Abstract
The complex, tetraploid genome structure of peanut (Arachis hypogaea) has obstructed advances in genetics and genomics in the species. The aim of this study is to understand the genome structure of Arachis by developing a high-density integrated consensus map. Three recombinant inbred line populations derived from crosses between the A genome diploid species, Arachis duranensis and Arachis stenosperma; the B genome diploid species, Arachis ipaënsis and Arachis magna; and between the AB genome tetraploids, A. hypogaea and an artificial amphidiploid (A. ipaënsis × A. duranensis)4×, were used to construct genetic linkage maps: 10 linkage groups (LGs) of 544 cM with 597 loci for the A genome; 10 LGs of 461 cM with 798 loci for the B genome; and 20 LGs of 1442 cM with 1469 loci for the AB genome. The resultant maps plus 13 published maps were integrated into a consensus map covering 2651 cM with 3693 marker loci which was anchored to 20 consensus LGs corresponding to the A and B genomes. The comparative genomics with genome sequences of Cajanus cajan, Glycine max, Lotus japonicus, and Medicago truncatula revealed that the Arachis genome has segmented synteny relationship to the other legumes. The comparative maps in legumes, integrated tetraploid consensus maps, and genome-specific diploid maps will increase the genetic and genomic understanding of Arachis and should facilitate molecular breeding.
Keywords: Arachis spp., comparative genomics, genetic linkage map, integrated consensus map, legume genome
1. Introduction
Fabaceae, or Leguminosae, is composed of ∼700 genera and 20 000 species.1 The family is classified into three major subfamilies Mimosoideae, Caesalpinioideae, and Papilionoideae.2 Papilionoideae is divided into four clades Genistoids, Dalbergioids, Phaseoloids, and Galegoids. Most tropical and cool season legumes used as crops and studied as model plants for symbiosis of rhizobium are members of the Phaseoloids, e.g. genera Cajanus, Glycine, Phaseolus, and Vigna, or the Galegoids, e.g. genera Lotus, Medicago, Pisum, Trifolium, and Vicia.2 Therefore, legume genetics and genomics, including genome sequencing and comparative genomics, have greatly advanced in the Phaseoloids and Galegoids.3–9
The Dalbergioids, which include the genus, Arachis, are more basal in their divergence within the Papilionoideae than the Phaseoloids and Galegoids. The genus Arachis comprises 80 species (2n = 2x = 18, 2n = 2x = 20, and 2n = 4x = 40) possessing A, B, C, D, E, F, K, R, and T genomes.10–12 As regarding the A genome diploids consisting of 15 wild species, they are morphologically similar, cross compatible, produce fertile hybrids with near normal chromosome pairing, and have a common karyotype structure.13 The B-genome diploids also have similar morphological and chromosome feature, cross-compatibility, bivalent formation at meiosis in interspecific hybrids, and the same karyotype structure.12 Peanut (Arachis hypogaea), or groundnut, is an autogamous allotetraploid legume (2n = 4x = 40) harbouring homoeologous A and B genomes that are derived from two diploids, most likely Arachis duranensis (A genome) and Arachis ipaënsis (B genome).14,15 While peanut is an important source of food and oil, the genetics and genomics of this legume lag far behind those of the Phaseoloids and Galegoids due to a complex genome structure with challenging features, such as tetraploidy, low genetic diversity, and a relatively large genome size of 2.8 Gb.16
Molecular genetic studies of Arachis initially progressed by using diploid rather than tetraploid peanut due to the greater simplicity of diploids as genetic models.17 The first genetic linkage map in genus Arachis was developed based on an interspecific crossing between diploids Arachis stenosperma (A genome) and Arachis cardenasii (also A genome),17 followed by a cross between A. duranensis and A. stenosperma (both A genomes)18,19 and between A. ipaënsis and Arachis magna (both B genomes).20 However, because of the small number of available DNA markers at the time, the number of mapped marker loci was limited to between 117 and 369. Recently, intraspecific maps for A-genome diploid (A. duranensis) and B-genome diploid (Arachi batizocoi) have been developed with 1724 and 449 marker loci, respectively, including single nucleotide polymorphisms and expressed sequence tag-simple sequence repeats (EST-SSRs) markers.21,22
Meanwhile, an artificial amphidiploid derived from a three-way cross between A. batizocoi (B genome) and a hybrid between A. cardenasii (A genome) and A. diogoi (A genome) has been crossed with A. hypogaea to introduce genetic diversity from wild diploid species into the tetraploid-cultivated peanut. The BC1 progenies of the cross have contributed to the generation of a tetraploid genetic linkage map with 370 restriction-fragment length polymorphisms.23 In addition, another artificial amphidiploid line, derived from a hybrid between A. duranensis and A. ipaënsis, was used to establish a genetic linkage map made up of 298 SSR marker loci.24 In the latter map, the linkage groups (LGs) were firstly anchored to the A and B genomes by identifying the genome origins of the mapped loci in the LGs based on the corresponding sizes of the DNA amplified from the original diploids. In the tetraploid-cultivated peanut (A. hypogaea), the population-specific linkage maps comprise 318 marker loci. Due to low genetic diversity,25–33 this is the maximum number of marker loci for population-specific linkage maps, although >6000 SSR markers have been developed for mapping.34 By integrating the 11 linkage maps of the artificial amphidiploids and cultivated tetraploid peanuts, a reference consensus map consisting of 897 marker loci was constructed and annotated with consensus nomenclature for the LGs (a01–a10 and b01–b10).35
Concurrent with the progress of the above genetic studies in Arachis spp., a high-density linkage map of cultivated peanut covering 2166 cM with 1114 marker loci has also been reported.36 Because most of the markers on this map were published after the development of the other Arachis maps,36–38 the markers do not appear on them.17–20,23–32 In addition, due to an insufficient number of commonly mapped loci in the published diploid maps, the LGs have not been assigned to either A or B genome types, even though homoeologous LGs have been identified.36 Anchoring the genetic LGs to the A and B genomes will enable enrichment of marker density on the consensus and diploid maps.19,20,35 This would contribute to a better understanding of genome evolution within Arachis and among the Fabaceae. In addition, it would serve to increase the efficiency of introgression of desirable alleles from diploid wild species into the tetraploid-cultivated peanut through marker-assisted selection.
Comparative analysis of the A genome diploid Arachis species, e.g. A. duranensis and A. stenosperma, and model legumes has previously been performed. This comparative analysis revealed candidate synteny blocks between the genomes of Lotus japonicus, Medicago truncatula, and Glycine max.21,39 To date, no comparative analysis of cultivated peanut and other legume species has been reported. Comparison of the tetraploid Arachis genome, represented by the high-density consensus map of the genomes of legumes Cajanus cajan, G. max, L. japonicus, and M. truncatula, would provide further insights into the legume genomes.
To address the above issues, it was considered that high-density linkage maps of the tetraploid and diploid species, on which common makers were mapped, would be required. Therefore, we tried to map the large number of DNA markers reported for the two cultivated peanut maps published by Shirasawa et al.36 onto the previously reported linkage maps of the A and B genomes,19,20 along with that of an artificial amphidiploid derived from a hybrid of diploid species. The established linkage maps were integrated with 13 reported tetraploid Arachis maps to increase the number of mapped loci on the previously published consensus map.35 Subsequently, the maps were subjected to comparative analysis with four legume genomes, C. cajan, G. max, L. japonicus, and M. truncatula, to clarify features of the genome structure of the genus Arachis.
2. Materials and methods
2.1. Plant materials
In previous studies, two F2 mapping populations, derived from interspecific crosses between the two A genome diploid species, A. duranensis ‘K7988’ and A. stenosperma ‘V10309’, and between the two B genome diploid species, A. ipaënsis ‘K30076’ and A. magna ‘K30097’, were used to construct genetic linkage maps.19,20 In the present study, the F5 and F6 progenies of the diploid A and B genome mapping populations, respectively, were generated by single seed descent, and used as recombinant inbred mapping populations. The A and B genome mapping populations were named as AF5 (n = 89) and BF6 (n = 94), respectively. In addition, other recombinant inbred lines were used to construct AB genome tetraploid (n = 91: population TF6) maps. This population consisted of F6 lines derived from a cross between A. hypogaea ‘Runner IAC 886’ and an artificial amphidiploid (A. ipaënsis × A. duranensis)4×, which was developed by hybridizing A. ipaënsis ‘K30076’ and A. duranensis ‘V14167’. Genomic DNA from each line was extracted from young leaflets essentially as described by Grattapaglia and Sederoff.40
2.2. Polymorphism analysis with DNA markers
A total of 3902 DNA markers comprising 1894 genomic SSRs A. hypogaea genomic SSR (AHGS),36 1571 EST-SSRs A. hypogaea EST-SSR (AHS),37 and 437 transposon-based markers A. hypogaea transposable element (AhTE)36,38 were screened against the parental lines of the populations AF5, BF6, and TF6 using an ABI-3730xl fluorescent fragment analyser (Applied Biosystems). Polymorphic markers were analysed on the three mapping populations using 10% polyacrylamide gels for markers exhibiting ≥10-bp allele size differences, or the fluorescent fragment analyser for markers exhibiting <10-bp allele size differences between the parental lines. PCR, electrophoresis, and data scoring were performed as described previously.36–38
2.3. Construction of linkage maps and the integrated map
Linkage analysis of segregated genotypic data obtained in this study was performed together with linkage analysis of the previously investigated genotypes with mapped markers on the diploid maps19,20 using JoinMap® version 4.41 The marker loci were roughly classified using the JoinMap® Grouping Module with logarithm of odds (LOD) scores of 4.0–10.0. The JoinMap® Combine Groups for Map Integration Module was used to integrate the linkage maps developed in this study and the 13 previously published maps into a tetraploid map, i.e. the cultivated peanut map25–32,36 and an artificial amphidiploid map.24 Marker order and genetic distance were calculated using a regression mapping algorithm with the following parameters: Haldane's mapping function, recombination frequency ≤0.30, and LOD score ≥2.0. The graphical linkage maps were drawn using the MapChart program.42
2.4. Comparative analysis of the Arachis maps with other legume genomes
BLASTN43 was used to conduct similarity searches (using the nucleotide sequences from which the mapped DNA markers were designed) against each pseudomolecule of the genome sequences of C. cajan,8 G. max (Glyma1),6 L. japonicus (build 2.5),5 and M. truncatula (Mt3.5v4),7 with a threshold E-value of 1e-20. The graphical comparative maps were drawn using the Circos program.44
3. Results
3.1. Polymorphism screening of the parental lines of the mapping populations and construction of the linkage maps
Prior to polymorphism analysis between the parental lines, a total of 3902 markers, i.e. 1894 AHGS, 1571 AHS, and 437 AhTE markers, were pre-selected from the 6680 AHGS, 3187 AHS, and 1039 AhTE markers that have been published.36–38 The markers were pre-selected according to the following criteria: (i) AHGS markers were selected if they were expected to detect polymorphism among the cultivated peanut lines according to the results of in silico polymorphism analysis;36 (ii) AHS markers were selected if they showed polymorphisms between cultivated lines and wild species;37 (iii) AhTE markers were selected if they amplified single or double bands.36,38
3.1.1. The AF5 population
A total of 582 markers (14.9% = 582/3902), including 450 AHGS (23.8% = 450/1894), 109 AHS (6.9% = 109/1571), and 23 AhTE (5.3% = 23/437) markers, were selected as polymorphism candidates between the parental lines of the AF5 population, A. duranensis ‘K7988’ and A. stenosperma ‘V10309’. Out of the 582 polymorphic markers, 395 generated a total of 437 segregation loci, since several markers detected more than one locus. Specifically, 359, 31, 4, and 1 marker(s) detected 1, 2, 3, and 4 polymorphic loci, respectively, which were suffixed with ‘_a1’ and ‘_a2’, e.g. AHGS1647_a1 and AHGS1647_a2, to distinguish the loci each other. The 437 polymorphic loci, 284 codominant, and 153 dominant loci, were successfully mapped onto 10 LGs covering 544 cM, together with 160 previously mapped polymorphic loci from 158 markers (Fig. 1, Table 1, and Supplementary Table S1).19 The average marker density of the map was 0.9 cM, and segregation distortions were observed in 65% (390 loci) of the mapped loci (Table 1).
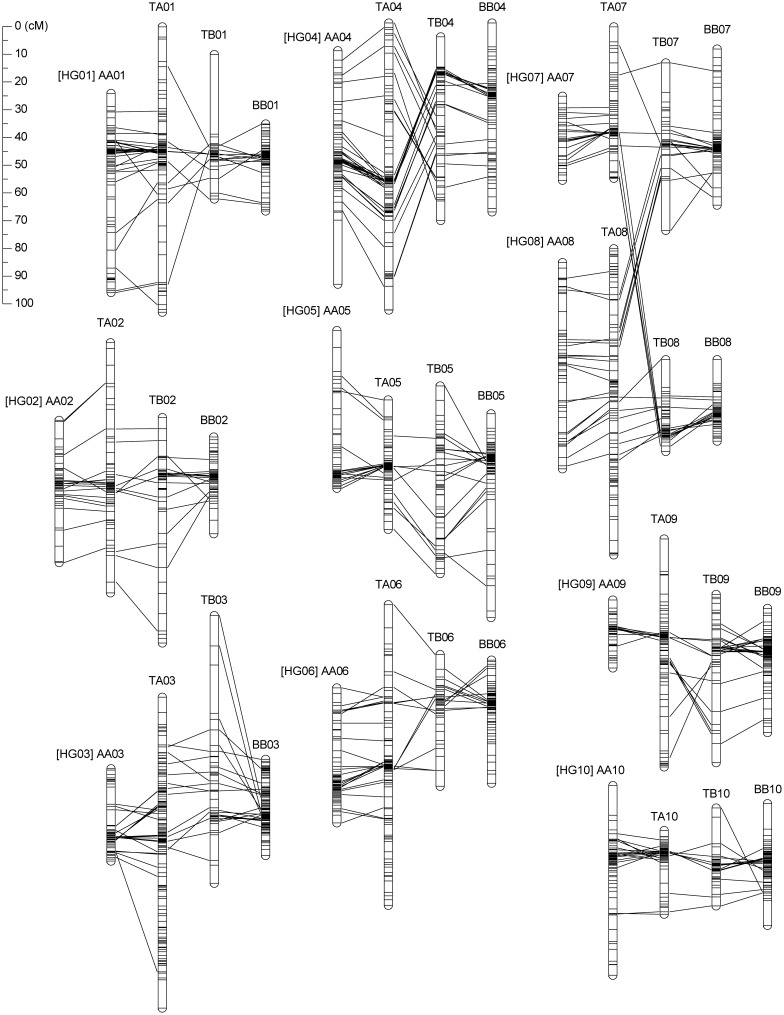
Figure 1.
Genetic linkage maps obtained for AF5, BF6, and TF6 populations. The linkage groups in the AF5, BF6, and TF6 maps are indicated by AA, BB, and TA or TB, respectively, and the homoeologous groups are prefixed with HGs. Homologous or homoeologous marker loci are connected by lines.
Table 1.
Descriptions of the genetic linkage maps AF5, BF6, and TF6
| LGs | AF5 map |
LGs | BF6 map |
LGs | TF6 map |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus no. | cM | cM/ locus | Percentages of loci with segregation distortiona | Locus no. | cM | cM/ locus | Percentages of loci with segregation distortiona | Locus no. | cM | cM/locus | Percentages of loci with segregation distortiona | |||
| AA01 | 84 | 71.8 | 0.9 | 69 | — | — | — | — | TA01 | 108 | 102.9 | 1.0 | 36 | |
| — | — | — | — | BB01 | 70 | 31.4 | 0.5 | 46 | TB01 | 47 | 52.1 | 1.1 | 17 | |
| AA02 | 55 | 51.3 | 1.0 | 20 | — | — | — | — | TA02 | 73 | 30.2 | 0.4 | 33 | |
| — | — | — | — | BB02 | 62 | 34.9 | 0.6 | 92 | TB02 | 47 | 35.3 | 0.8 | 36 | |
| AA03 | 63 | 33.3 | 0.5 | 78 | — | — | — | — | TA03 | 154 | 112.1 | 0.7 | 4 | |
| — | — | — | — | BB03 | 113 | 34.7 | 0.3 | 8 | TB03 | 50 | 96.5 | 2.0 | 26 | |
| AA04 | 75 | 84.2 | 1.1 | 97 | — | — | — | — | TA04 | 105 | 103.5 | 1.0 | 7 | |
| — | — | — | — | BB04 | 85 | 68.1 | 0.8 | 48 | TB04 | 62 | 66.2 | 1.1 | 23 | |
| AA05 | 56 | 56.9 | 1.0 | 82 | — | — | — | — | TA05 | 81 | 46.7 | 0.6 | 23 | |
| — | — | — | — | BB05 | 75 | 73.5 | 1.0 | 13 | TB05 | 60 | 67.7 | 1.1 | 8 | |
| AA06 | 63 | 48.7 | 0.8 | 65 | — | — | — | — | TA06 | 67 | 90.2 | 1.4 | 7 | |
| — | — | — | — | BB06 | 84 | 44.4 | 0.5 | 35 | TB06 | 49 | 81.3 | 1.7 | 55 | |
| AA07 | 31 | 30.3 | 1.0 | 16 | — | — | — | — | TA07 | 70 | 54.6 | 0.8 | 20 | |
| — | — | — | — | BB07 | 78 | 56.3 | 0.7 | 31 | TB07 | 40 | 60.4 | 1.5 | 20 | |
| AA08 | 54 | 74.4 | 1.4 | 28 | — | — | — | — | TA08 | 98 | 110.4 | 1.1 | 16 | |
| — | — | — | — | BB08 | 61 | 29.4 | 0.5 | 11 | TB08 | 50 | 33.1 | 0.7 | 14 | |
| AA09 | 48 | 24.5 | 0.5 | 100 | — | — | — | — | TA09 | 96 | 82.2 | 0.9 | 3 | |
| — | — | — | — | BB09 | 91 | 44.8 | 0.5 | 35 | TB09 | 56 | 60.7 | 1.1 | 18 | |
| AA10 | 68 | 68.4 | 1.0 | 65 | — | — | — | — | TA10 | 109 | 108.4 | 1.0 | 39 | |
| — | — | — | — | BB10 | 79 | 43.9 | 0.6 | 11 | TB10 | 47 | 47.5 | 1.0 | 2 | |
| Total | 597 | 543.8 | 0.9 | 65 | Total | 798 | 461.4 | 0.6 | 31 | Total | 1469 | 1441.8 | 1.0 | 19 |
aPercentages of loci that showed segregation distortion (P < 0.01).
3.1.2. The BF6 population
A total of 862 markers (22.1% = 862/3902), including 513 AHGS (27.1% = 513/1894), 279 AHS (17.8% = 279/1571), and 70 AhTE (16.0% = 70/437) markers, were selected as polymorphism candidates between the parental lines of the BF6 population, A. ipaënsis ‘K30076’ and A. magna ‘K30097’. Out of the 862 polymorphic markers, 624 generated a total of 680 segregation loci, since, as with the AF5 population, several of these markers detected more than one locus. Specifically, 579, 35, 9, and 1 marker(s) detected 1, 2, 3, and 4 polymorphic loci, respectively, which were suffixed with ‘_b1’ and ‘_b2’, e.g. AHGS1478_b1 and AHGS1478_b2, to distinguish the loci each other. The 680 polymorphic loci, 549 codominant, and 131 dominant, were successfully mapped onto 10 LGs covering 461 cM together with 118 previously mapped loci from 116 markers (Fig. 1, Table 1, and Supplementary Table S1).20 The average marker density of the map was 0.6 cM, and segregation distortions were observed in 31% (250 loci) of the mapped loci (Table 1).
3.1.3. The TF6 population
Together with the parental lines of TF6, the two original diploids of the artificial amphidiploid, A. ipaënsis ‘K30076’ and A. duranensis ‘V14167’, were also genotyped to identify the genome origin (A or B genome) of the polymorphic DNA fragments. A total of 1144 markers (29.3% = 1144/3902), including 664 AHGS (35.1% = 664/1894), 257 AHS (16.4% = 257/1571), and 223 AhTE (51.0% = 223/437) were selected as polymorphism candidates between the parental lines of the TF6 population, A. hypogaea ‘Runner IAC 886’ and an artificial amphidiploid (A. ipaënsis ‘K30076’ × A. duranensis ‘V14167’)4×. Out of the 1144 polymorphic markers, 1055 generated a total of 1261 segregation loci, since, as with the AF5 and BF6 populations, several of these markers detected more than one locus. Specifically, 879, 150, 22, and 4 markers detected 1, 2, 3, and 4 polymorphic loci, respectively, which were suffixed with ‘_t1’ and ‘_t2’, e.g. AHGS1991_t1 and AHGS1991_t2, to distinguish the loci each other. The 1261 polymorphic loci, 970 codominant, and 291 dominant, were successfully mapped onto 20 LGs covering 1442 cM together with 208 loci from 180 markers (Moretzsohn et al. unpublished data). The 20 LGs were classified into 10 pairs of A and B genomes, according to the DNA fragment size of mapped loci corresponding to ‘K30076’ or ‘V14167’ (Fig. 1, Table 1, and Supplementary Table S1). The average marker density of the map was 1.0 cM, and segregation distortions were observed in 19% (285 loci) of the mapped loci (Table 1).
In total, the number of non-redundant AHGS, AHS, and AhTE markers mapped on the three maps were 890, 446, and 211, respectively. Out of the total 1547 markers, 73 (67 AHGSs and six AHSs) and 381 (298 AHGSs, 59 AHSs, and 24 AhTEs) were mapped on three and two maps, respectively, while 1093 (525 AHGSs, 381 AHSs, and 187 AhTEs) were mapped on just one map; the AF5, BF6, or TF6 map (Supplementary Table S1). Comparing the three maps, 203 of the 597 mapped loci on the AF5 map, and 157 out of 798 loci on the BF6 map, were also located on the TF6 map (Supplementary Table S1).
The LGs were designated AA, BB, and TA or TB in the AF5, BF6 and TF6 maps, respectively. The nomenclature of the LGs of the diploid maps was tentatively determined according to the commonly mapped marker loci between the present and the previously constructed maps.19,20 Eight LGs of the AF5 map (AA01 and AA03–AA09) and all 10 of the LGs of the BF6 map (BB01–BB10) were assigned to the corresponding LGs on the previous maps (Supplementary Table S2). The two LGs of the AF5 map were assigned to ‘Group 02’ in the previous map,19 and they were tentatively named AA02 and AA10 (Table 1 and Supplementary Table S2). ‘Group 10’ in the previous map19 was disassembled on the AF5 map. On the other hand, in the TF6 map, TA01, TA03–TA09, and TB01–TB10 were assigned to the corresponding LGs of the AF5 and BF6 maps, and TA02 and TA10 were assigned to the AA02 and AA10 LGs, respectively (Table 1 and Supplementary Table S2). After integrating all of these data, 10 homoeologous groups (HGs) were identified on the TF6 map.
The marker order was almost completely conserved between the HGs except for HG04, HG07, and HG08 (Fig. 1). In HG04, the lower part of TA04 corresponded to the upper part of TB04 (Fig. 1). In HG07, the loci mapped onto TB07 corresponded to those of TA07 and the upper part of TA08. Moreover, corresponding loci on TB08 were observed on the lower parts of TA08 and TA07 (Fig. 1), indicating a translocation between the A and B genomes as previously suggested.20,22,24
3.2. Integration of the genetic linkage maps into a consensus map
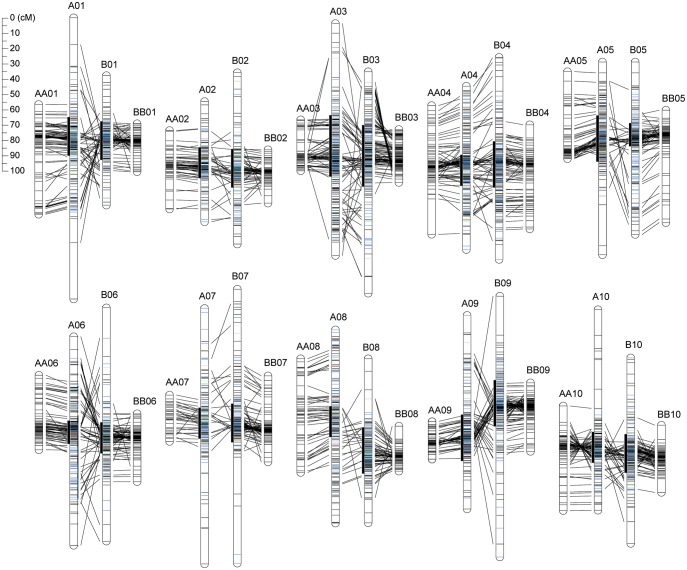
An integrated consensus map was constructed based on the segregation genotypes of 16 populations: three from this study and 13 from previous studies (Supplementary Table S2).19,20,24–32,36 The integrated consensus map was 2651 cM in total length and comprised 20 LGs, on which 3693 loci, including 1564 AHGS, 569 AHS, 450 AhTE, and 1110 other loci, were mapped (Table 2, Fig. 2, and Supplementary Table S3), out of which markers mapped on mote than two loci were suffixed with ‘_c1’ and ‘_c2’, e.g. AHGS1403_c1 and AHGS1403_c2, to distinguish each other. The average marker density of this map was 0.7 cM (Table 2). On the consensus map, 391 marker loci were commonly mapped onto pairs of LGs in each HG, and the order of the markers was roughly conserved (Fig. 2).
Table 2.
Descriptions of the integrated consensus map and integrated LGs of the population-specific maps
| LGs | Locus no. | cM | cM/locus | Length of the locus clustera | Locus no. in the marker cluster |
|---|---|---|---|---|---|
| A01 | 238 | 183.8 | 0.8 | 25 | 158 |
| B01 | 168 | 84.7 | 0.5 | 25 | 129 |
| A02 | 126 | 78.6 | 0.6 | 20 | 86 |
| B02 | 154 | 112.0 | 0.7 | 25 | 104 |
| A03 | 272 | 151.6 | 0.6 | 40 | 175 |
| B03 | 234 | 144.8 | 0.6 | 40 | 167 |
| A04 | 213 | 106.8 | 0.5 | 20 | 106 |
| B04 | 208 | 132.1 | 0.6 | 30 | 122 |
| A05 | 189 | 126.0 | 0.7 | 30 | 135 |
| B05 | 169 | 112.7 | 0.7 | 15 | 77 |
| A06 | 187 | 136.4 | 0.7 | 15 | 85 |
| B06 | 176 | 152.5 | 0.9 | 20 | 109 |
| A07 | 145 | 167.0 | 1.2 | 20 | 76 |
| B07 | 162 | 179.1 | 1.1 | 25 | 105 |
| A08 | 167 | 126.3 | 0.8 | 20 | 53 |
| B08 | 176 | 107.3 | 0.6 | 30 | 138 |
| A09 | 194 | 126.5 | 0.7 | 30 | 141 |
| B09 | 179 | 170.5 | 1.0 | 30 | 121 |
| A10 | 172 | 131.1 | 0.8 | 20 | 117 |
| B10 | 164 | 121.3 | 0.7 | 25 | 115 |
| Total | 3693 | 2651.1 | 0.7 | 505 | 2319 |
aRegions in which ≥10 loci were mapped in 5 cM windows.
Figure 2.
Comparison of the integrated consensus tetraploid Arachis map with diploid maps. The abbreviations AA and BB indicate LGs on the AF5 and BF6 maps, respectively, and the prefixes A and B show LGs on the integrated consensus map. Horizontal lines on the LGs indicate the positions of the mapped loci; the loci derived from one, two, three, four, five, and more than five genetic linkage maps are shown in black, blue, light blue, green, yellow, and red. Vertical bars on the left side of the LGs indicate the locus clusters. Homologous and homoeologous loci are connected by black lines.
In the present study, locus clusters were defined as regions in which ≥10 loci were mapped in 5 cM windows. They were observed in all of the LGs (Fig. 2, Table 2, and Supplementary Table S3). The total length of the locus clusters was 505 cM (101 clusters), varying from 15 cM (three clusters) in A06 and B05 to 40 cM (eight clusters) in A03 and B03. A total of 2319 loci, representing 62.8% of the mapped loci, were located in 101 clusters, which varied in loci content from 53 (A08) to 175 (A03) loci.
3.3. Comparative mapping between the Arachis spp. and the four comparison legumes, C. cajan, G. max, L. japonicus, and M. truncatula
Corresponding sequences were available for 3473 of the 3693 mapped loci (Supplementary Table S3). Of these 3473 sequences, 869 showed significant similarity to one of the four legume genome sequences (C. cajan, G. max, L. japonicus, and M. truncatula). For each legume species, 515, 781, 436, and 419 sequences showed similarities to sequences in the C. cajan, G. max, L. japonicus, and M. truncatula genomes, respectively, and 170 sequences showed similarity to sequences in all four legume genomes.
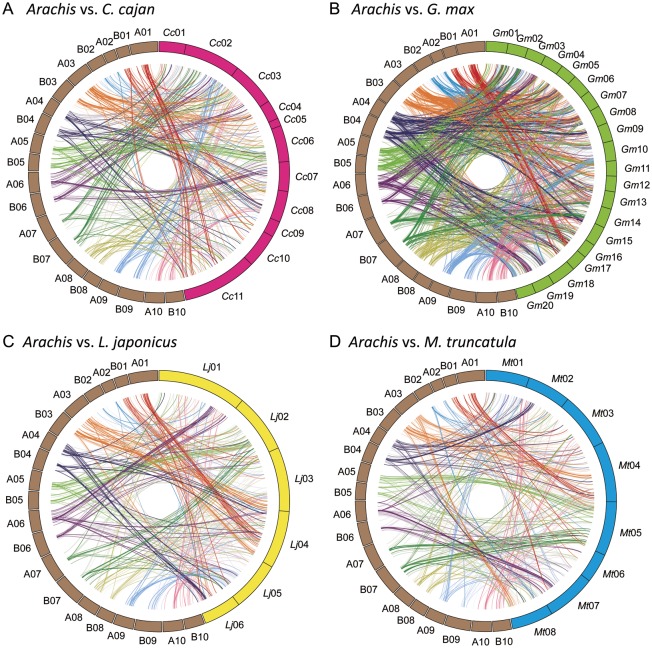
Segmental syntenic regions were observed between the Arachis HGs and the genomes of the four comparison legumes (Fig. 3, Table 3, and Supplementary Table S3). Most of the LG pairs in each HG showed similar synteny relationships to those in the four comparison legumes. Between the four comparison legume species, the genome of G. max showed the densest synteny of LGs, corresponding to those of Arachis. In addition, most of the LGs in Arachis were identified as shared syntenies with LGs on each of two chromosomes of G. max. This might reflect the palaeopolyploid nature of the genomes of these species. Comparing Arachis with the other three legume genomes, clearly syntenic segments were observed as follows: Arachis HG01 shared synteny with C. cajan chromosome 11 (Cc11) and M. truncatula chromosome 4 (Mt04); Arachis HG04 shared synteny with L. japonicus chromosome 4 (Lj04); Arachis HG05 shared synteny with Cc06, Lj02, and Mt05; Arachis HG06 shared synteny with Lj01 and Mt07; Arachis HG07 shared synteny with Cc02, Lj02, and Mt05; Arachis HG08 shared synteny with Cc03; Arachis HG09 shared synteny with Cc02, Lj05, and Mt01; and Arachis HG10 shared synteny with Lj01. On Arachis HG03, segmental blocks were identified that showed shared synteny between the comparison legume genomes, while no obvious synteny was observed between Arachis HG02 and the genomes of C. cajan, L. japonicus, and M. truncatula.
Figure 3.
Comparative maps of Arachis and four other legumes, C. cajan, G. max, L. japonicus, and M. truncatula. Circled bars in brown, magenta, green, yellow, and blue indicate the genomes of Arachis, C. cajan (A), G. max (B), L. japonicus (C), and M. truncatula (D), respectively. Homologous loci are connected by lines.
Table 3.
Similarity of other legume chromosomes with that of Arachis homoeologous groups
| Arachis | C. cajan | G. max | L. japonicus | M. truncatula |
|---|---|---|---|---|
| HG01a | 11 | 5, 7, 13, 17 | 4, 6 | 4 |
| HG02 | — | 3, 7 | — | — |
| HG03 | 4, 5, 8, 9, 10 | 4, 5, 6, 8, 9, 11, 12, 18 | 3, 4 | 4, 8 |
| HG04 | 7, 8 | 3, 7, 8, 9, 12, 13, 15, 18, 20 | 6 | 2, 7 |
| HG05 | 6 | 1, 9, 11, 16 | 2 | 5 |
| HG06 | 2, 3, 7 | 2, 3, 16, 19 | 1 | 7 |
| HG07 | 2 | 2, 6, 13 | 2 | 5 |
| HG08 | 3 | 4, 6, 8, 12, 13 | 1, 3 | 2, 3 |
| HG09 | 2 | 7, 10, 20 | 5 | 1 |
| HG10 | 1, 8 | 10, 15, 19 | 1 | 2, 3 |
aHG indicates homologous group.
4. Discussion
4.1. Polymorphism potential of the DNA markers mapped on the genetic linkage map
The polymorphism potential of DNA markers in Arachis is usually evaluated on the basis of the polymorphism information content (PIC) value.34 While the PIC values of the AHS were estimated from genotyping data from 16 Arachis lines,37 those of the AHGS and AhTE have not been investigated thoroughly.36,38 In the present study and in previous studies,36 968 (416 AHGS, 395 AHS, and 157 AhTE), 666 (392 AHGS, 65 AHS, and 209 AhTE), 245 (197 AHGS, 10 AHS, and 38 AhTE), 73 (72 AHGS and 1 AhTE), and 3 (AHGS only) markers were mapped on single, double, triple, quadruple, and quintuple maps, respectively. Of these, the PIC values of the 395, 65, and 10 AHS markers were estimated to be, on average, 0.23, 0.28, and 0.36, respectively, from the results of Koilkonda et al.37 A positive correlation was observed between the PIC values and the number of maps on which the AHS was mapped. Therefore, if this estimation can be generalized, the AHGS and AhTE markers mapped on multiple maps can be considered as potential sources for obtaining polymorphic markers in other mapping populations.
In the screening of polymorphic markers between the parental lines of the mapping populations, 582, 862, and 1144 markers were selected as polymorphism candidates, but 187, 238, and 89 were excluded from the mapping analysis. The excluded markers might amplify DNAs from not the target locus but non-allelic repetitive and putative paralogous sequences in the Arachis genome,38,45,46 which are not suitable for segregation and linkage analysis.
4.2. Genetic linkage maps for three mapping populations
Comparison of the AF5, BF6, and TF6 maps revealed that the marker order was almost perfectly conserved between the HGs, with the exception of HG04, HG07, and HG08 (Fig. 1 and Supplementary Fig. S1). This result indicated that the A and B genomes of Arachis evolved from a common ancestor species. Exceptions to this shared marker order were found within HG04, and between HG07 and HG08. The former was observed for the first time in this study, while the latter confirms previous observations.20,24 These findings suggest that the chromosome translocations might have occurred at, or after, the divergence of the A and B genomes in Arachis.
The AF5 and BF6 linkage maps have been improved by subsequent mapping of the newly developed markers after they were originally published.19,20 However, despite the increase in the number of mapped loci from 170 to 597 in the AF5 map and from 149 to 798 in the BF6 map, the total map length decreased from 1231 to 544 cM in the AF5 map and from 1294 to 461 cM in the BF6 map. In the maps generated in the present study, the marker loci that mapped to the ends of LGs in previous studies19,20 were excluded, and the intervals between the markers were shorter than those in previous maps. Mapping telomere-based makers will clarify whether the maps from this study have been saturated or not.
In the linkage map for the TF6 population, the total length of the map was 1442 cM, which was shorter than that of the maps reported for Arachis tetraploids (2210 cM with 370 loci;23 2166 cM with 1114 loci;36 1844 cM with 298 loci;24 1785 cM with 191 loci29). Nagy et al.21 reported that the length of the genetic linkage map in A. duranensis ranged from 1081.1 to 2056.5 cM (depending on the mapping program used) when 1054 segregation loci were used for the linkage analysis. Therefore, the different programs, algorithms, and functions used for mapping may explain the differences in map length reported by the present and previous studies. Our preliminary analysis, in which MapMaker program47 gave longer map lengths than the JoinMap, also supported these explanations. Alternatively, as Sim et al.48 suggested (based on linkage analysis in tomato), different ratios between the number of markers and population size in each population might have resulted in the length differences.
Segregation distortions were observed in the three maps, the AF5, BF6, and TF6, and the ratios were different among the populations and LGs (Table 1). Similar observations were found in our previous studies on tomato on which high and low ratios of segregation distortion were observed in inter- and intraspecific maps, respectively.49,50 Segregation distortion is generally caused by the chromosomal structural differences or the presence of transmission ration distorter factors on some chromosome.51 Our findings will contribute to identify factors for the segregation distortion.
4.3. Characterization of the cultivated peanut genome using the Arachis consensus map
The consensus map was constructed using segregation data from 16 populations, in which five species, A. duranensis (A), A. stenosperma (A), A. ipaënsis (B), A. magna (B), and A. hypogaea (AB), were involved (Supplementary Table S2). A total of 889 loci derived from 421 markers were commonly mapped onto all of the paired LGs as HGs; however, the locus orders were not always conserved. The regions in which mapped loci showed non-corresponding orders between the paired LGs in each HG were considered as candidate regions having A or B genome-specific structures. Most of the locus orders were conserved between AA (diploid) and A (tetraploid) LGs, or between BB (diploid) and B (tetraploid) LGs (Fig. 2). However, several regions showed evidence of rearrangement, such as between BB03 and B03, BB09 and B09, and AA10 and A10. Arachis duranensis and A. ipaënsis are considered to be the most probable ancestors of A. hypogaea, because of similar karyotypes of A. duranensis and A. ipaënsis to that of A. hypogaea.12–15 By contrast, even though the structures of A genome of different species are well conserved, the A. stenosperma genome is distinguishable from that of A. duranensis on the basis of the variability observed in the heterochromatin and 18S–26S rRNA loci.13 It is considered the possibility that such differences in the genome structure of the parental lines of the mapping populations might disrupt the marker order between diploid and tetraploid consensus maps.
The average marker density of the consensus map was 0.7 cM/locus (Table 2). Because the genome size of A. hypogaea is estimated to be ∼2.8 Gb,16 the DNA markers were located at <760 kb intervals on average. In other words, a map-based cloning strategy has become a realistic approach for molecular genetics in Arachis spp., because a candidate region for the location of a target gene can, on average, be identified to within 760 kb. Furthermore, this high-density map should aid in genome sequencing analysis in Arachis spp., because the locus order can work as a reference for the orientation of sequence contigs and scaffolds. Moreover, the marker clusters were observed in all of the LGs (Fig. 2 and Supplementary Table S3). Because chromosomal crossover, and the resulting genetic recombination, are severely suppressed in heterochromatin,6,49 the clustered regions were speculated to be located in heterochromatin, which makes up 10–20% of the Arachis chromosomes in somatic metaphase.13,15 In the case of soybean, broad pericentromeric regions show markedly diminished recombination (4.2 Mb/cM), while the euchromatic regions at the chromosome ends maintain consistent levels of recombination (193 kb/cM).6 This pattern is also observed in tomato, with 6042 and 172 kb/cM in heterochromatic and euchromatic regions, respectively.49 As with soybean and tomato, it was predicted that the physical marker-to-marker distance intervals in the peanut genome were different between heterochromatic and euchromatic regions.
4.4. Comparative genomics of Arachis spp. with C. cajan, G. max, L. japonicus, and M. truncatula
All of the Arachis HGs showed similarities to the genomes of the four legumes (Fig. 3, Table 3, and Supplementary Table S2). The genetic composition of Arachis corresponded to that of the C. cajan, L. japonicus, and M. truncatula genomes on a one-on-one basis, but was doubled when compared with the G. max genome. These observations support the estimation that a whole-genome duplication occurred in legumes ∼58 million years ago (mya), at the time of the Papilionoid origin,52 whereas the soybean-specific duplication is estimated to have occurred <13 mya.6 These insights are based on a comparison of the A genome species between Arachis and L. japonicus and M. truncatula.39
At the chromosome level, each chromosome of Arachis, which belongs to the Dalbergioids, might have evolved differently and show different rearrangements from those in the ancestral chromosomes of the legumes after isolation from the other subfamilies, the Phaseoloids and Galegoids, ∼55 mya.52 Of the 10 Arachis HGs, HG01, 05, 07 and 09 showed microsyntenies with the chromosomes of C. cajan, L. japonicus, and M. truncatula. This suggested the possibility that the major chromosome rearrangements did not occur after the time of divergence of subfamilies in the Papilionoideae. By contrast, HG02 showed the most disparate syntenies with the genomes of C. cajan, G. max, L. japonicus, and M. truncatula (Table 3), which suggested frequent chromosome rearrangement.
5. Conclusion
In this study, we developed three linkage maps for Arachis spp., and anchored the LGs to the A and B genomes. Integration of the tetraploid maps developed in this study with the 13 previously published studies generated a high-density consensus map of tetraploid Arachis. The developed maps identified structural features within the Arachis genome using comparative genomic analysis, and also identified differences between the Arachis genome and that of other legumes, C. cajan, G. max, L. japonicus, and M. truncatula. The results obtained in this study will bridge the gaps in our knowledge regarding the genomes of Arachis and other legumes, and will further the genetic/genomic study and molecular breeding of Arachis.
5.1. Availability
Information for the genetic linkage maps and the integrated consensus map is available at http://marker.kazusa.or.jp.
Funding
This work was supported by the Kazusa DNA Research Institute Foundation.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Acknowledgements
We are grateful to S. Sasamoto, S. Nakayama, A. Watanabe, T. Fujishiro, M. Kato, Y. Kishida, M. Kohara, C. Minami, K. Nanri, C. Takahashi, and H. Tsuruoka in the Kazusa DNA Research Institute for their technical assistance.
Footnotes
Edited by Kazuhiro Sato
References
- 1.Doyle J.J., Luckow M.A. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003;131:900–10. doi: 10.1104/pp.102.018150. doi:10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wojciechowski M.F., Lavin M., Sanderson M.J. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 2004;91:1846–62. doi: 10.3732/ajb.91.11.1846. doi:10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 3.Choi H.K., Mun J.H., Kim D.J., et al. Estimating genome conservation between crop and model legume species. Proc. Natl. Acad. Sci. USA. 2004;101:15289–94. doi: 10.1073/pnas.0402251101. doi:10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isobe S.N., Hisano H., Sato S., et al. Comparative genetic mapping and discovery of linkage disequilibrium across linkage groups in white clover (Trifolium repens L.) G3. 2012;2:607–17. doi: 10.1534/g3.112.002600. doi:10.1534/g3.112.002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. doi:10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 7.Young N.D., Debellé F., Oldroyd G.E., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–4. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshney R.K., Chen W., Li Y., et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012;30:83–9. doi: 10.1038/nbt.2022. doi:10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H., Choi H.K., Cook D.R., Shoemaker R.C. Bridging model and crop legumes through comparative genomics. Plant Physiol. 2005;137:1189–96. doi: 10.1104/pp.104.058891. doi:10.1104/pp.104.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krapovickas A., Gregory W. Taxonomia del genero Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- 11.Stalker H.T. A new species in section Arachis of peanuts with a D genome. Am. J. Bot. 1991;78:630–7. doi:10.2307/2445084. [Google Scholar]
- 12.Robledo G., Seijo G. Species relationships among the wild B genome of Arachis species (section Arachis) based on FISH mapping of rDNA loci and heterochromatin detection: a new proposal for genome arrangement. Theor. Appl. Genet. 2010;121:1033–46. doi: 10.1007/s00122-010-1369-7. doi:10.1007/s00122-010-1369-7. [DOI] [PubMed] [Google Scholar]
- 13.Robledo G., Lavia G.I., Seijo G. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theor. Appl. Genet. 2009;118:1295–307. doi: 10.1007/s00122-009-0981-x. doi:10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- 14.Seijo G., Lavia G.I., Fernandez A., et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 2007;94:1963–71. doi: 10.3732/ajb.94.12.1963. doi:10.3732/ajb.94.12.1963. [DOI] [PubMed] [Google Scholar]
- 15.Seijo J.G., Lavia G.I., Fernandez A., Krapovickas A., Ducasse D., Moscone E.A. Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaënsis are the wild diploid progenitors of A. hypogaea (Leguminosae) Am. J. Bot. 2004;91:1294–303. doi: 10.3732/ajb.91.9.1294. doi:10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- 16.Temsch E.M., Greilhuber J. Genome size variation in Arachis hypogaea and A. monticola re-evaluated. Genome. 2000;43:449–51. [PubMed] [Google Scholar]
- 17.Halward T., Stalker H.T., Kochert G. Development of an RFLP linkaeg map in diploid peanut species. Theor. Appl. Genet. 1993;87:379–84. doi: 10.1007/BF01184927. doi:10.1007/BF01184927. [DOI] [PubMed] [Google Scholar]
- 18.Leal-Bertioli S.C., Jose A.C., Alves-Freitas D.M., et al. Identification of candidate genome regions controlling disease resistance in Arachis. BMC Plant Biol. 2009;9:112. doi: 10.1186/1471-2229-9-112. doi:10.1186/1471-2229-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretzsohn M.C., Leoi L., Proite K., et al. A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae) Theor. Appl. Genet. 2005;111:1060–71. doi: 10.1007/s00122-005-0028-x. doi:10.1007/s00122-005-0028-x. [DOI] [PubMed] [Google Scholar]
- 20.Moretzsohn M.C., Barbosa A.V., Alves-Freitas D.M., et al. A linkage map for the B-genome of Arachis (Fabaceae) and its synteny to the A-genome. BMC Plant Biol. 2009;9:40. doi: 10.1186/1471-2229-9-40. doi:10.1186/1471-2229-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy E.D., Guo Y., Tang S., et al. A high-density genetic map of Arachis duranensis, a diploid ancestor of cultivated peanut. BMC Genomics. 2012;13:469. doi: 10.1186/1471-2164-13-469. doi:10.1186/1471-2164-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y., Khanal S., Tang S., et al. Comparative mapping in intraspecific populations uncovers a high degree of macrosynteny between A- and B-genome diploid species of peanut. BMC Genomics. 2012;13:608. doi: 10.1186/1471-2164-13-608. doi:10.1186/1471-2164-13-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burow M.D., Simpson C.E., Starr J.L., Paterson A.H. Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics. 2001;159:823–37. doi: 10.1093/genetics/159.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foncéka D., Hodo-Abalo T., Rivallan R., et al. Genetic mapping of wild introgressions into cultivated peanut: a way toward enlarging the genetic basis of a recent allotetraploid. BMC Plant Biol. 2009;9:103. doi: 10.1186/1471-2229-9-103. doi:10.1186/1471-2229-9-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautami B., Pandey M.K., Vadez V., et al. Quantitative trait locus analysis and construction of consensus genetic map for drought tolerance traits based on three recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.) Mol. Breed. 2012;30:773–88. doi: 10.1007/s11032-011-9660-0. doi:10.1007/s11032-011-9661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Y., Chen X., Liang X., et al. A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol. 2010;10:17. doi: 10.1186/1471-2229-10-17. doi:10.1186/1471-2229-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khedikar Y.P., Gowda M.V., Sarvamangala C., Patgar K.V., Upadhyaya H.D., Varshney R.K. A QTL study on late leaf spot and rust revealed one major QTL for molecular breeding for rust resistance in groundnut (Arachis hypogaea L.) Theor. Appl. Genet. 2010;121:971–84. doi: 10.1007/s00122-010-1366-x. doi:10.1007/s00122-010-1366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H., Feng S., Chen C., et al. An integrated genetic linkage map of cultivated peanut (Arachis hypogaea L.) constructed from two RIL populations. Theor. Appl. Genet. 2012;124:653–64. doi: 10.1007/s00122-011-1737-y. doi:10.1007/s00122-011-1737-y. [DOI] [PubMed] [Google Scholar]
- 29.Ravi K., Vadez V., Isobe S., et al. Identification of several small main-effect QTLs and a large number of epistatic QTLs for drought tolerance related traits in groundnut (Arachis hypogaea L.) Theor. Appl. Genet. 2011;122:1119–32. doi: 10.1007/s00122-010-1517-0. doi:10.1007/s00122-010-1517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarvamangala C., Gowda M.V.C., Varshney R.K. Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.) Field Crops Res. 2011;122:49–59. doi:10.1016/j.fcr.2011.02.010. [Google Scholar]
- 31.Sujay V., Gowda M.V., Pandey M.K., et al. Quantitative trait locus analysis and construction of consensus genetic map for foliar disease resistance based on two recombinant inbred line populations in cultivated groundnut (Arachis hypogaea L.) Mol. Breed. 2012;30:773–88. doi: 10.1007/s11032-011-9661-z. doi:10.1007/s11032-011-9661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshney R.K., Bertioli D.J., Moretzsohn M.C., et al. The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.) Theor. Appl. Genet. 2009;118:729–39. doi: 10.1007/s00122-008-0933-x. doi:10.1007/s00122-008-0933-x. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., Penmetsa R.V., Yuan M., et al. Development and characterization of BAC-end sequence derived SSRs, and their incorporation into a new higher density genetic map for cultivated peanut (Arachis hypogaea L.) BMC Plant Biol. 2012;12:10. doi: 10.1186/1471-2229-12-10. doi:10.1186/1471-2229-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey M.K., Monyo E., Ozias-Akins P., et al. Advances in Arachis genomics for peanut improvement. Biotechnol. Adv. 2012;30:639–51. doi: 10.1016/j.biotechadv.2011.11.001. doi:10.1016/j.biotechadv.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Gautami B., Fonceka D., Pandey M.K., et al. An international reference consensus genetic map with 897 marker loci based on 11 mapping populations for tetraploid groundnut (Arachis hypogaea L.) PLoS One. 2012;7:e41213. doi: 10.1371/journal.pone.0041213. doi:10.1371/journal.pone.0041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirasawa K., Koilkonda P., Aoki K., et al. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol. 2012;12:80. doi: 10.1186/1471-2229-12-80. doi:10.1186/1471-2229-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koilkonda P., Sato S., Tabata S., et al. Large-scale development of expressed sequence tag-derived simple sequence repeat markers and diversity analysis in Arachis spp. Mol. Breed. 2012;30:125–38. doi: 10.1007/s11032-011-9604-8. doi:10.1007/s11032-011-9604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirasawa K., Hirakawa H., Tabata S., et al. Characterization of active miniature inverted-repeat transposable elements in the peanut genome. Theor. Appl. Genet. 2012;124:1429–38. doi: 10.1007/s00122-012-1798-6. doi:10.1007/s00122-012-1798-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertioli D.J., Moretzsohn M.C., Madsen L.H., et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10:45. doi: 10.1186/1471-2164-10-45. doi:10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grattapaglia D., Sederoff R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137:1121–37. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Ooijen J.W. JoinMap®4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen, The Netherlands: Kyazma. BV; 2006. [Google Scholar]
- 42.Voorrips R.E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–8. doi: 10.1093/jhered/93.1.77. doi:10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S.F., Madden T.L., Schaffer A.A., et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krzywinski M., Schein J., Birol I., et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. doi:10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielen S., Campos-Fonseca F., Leal-Bertioli S., et al. FIDEL-a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut. Chromosome Res. 2010;18:227–46. doi: 10.1007/s10577-009-9109-z. doi:10.1007/s10577-009-9109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielen S., Vidigal B.S., Leal-Bertioli S.C., et al. Matita, a new retroelement from peanut: characterization and evolutionary context in the light of the Arachis A-B genome divergence. Mol. Genet. Genomics. 2012;287:21–38. doi: 10.1007/s00438-011-0656-6. doi:10.1007/s00438-011-0656-6. [DOI] [PubMed] [Google Scholar]
- 47.Lander E.S., Green P., Abrahamson J., Barlow A., Daly M.J., Lincoln S.E., Newberg L.A. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–81. doi: 10.1016/0888-7543(87)90010-3. doi:10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 48.Sim S.C., Durstewitz G., Plieske J., et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE. 2012;7:e40563. doi: 10.1371/journal.pone.0040563. doi:10.1371/journal.pone.0040563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirasawa K., Asamizu E., Fukuoka H., et al. An interspecific linkage map of SSR and intronic polymorphism markers in tomato. Theor. Appl. Genet. 2010;121:731–9. doi: 10.1007/s00122-010-1344-3. doi:10.1007/s00122-010-1344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirasawa K., Isobe S., Hirakawa H., et al. SNP discovery and linkage map construction in cultivated tomato. DNA Res. 2010;17:381–91. doi: 10.1093/dnares/dsq024. doi:10.1093/dnares/dsq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rieseberg L.H., Linder C.L., Seiler G.J. Chromosomal and genic barriers to introgression in Helianthus. Genetics. 1995;141:1163–71. doi: 10.1093/genetics/141.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lavin M., Herendeen P.S., Wojciechowski M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005;54:575. doi: 10.1080/10635150590947131. doi:10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.