10529-010-0475-6. doi:10.1007/s10529-010-0475-6. [DOI] [PubMed] [Google Scholar]
- 10.Reddy A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant. Biol. 2007;58:267–94. doi: 10.1146/annurev.arplant.58.032806.103754. doi:10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 11.Mastrangelo A.M., Marone D., Laidò G., De Leonardis A.M., De Vita P. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185–186:40–9. doi: 10.1016/j.plantsci.2011.09.006. doi:10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Filichkin S.A., Priest H.D., Givan S.A., et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. doi:10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filichkin S.A., Mockler T.C. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol. Direct. 2012;7:20. doi: 10.1186/1745-6150-7-20. doi:10.1186/1745-6150-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed N.H., Kalyna M., Marquez Y., Barta A., Brown J.W. Alternative splicing in plants – coming of age. Trends Plant Sci. 2012;17:616–23. doi: 10.1016/j.tplants.2012.06.001. doi:10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy A.S.N., Rogers M.F., Richardson D.N., Hamilton M., Ben-Hur A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 2012;3:18. doi: 10.3389/fpls.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel J., Bragg J. Brachypodium distachyon, a new model for the Triticeae. In: Feuillet C., Muehlbauer G., editors. Genetics and Genomics of the Triticeae. New York: Springer; 2009. pp. 427–49. [Google Scholar]
- 17.Higgins J.A., Bailey P.C., Laurie D.A. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS ONE. 2010;5:e10065. doi: 10.1371/journal.pone.0010065. doi:10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brkljacic J., Grotewold E., Scholl R., et al. Brachypodium as a model for the grasses: today and the future. Plant Physiol. 2011;157:3–13. doi: 10.1104/pp.111.179531. doi:10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bevan M.W., Garvin D.F., Vogel J.P. Brachypodium distachyon genomics for sustainable food and fuel production. Curr. Opin. Biotechnol. 2010;21:211–7. doi: 10.1016/j.copbio.2010.03.006. doi:10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 20.The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–8. doi: 10.1038/nature08747. doi:10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S., Mohan A., Balyan H.S., Gupta P.K. Orthology between genomes of Brachypodium, wheat and rice , BMC Res. Notes. 2009;2:93. doi: 10.1186/1756-0500-2-93. doi:10.1186/1756-0500-2-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel J.P., Gu Y.Q., Twigg P., et al. EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor. Appl. Genet. 2006;113:186–95. doi: 10.1007/s00122-006-0285-3. doi:10.1007/s00122-006-0285-3. [DOI] [PubMed] [Google Scholar]
- 23.Mochida K., Uehara Y., Takahashi F., Yoshida T., Sakurai T., Shinozaki K. Large-scale analysis of full-length cDNAs of Brachypodium distachyon. Plant Animal Genome. 2012;XX:P0017. doi: 10.1371/journal.pone.0075265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duvick J., Fu A., Muppirala U., et al. PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res. 2008;36:D959–65. doi: 10.1093/nar/gkm1041. doi:10.1093/nar/gkm1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X., Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–77. doi: 10.1101/gr.9.9.868. doi:10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min X.J., Butler G., Storms R., Tsang A. Comparative assessment of DNA assemblers for assembling expressed sequence tags. 2009 Ohio Collaborative Conference on Bioinformatics; Cleveland, Ohio.: The IEEE Computer Society; 2009. pp. 79–82. [Google Scholar]
- 27.Goodstein D.M., Shu S., Howson R., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86. doi: 10.1093/nar/gkr944. doi:10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min X.J. ASFinder: a tool for genome-wide identification of alternatively spliced transcripts from EST-derived sequences. Int. J. Bioinform. Res. Appl. 2013 doi: 10.1504/IJBRA.2013.053603. in press. [DOI] [PubMed] [Google Scholar]
- 29.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–74. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foissac S., Sammeth M. ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007;35:W297–9. doi: 10.1093/nar/gkm311. doi:10.1093/nar/gkm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ner-Gaon H., Leviatan N., Rubin E., Fluhr R. Comparative cross-species alternative splicing in plants. Plant Physiol. 2007;144:1632–41. doi: 10.1104/pp.107.098640. doi:10.1104/pp.107.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchler-Bauer A., Anderson J.B., Chitsaz F., et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–10. doi: 10.1093/nar/gkn845. doi:10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy F.M., Wang N., Magee G.B., Williams W.P., Luthe D.S., Burgess S.C. AgBase: a functional genomics resource for agriculture. BMC Genomics. 2006;7:229. doi: 10.1186/1471-2164-7-229. doi:10.1186/1471-2164-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Min X.J., Butler G., Storms R., Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33:W677–80. doi: 10.1093/nar/gki394. doi:10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baek J.M., Han P., Iandolino A., Cook D.R. Characterization and comparison of intron structure and alternative splicing between Medicago truncatula, Populus trichocarpa, Arabidopsis and rice. Plant Mol. Biol. 2008;67:499–510. doi: 10.1007/s11103-008-9334-4. doi:10.1007/s11103-008-9334-4. [DOI] [PubMed] [Google Scholar]
- 36.Labadorf A., Link A., Rogers M.F., Thomas J., Reddy A.S.N., Ben-Hur A. Genome-wide analysis of alternative splicing in Chlamydomonas reinhardtii. BMC Genomics. 2010;111:14. doi: 10.1186/1471-2164-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koralewski T.E., Krutovsky K.V. Evolution of exon-intron structure and alternative splicing. PLoS ONE. 2011;6:e18055. doi: 10.1371/journal.pone.0018055. doi:10.1371/journal.pone.0018055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H., Ouyang S., Egan A., et al. Characterization of paralogous protein families in rice. BMC Plant Biol. 2008;8:18. doi: 10.1186/1471-2229-8-18. doi:10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Y., Chung J.D., Fu X., et al. Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:22020–5. doi: 10.1073/pnas.0906869106. doi:10.1073/pnas.0906869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P.G., Huang S.Z., Pin A.L., Adams K.L. Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis. Mol. Biol. Evol. 2010;27:1686–97. doi: 10.1093/molbev/msq054. doi:10.1093/molbev/msq054. [DOI] [PubMed] [Google Scholar]
- 41.English A.C., Patel K.S., Loraine A.E. Prevalence of alternative splicing choices in Arabidopsis thaliana. BMC Plant Biol. 2010;10:102. doi: 10.1186/1471-2229-10-102. doi:10.1186/1471-2229-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morello L., Breviario D. Plant spliceosomal introns: not only cut and paste. Curr. Genomics. 2008;9:227–38. doi: 10.2174/138920208784533629. doi:10.2174/138920208784533629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyna M., Simpson C.G., Syed N.H., et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–69. doi: 10.1093/nar/gkr932. doi:10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Severing E.I., van Dijk A.D.J., Morabito G., Busscher-Lange J., Immink R.G.H., van Ham R.C. Predicting the impact of alternative splicing on plant Mads domain protein function. PLoS ONE. 2012;7:e30524. doi: 10.1371/journal.pone.0030524. doi:10.1371/journal.pone.0030524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day I.S., Golovkin M., Palusa S.G., et al. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: insights into regulated splicing. Plant J. 2012;71:936–47. doi: 10.1111/j.1365-313X.2012.05042.x. doi:10.1111/j.1365-313X.2012.05042.x. [DOI] [PubMed] [Google Scholar]
- 46.Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–88. doi: 10.1105/tpc.010456. doi:10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo P.J., Kim M.J., Ryu J.Y., Jeong E.Y., Park C.M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2011;2:303. doi: 10.1038/ncomms1303. doi:10.1038/ncomms1303. [DOI] [PubMed] [Google Scholar]
- 48.Seo P.J., Park M.J., Lim M.H., et al. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–42. doi: 10.1105/tpc.112.098723. doi:10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan K., Liu P., Wu C.A., et al. Stress-Induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell. 2012;48:521–31. doi: 10.1016/j.molcel.2012.08.032. doi:10.1016/j.molcel.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 50.Kriechbaumer V., Wang P., Hawes C., Abell B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012;70:292–302. doi: 10.1111/j.1365-313X.2011.04866.x. doi:10.1111/j.1365-313X.2011.04866.x. [DOI] [PubMed] [Google Scholar]
- 51.Novák O., Hényková E., Sairanen I., Kowalczyk M., Pospíšil T., Ljung K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012;72:523–36. doi: 10.1111/j.1365-313X.2012.05085.x. doi:10.1111/j.1365-313X.2012.05085.x. [DOI] [PubMed] [Google Scholar]
- 52.Barbazuk W.B., Fu Y., McGinnis K.M. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–92. doi: 10.1101/gr.053678.106. doi:10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 53.Barbazuk W.B. A conserved alternative splicing event in plants reveals an ancient exonization of 5S rRNA that regulates TFIIIA. RNA Biol. 2010;7:397–402. doi: 10.4161/rna.7.4.12684. doi:10.4161/rna.7.4.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iida K., Go M. Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol. Biol. Evol. 2006;23:1085–94. doi: 10.1093/molbev/msj118. doi:10.1093/molbev/msj118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.