The kinetochore localization of MPS1 is necessary for mitotic checkpoint activity and requires the microtubule-binding domain of HEC1 and the Aurora B–dependent regulation of the TPR domain.
Abstract
The mitotic checkpoint ensures correct chromosome segregation by delaying cell cycle progression until all kinetochores have attached to the mitotic spindle. In this paper, we show that the mitotic checkpoint kinase MPS1 contains an N-terminal localization module, organized in an N-terminal extension (NTE) and a tetratricopeptide repeat (TPR) domain, for which we have determined the crystal structure. Although the module was necessary for kinetochore localization of MPS1 and essential for the mitotic checkpoint, the predominant kinetochore binding activity resided within the NTE. MPS1 localization further required HEC1 and Aurora B activity. We show that MPS1 localization to kinetochores depended on the calponin homology domain of HEC1 but not on Aurora B–dependent phosphorylation of the HEC1 tail. Rather, the TPR domain was the critical mediator of Aurora B control over MPS1 localization, as its deletion rendered MPS1 localization insensitive to Aurora B inhibition. These data are consistent with a model in which Aurora B activity relieves a TPR-dependent inhibitory constraint on MPS1 localization.
Introduction
Faithful chromosome segregation is essential to maintain genomic stability. A mitotic checkpoint has evolved to prevent the onset of anaphase until all chromosomes have attached to spindle microtubules, a prerequisite for error-free chromosome segregation (Vleugel et al., 2012). Components of the mitotic checkpoint, such as MAD1 and MAD2, are recruited specifically to kinetochores devoid of microtubules, whereas microtubule attachments to kinetochores cause removal of these components and local silencing of the checkpoint signal (Kops and Shah, 2012).
Unattached kinetochores elicit a checkpoint response by recruiting various checkpoint proteins, including MAD1/MAD2 heterotetramers. This subsequently culminates in the production of an anaphase inhibitor consisting of BUBR1, BUB3, and MAD2 (Hardwick et al., 2000; Sudakin et al., 2001; Chao et al., 2012). This inhibitor, known as the mitotic checkpoint complex, prevents premature activation of the anaphase-promoting complex/cyclosome–CDC20 complex that triggers anaphase by licensing Cyclin B and Securin for proteasomal degradation (Musacchio and Salmon, 2007). Unattached kinetochores also recruit and activate the mitotic kinase MPS1 that simultaneously promotes efficient activation of the error correction and mitotic checkpoint machineries (Lan and Cleveland, 2010). MPS1 is required for kinetochore localization of at least MAD1, MAD2, CDC20, and BUB1 (Lan and Cleveland, 2010). Although not required in vitro (Vink et al., 2006), MPS1 is needed for MAD2 dimerization in cells (Hewitt et al., 2010). Once activated, MPS1 also promotes its own dissociation from kinetochores, a process that permits removal of the MAD1–MAD2 complexes and checkpoint silencing when kinetochores have properly bioriented (Jelluma et al., 2010). Consequently, loss of MPS1 activity results in failure to delay mitosis when unattached kinetochores persist, in a dramatic shortening of mitosis and in anaphases with severe chromosome missegregations that can culminate in chromosomal translocations (Jelluma et al., 2008b; Tighe et al., 2008; Maciejowski et al., 2010; Sliedrecht et al., 2010; Janssen et al., 2011).
Localization of MPS1 to unattached kinetochores at the onset of mitosis depends on the outer kinetochore proteins HEC1 and NUF2 (Martin-Lluesma et al., 2002; Stucke et al., 2002; Meraldi et al., 2004) and is regulated by the Aurora B kinase (Santaguida et al., 2011; Saurin et al., 2011). These proteins operate in one pathway, as the ability of centromere-tethered Aurora B to recruit MPS1 in G2-phase cells depends on HEC1 (Saurin et al., 2011). The Aurora B–HEC1–MPS1 pathway is critical for rapid establishment of mitotic checkpoint activity at the onset of mitosis (Saurin et al., 2011).
We sought to examine the molecular mechanism of MPS1 kinetochore binding and regulation thereof. Here, we present the crystal structure of a tetratricopeptide repeat (TPR) domain in the kinetochore-binding region of MPS1 and provide evidence that association of MPS1 with kinetochores is essential for mitotic checkpoint activity. This association depends on the microtubule-binding domain of HEC1 and is regulated by the TPR domain in an Aurora B–dependent manner.
Results
Crystal structure of a TPR-like fold in the kinetochore-targeting region of MPS1
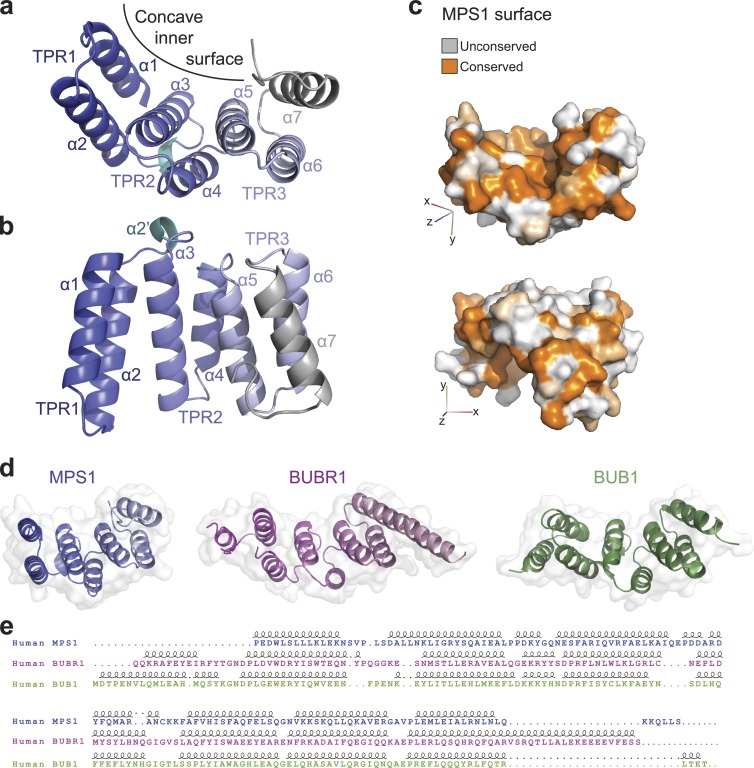
The N-terminal 301 amino acids of MPS1 are sufficient for localization of the kinase to kinetochores during mitosis (Liu et al., 2003; Stucke et al., 2004), whereas the N-terminal 100 amino acids, although not sufficient, are essential for MPS1 kinetochore binding (Stucke et al., 2004; Maciejowski et al., 2010). Sequence similarity searches using PSI-BLAST (Position-Specific Iterated Basic Local Alignment Search Tool) suggest that the MPS1 N-terminal region has significant similarity with the TPR domains in BUB1 (Bolanos-Garcia et al., 2009) and BUBR1 (Bolanos-Garcia et al., 2005; Beaufils et al., 2008; D’Arcy et al., 2010), as recently modeled (Lee et al., 2012). To understand the molecular mechanism by which the TPR-containing N-terminal region of MPS1 regulates binding to kinetochores, we determined its three-dimensional structure. Several MPS1 protein fragments were expressed, purified, and screened for crystallization. The best diffracting crystals were obtained from a construct consisting of residues 62–239, MPS162–239. The structure was determined to 2.2-Å resolution by single-wavelength anomalous dispersion using selenomethionine-substituted protein and was refined to an Rfree of 18.6% without any Ramachandran plot outliers (for crystallographic details see Materials and methods and Table 1). The asymmetric unit contained four molecules, which were all well ordered with the exception of the 40 C-terminal residues that were not visible in the electron density and were not included in the model. The structure was formed by seven helices, the first six of which are arranged in three TPR repeats (TPR1–3) that fold together to produce a concave “C”-shaped cross section (Fig. 1, a–c). The inner concave surface, the typical ligand binding site for many TPR domains, is well conserved, but surface patches with good sequence conservation are also clearly present in the outer convex surface (Fig. 1 c).
Table 1.
X-ray data statistics and model refinement parameters
| Parameters | Values |
| Diffraction data | |
| Space group | P212121 |
| Unit cell: a, b, c (Å) | 79.9, 80.1, 142.2 |
| Molecules (a.u.)/solvent content | 4/61% |
| Resolution (Å) | 44.28–2.2 (2.32–2.20) |
| Completeness (%) | 98.8 (92.7) |
| Unique reflections | 46,558 (6,239) |
| Rmerge | 0.07 (0.45) |
| <(I)/σ(I)> | 14.1 (2.8) |
| Multiplicity | 5.8 (3.7) |
| Wilson B factor (Å2) | 41.5 |
| Model statistics | |
| R-factor (%) | 17.0 |
| Rfree (%) | 18.6 |
| Ramachandran plot favored (%) | 99.1 |
| Ramachandran plot outliers (%) | 0.0 |
| Protein atoms number | 4,475 |
| Ligand atom number | 365 |
| Water atom number | 232 |
| Protein B factor | 50 |
| Ligand B factor | 68 |
| Water B factor | 46 |
| RMSD bond lengths (Å) | 0.01 |
| RMSD bond angles (°) | 0.97 |
The Rfree set comprised 2,362 reflections corresponding to 5% of the total data. Numbers in parentheses denote high resolution statistics. a.u., asymmetric unit; RMSD, root-mean-square deviation.
Figure 1.
Crystal structure of the MPS1 TPR domain. (a) Crystal structure of the TPR domain. A cartoon diagram of the three TPR1–3 helical doublets forming the concave surface is shown in blue shades that fade toward gray form the N toward the C terminus; the C-terminal helix is in gray, and the α2′ short helix between TPR1 and TPR2 is in cyan. (b) A side view of the TPR domain. (c) A surface representation of the TPR domain colored by sequence conservation among vertebrate MPS1 TPR domains; the top view emphasizes the conservation of the concave inner surface, and the bottom view shows some conserved patches on the generally unconserved outer surface. (d) The TPR domains of MPS1, BUBR1, and BUB1 are shown in the same orientation after structural superposition, as cartoon diagrams within a transparent surface. (e) The sequence alignment resulting from the structural superposition of the three TPR domains above is shown together with secondary structure elements. Dots indicate gaps. Loops indicate helices.
Evolutionary conservation of the MPS1 TPR domain and similarities with the BUB family of TPR domains
Structure similarity searches using Dali (Holm and Rosenström, 2010) show that the MPS1 TPR domain is most similar to the N-terminal TPR domains of BUBR1 (Protein Data Bank accession no. 2WVI) and BUB1 (Protein Data Bank accession no. 4A1G; Fig. 1 D). Although the structure-based sequence alignment of MPS1, BUBR1, and BUB1 shows limited sequence similarity (Fig. 1 E), the MPS1 TPR domain should also be considered a member of this family. Some differences between the three TPR domains are notable. Whereas in BUB1 the residues following the C-terminal helix point away from the inner concave surface of the domain, the first few residues following the C-terminal capping helix in the MPS1 structure turn toward the inner concave surface of the domain, extending it (Fig. 1, a and b). The 310 helix connecting the first two TPR motifs in BUB1 and BUBR1 is substituted by a single-turn α helix in MPS1 (Fig. 1 b, α2′). Similarly, both the GIG and G(N/D)D motifs connecting the last two TPR repeats in BUB1 and BUBR1, which have been shown to be important for structural integrity, are missing in MPS1, but the overall arrangement of the domain is retained. Both the BUB1 and the BUBR1 TPR domains bind KNL1 through a characteristic depression in their convex surface (Bolanos-Garcia et al., 2011; Krenn et al., 2012). That exact mode of binding is unlikely to be conserved in the MPS1 TPR domain, as this surface depression is not present (Figs. 1 c and S1 a). However, ligand binding on the convex surface of the MPS1 TPR domain remains a possibility, for instance, through other conserved patches (Fig. 1 c). Finally, the BUB1 TPR domain dimerizes in solution and in the crystal structure, which is mediated by contacts made through a short loop between the N-terminal helix (absent in our MPS1 structure) and the first helix of TPR1 (Bolanos-Garcia et al., 2009). Although MPS1 forms dimers in cells (Hewitt et al., 2010; Lee et al., 2012), dimerization is unlikely to be mediated by the TPR domain or the N-terminal region of MPS1 that includes the TPR domain. First, in vitro, four different MPS1 constructs containing various regions of the N terminus (MPS11–196, MPS19–255, MPS162–239, and MPS11–239) were monomers in solution as shown by multiangle laser light scattering (Fig. S1 b). Second, immunoprecipitation experiments using mitotic 293T cells showed that MPS1 dimerization in cells did not rely on the N-terminal 192 amino acids of MPS1 (Fig. S1 c).
Given the strong conservation of the BUB (BUB1 and BUBR1) TPR domains (Suijkerbuijk et al., 2012) and their similarity to the TPR domain of human MPS1, we examined the origin and evolution of the TPR-fold sequence in eukaryotic MPS1 homologues (Vleugel et al., 2012). An hidden Markov model profile constructed from the TPR domain sequences of human MPS1 homologues could identify additional TPR domain sequence homology only in vertebrates and in some distantly related eukaryotes, such as green algae and choanoflagellates (Fig. S2). These homologous sequences were all predicted to fold into helical arrays, consistent with the TPR-like fold. Given the presence of a TPR domain in early branching species and loss in several later branching species, we infer the presence of a MPS1 with an N-terminal TPR domain in the common ancestor of all eukaryotes (last eukaryotic common ancestor) and subsequent parallel loss in distinct eukaryotic lineages. Although the TPR domain of MPS1 belongs to the same structural family as BUB1 and BUBR1 TPR domains, parallel gain of the MPS1 TPR domain from BUB-like sequences is highly unlikely because both groups of TPR domains show monophyletic clustering in a tree of the TPR domains. Finally, the patchy phyletic distribution of the TPR domain is not the result of horizontal gene transfer because the kinase tree for MPS1 orthologues is consistent with the species tree. In summary, the MPS1 TPR domain is likely ancient but maintained in only few branches of the eukaryotic tree of life.
The N-terminal region of MPS1 harbors a localization module required for checkpoint function
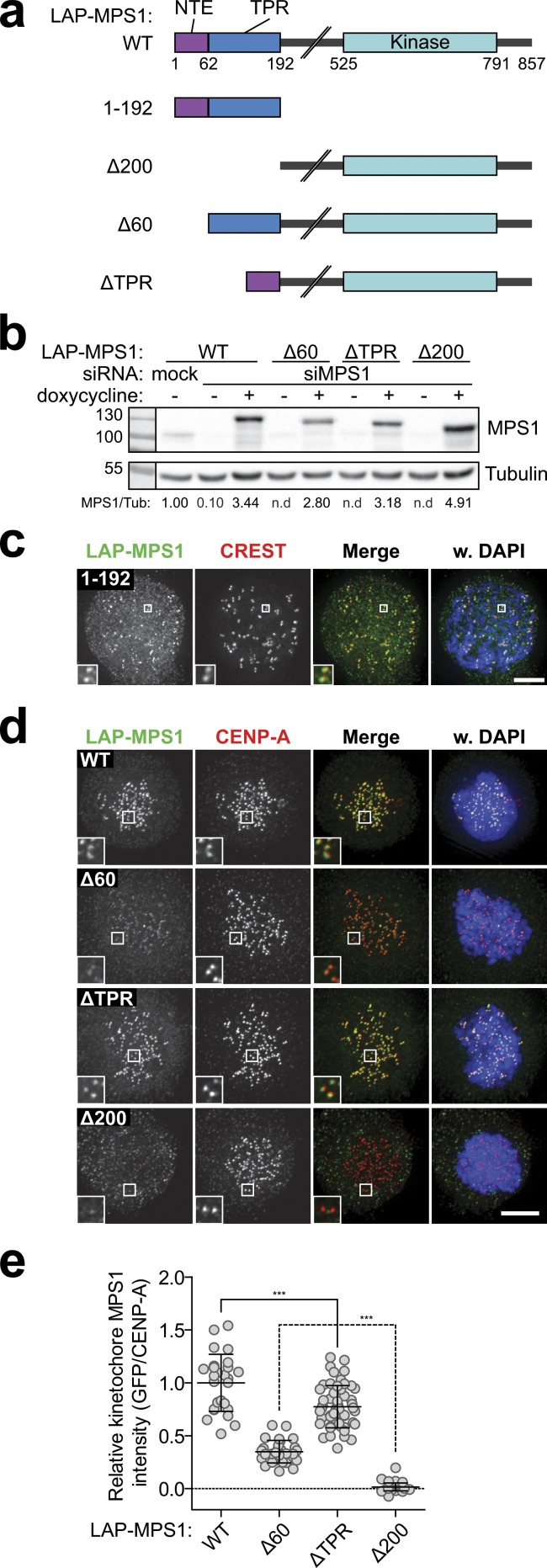
To examine the functional significance of the MPS1 TPR domain, we designed various MPS1 mutants based on the structure and generated cell lines stably expressing them (Figs. 2, a and b; and S3 a) from a doxycycline-inducible promoter in a single integration site to ensure comparable genetic background and expression levels (Klebig et al., 2009). The localization of localization and affinity purification (LAP)–tagged MPS1 proteins was assayed in cells depleted of endogenous MPS1 to prevent confounding effects of dimerization or competition for kinetochore ligands (Fig. S4, a and b) and in the presence of the small molecule MPS1 inhibitor reversine (Santaguida et al., 2010) to prevent indirect effects on localization by changes in MPS1 activity (Hewitt et al., 2010; Jelluma et al., 2010). These experiments showed that the N-terminal region of MPS1 that encompasses the TPR domain (MPS11–192) localized weakly but reproducibly to kinetochores during prophase (Fig. 2 c), which is when maximal kinetochore enrichment of MPS1 is normally observed (Saurin et al., 2011). The inefficient prophase localization and the absent prometaphase localization of MPS11–192 compared with wild-type MPS1 (MPS1WT) further suggested that additional, yet undefined, residues in MPS1 contribute to efficient MPS1 kinetochore binding. Consistently, whereas MPS1WT localized to kinetochores efficiently, a truncated MPS1 mutant lacking this N-terminal region (MPS1Δ200) was undetectable at kinetochores (Fig. 2, d and e). Thus, the MPS1 N-terminal region that encompasses the TPR domain is necessary for kinetochore binding. Surprisingly, however, deletion of the TPR domain (aa 61–192; MPS1ΔTPR) did not potently disturb localization of MPS1 to kinetochores (Fig. 2, d and e). The difference in localization between MPS1Δ200 and MPS1ΔTPR suggested that the 60 amino acids preceding the TPR domain are crucial for localizing MPS1. In support of this, a mutant that lacks this N-terminal extension (NTE; MPS1Δ60) showed strongly reduced kinetochore binding compared with both MPS1WT and MPS1ΔTPR (Fig. 2, d and e). Quantitation of the signal revealed that kinetochore levels of MPS1Δ60 were significantly higher than those of MPS1Δ200, which was undetectable at kinetochores. MPS1Δ60 therefore retains residual low affinity for kinetochores, which is provided by the TPR domain.
Figure 2.
MPS1 kinetochore localization is mediated by the NTE-TPR module. (a) Schematic representation of the domain organization of various MPS1 proteins used throughout this study. (b) Immunoblot of whole-cell lysates from mitotic HeLa Flp-in LAP-MPS1 cell lines that were transfected with mock or MPS1 siRNA and induced (+ doxycycline) to express the indicated LAP-MPS1 proteins; band intensity of MPS1/tubulin relative to mock is indicated. (c) Immunolocalization of LAP-MPS11–192 and centromeres (CREST) in nocodazole-treated, MPS1-depleted HeLaK FRT TetR cells. Cells were imaged for prophase figures. DNA (DAPI) is shown in blue. Insets show magnification of the boxed regions. (d and e) Representative images (d) and quantification (e) of immunolocalization of the various LAP-MPS1 proteins and centromeres (CENP-A) in nocodazole, 500 nM reversine, and MG132-treated, MPS1-depleted Flp-in HeLa cells. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graph in e displays total kinetochore intensities (±SD) of the indicated LAP-MPS1 proteins relative to centromeres (CENP-A) in cells treated as in d. Data are representative of three experiments. Ratios for LAP-MPS1WT are set to 1. One dot represents one cell. Line indicates means ± SD. ***, P < 0.0001; significant (Student’s t test, unpaired). Bars, 5 µm. WT, wild type; Tub, tubulin.
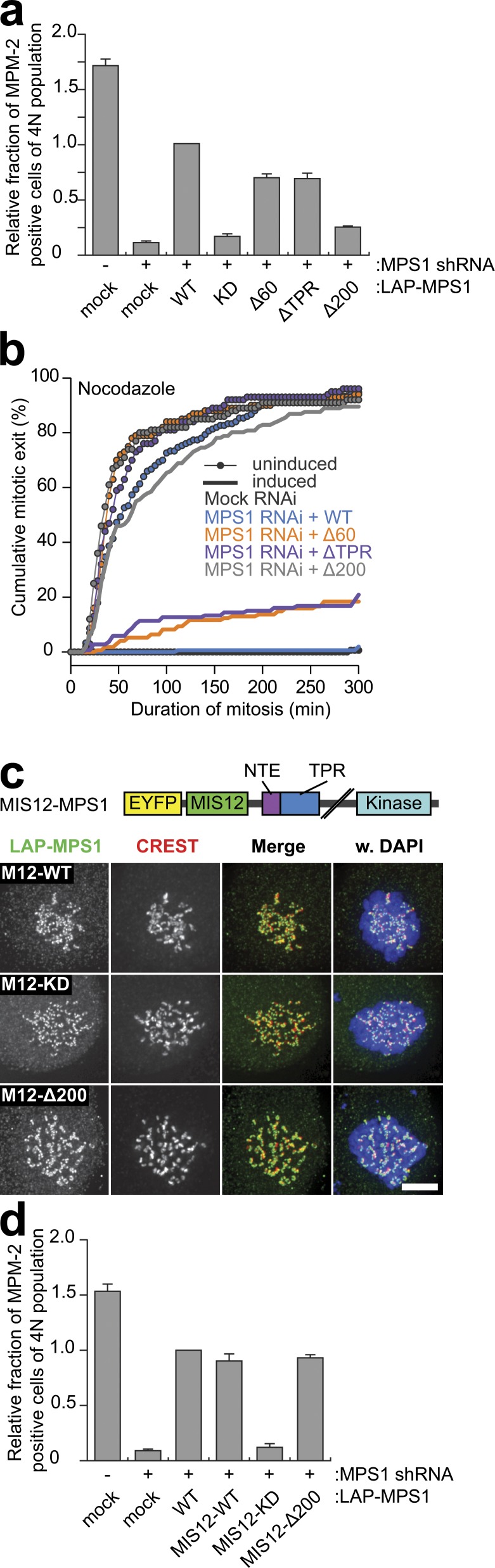
We next assessed whether the NTE and the TPR are needed for MPS1 function. Cells depleted of endogenous MPS1 and expressing the various RNAi-resistant mutants (Jelluma et al., 2008b) were examined for mitotic checkpoint activity by measuring mitotic index upon treatment of cells with the spindle-depolymerizing drug nocodazole and by real-time imaging of mitotic delay in nocodazole-treated cells. As expected, cells depleted of MPS1 failed to accumulate in mitosis in response to nocodazole (Fig. 3, a and b). This was largely rescued by expression of LAP-tagged RNAi-resistant MPS1WT but not by kinase-deficient MPS1D664A (Jelluma et al., 2008b). In accordance with its observed inability to localize to kinetochores, MPS1Δ200 could not restore mitotic checkpoint function in either assay (Fig. 3, a and b). In contrast, both MPS1ΔTPR and MPS1Δ60 displayed weakened checkpoint function. Mitotic index in nocodazole-treated cells expressing MPS1ΔTPR or MPS1Δ60 was reduced by ∼30% relative to MPS1WT. In addition, 28 and 22% of MPS1ΔTPR- and MPS1Δ60-expressing cells, respectively, were unable to maintain a mitotic delay for 5 h (Fig. 3, a and b). Fluorescence recovery after photobleaching showed that kinetochore-bound LAP-MPS1ΔTPR in nocodazole-treated cells had similar rapid turnover as MPS1WT (Fig. S4, c and d), and analysis of in vitro kinase activity of the various mutants immunoprecipitated from mitotic HEK 293T cells showed that none of the mutants suffered from compromised kinase activity (Fig. S4, e and f). Of note, MPS1ΔTPR displayed elevated levels of autophosphorylation (approximately twofold higher than MPS1WT), indicating that the TPR domain may be involved in regulating kinase activity, which could somehow contribute to compromised checkpoint function in MPS1ΔTPR-expressing cells. Finally, artificial tethering of localization-deficient MPS1 by fusion to the constitutive kinetochore protein MIS12 (Jelluma et al., 2010) was able to restore mitotic checkpoint activity (Fig. 3 d). Collectively, these data support the hypothesis that functional defects of MPS1 N-terminal truncation/deletion mutants are caused primarily by their inability to efficiently bind kinetochores.
Figure 3.
The NTE-TPR module is essential for mitotic checkpoint activity. (a) Mitotic index from flow cytometric analysis of MPM-2 positivity within a population of cells transfected with mock or MPS1 shRNA plasmids along with the indicated RNAi-resistant MPS1 alleles and treated with nocodazole for 16 h. Graph represents means of at least five independent experiments (±SEM); mean for LAP-MPS1WT reconstitution is set to 1. (b) Time-lapse analysis of duration of mitotic arrest in nocodazole-treated Flp-in HeLa cells transfected with mock or MPS1 siRNA and expressing the indicated LAP-MPS1 proteins (induced). Data indicate cumulative percentage of cells (from a total of ≥100 cells) that exit mitosis (scored as cell flattening) at the indicated times after nuclear envelope breakdown (NEB) and are representative of at least two independent experiments. Data for mock siRNA–treated cells and MPS1 siRNA–treated cells expressing LAP-MPS1WT overlap. (c) Immunolocalization of the indicated LAP-MIS12-MPS1 proteins and centromeres (CREST) in nocodazole-treated HeLa cells transfected with MPS1 siRNA for 48 h. M12, MIS12. DNA (DAPI) is shown in blue. Bar, 5 µm. A schematic representation of the LAP-MIS12-MPS1 protein is depicted. (d) Mitotic index from flow cytometric analysis as in a. Graph represents means of at least two independent experiments (±SEM); mean for LAP-MPS1WT reconstitution is set to 1. WT, wild type; KD, kinase dead.
Interestingly, MPS1Δ200 was readily detectable at kinetochores of cells containing normal levels of endogenous MPS1 (Fig. S4, a and b), in contrast to cells in which endogenous MPS1 was depleted (Fig. 2 d). Similar observations using mouse oocytes have been reported (Hached et al., 2011). Dimerization to kinetochore-localized forms of MPS1 may thus endow N-terminal truncation mutants with some kinetochore localization and function, possibly explaining why a recent study reported significant mitotic checkpoint signaling in cells expressing MPS1Δ100 (Maciejowski et al., 2010).
NTE-mediated kinetochore localization of MPS1 requires the microtubule-binding domain of HEC1
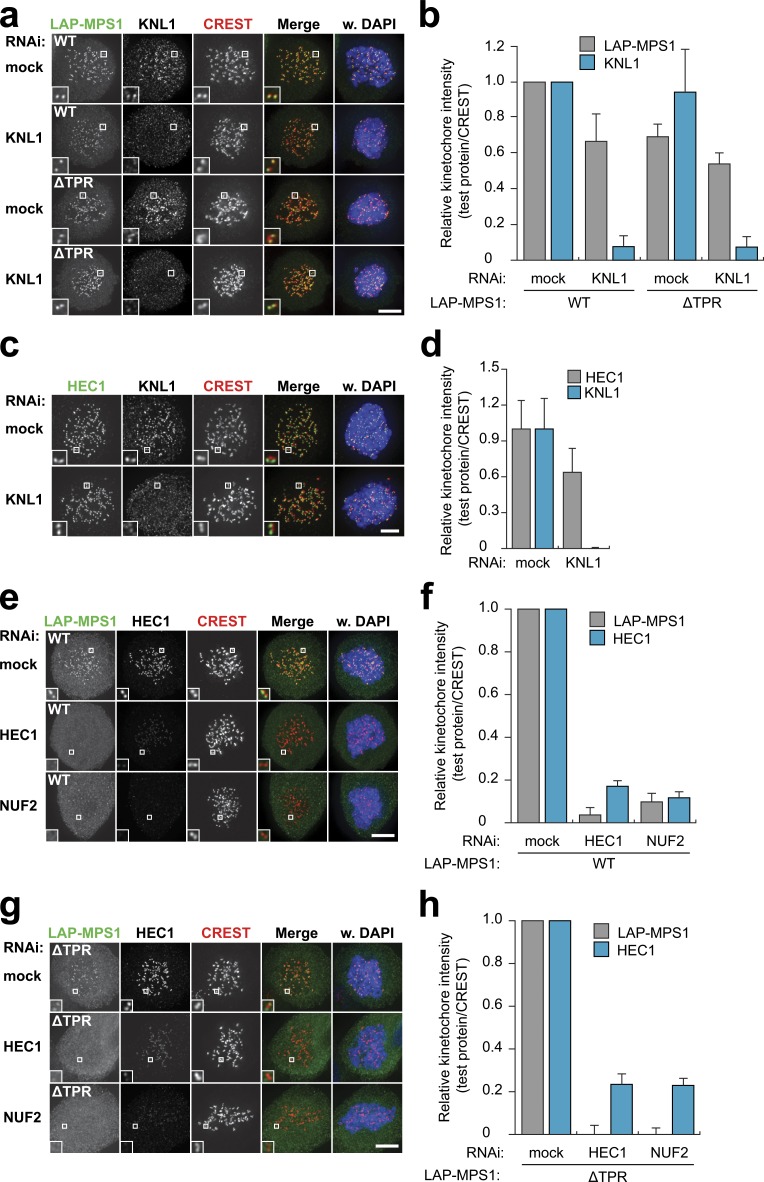
Having established that the primary localization signal in MPS1 resides in the N-terminal 192 amino acids with a dominant contribution from the NTE, we next wished to investigate the kinetochore requirements for MPS1 localization. As predicted by our structural analysis, KNL1 did not seem to contribute significantly to MPS1 kinetochore binding: depletion of KNL1 reduced MPS1 localization only slightly (Fig. 4, a and b), a reduction that is explained by a similar reduction in kinetochore HEC1 levels (Fig. 4, c–h).
Figure 4.
NTE-mediated MPS1 localization depends on the NDC80 complex. (a, b, and e–h) Representative images (a, e, and g) and quantification (b, f, and h) of immunolocalization of LAP-MPS1WT or LAP-MPS1ΔTPR and centromeres (CREST) in Flp-in HeLa cells transfected with siRNAs to MPS1 and luciferase (mock), HEC1, NUF2, or KNL1 and treated with nocodazole and reversine. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graphs display total kinetochore intensities (±SEM) of the indicated proteins relative to centromeres (CREST). Data are from ≥21 cells from at least two independent experiments. Ratios for mock RNAi–treated cells are set to 1. (c and d) Representative images (c) and quantification (d) of immunolocalization of HEC1, KNL1, and centromeres (CREST) in HeLa cells transfected with mock or KNL1 siRNAs and treated with nocodazole. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graph in b shows total kinetochore intensities (±SD) of HEC1 and KNL1 relative to centromeres. Data are from ≥13 cells and are representative of three experiments. Ratios for mock RNAi–treated cells are set to 1. Bars, 5 µm. WT, wild type.
The localization of MPS1 to kinetochores depends on the NDC80 complex members HEC1 and its obligate binding partner NUF2 (Martin-Lluesma et al., 2002; Meraldi et al., 2004). In agreement with this, localization of MPS1WT to unattached kinetochores in our inducible stable cell lines was lost upon depletion of HEC1 or NUF2 (Fig. 4, e and f). Similar results were obtained when examining localization of MPS1ΔTPR (Fig. 4, g and h), showing that the affinity of the NTE for kinetochores relies on the presence of the NDC80 complex.
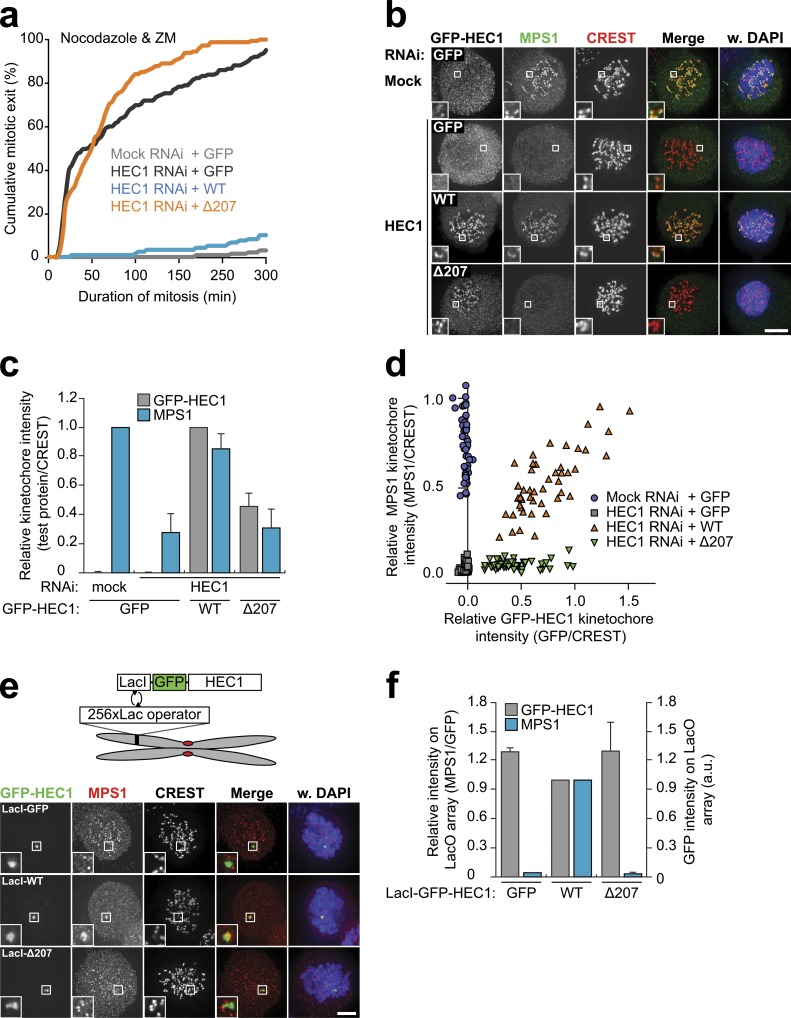
Full-length Saccharomyces cerevisiae MPS1 interacts with amino acids 1–257 of ScNdc80 (the N-terminal tail and the calponin homology [CH] domain) when coexpressed in Escherichia coli (Kemmler et al., 2009). In addition, PtK1 cells expressing an HEC1Δ1–207 protein (which lacks both the tail and the CH domain) have reduced ability to delay mitosis in the absence of kinetochore–microtubule attachments (Guimaraes et al., 2008). Incomplete HEC1 depletion does not prevent checkpoint activation in human cells (Meraldi et al., 2004), likely a result of insufficient penetrance of MPS1 displacement (Saurin et al., 2011). It does, however, sensitize the checkpoint to slight reductions in MPS1 activity or inhibition of Aurora B (Santaguida et al., 2011; Saurin et al., 2011). We wished to use this sensitization to ask whether the CH domain and tail of HEC1 are involved in the mitotic checkpoint. To this end, we created a set of stable, isogenic cell lines that inducibly express GFP-HEC1WT, GFP-HEC1Δ207, or GFP (Fig. S3 b). Although nocodazole-treated cells depleted of HEC1 or treated with the Aurora B inhibitor ZM447439 (Ditchfield et al., 2003) maintained mitotic arrest for many hours, addition of ZM447439 to HEC1-depleted cells caused rapid mitotic exit (Fig. 5 a; Saurin et al., 2011). This phenotype was rescued by expression of RNAi-insensitive wild-type GFP-HEC1WT but not by GFP-HEC1Δ207 (Fig. 5 a). In agreement with this, HEC1 depletion delocalized MPS1 from kinetochores, which was recovered by expression of GFP-HEC1WT (Fig. 5, b–d). The amount of MPS1 recruited to kinetochores correlated with the amount of kinetochore HEC1 (Fig. 5 d). Consistently, expression of GFP-HEC1Δ207, which was unable to reinstate a robust checkpoint response (Fig. 5 a), could not recover MPS1 localization (Fig. 5, b–d). Although GFP-HEC1Δ207 was generally incorporated less efficiently than wild-type GFP-HEC1, even kinetochores containing high levels of GFP-HEC1Δ207 were devoid of MPS1 (Fig. 5 d).
Figure 5.
The microtubule-binding domain of HEC1 directs MPS1 localization and function. (A) Time-lapse analysis of duration of mitotic arrest in nocodazole- and ZM447439 (ZM)-treated Flp-in HeLa cells transfected with mock or HEC1 siRNA and expressing the indicated GFP-HEC1 proteins. Data indicate cumulative percentages of cells (from a total of ≥125 cells per treatment) that exit mitosis (scored as cell flattening) at the indicated times after NEB and are representative of three independent experiments. (b–d) Representative images (b) and quantification (c and d) of immunolocalization of MPS1, the indicated GFP-HEC1 proteins, and centromeres (CREST) in nocodazole-treated Flp-in HeLa cells transfected with mock or HEC1 siRNA. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graph in c displays total kinetochore intensities (±SEM) of the indicated proteins relative to centromeres (CREST). Data are from a total of ≥103 cells per treatment from two experiments. Ratios are set to 1 for mock RNAi–treated cells (MPS1) and for GFP-HEC1WT–expressing cells (GFP-HEC1). Graph in d displays total kinetochore intensities of the indicated proteins relative to centromeres (CREST) for all cells of a single experiment. (e and f) Representative images (e) and quantification (f) of immunolocalization of MPS1, the indicated LacI-GFP-HEC1 proteins, and centromeres (CREST) in nocodazole-treated U2OS-LacO cells. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graph in f displays total intensities (±SEM) of MPS1 at LacO arrays relative to LacI-GFP-HEC1 (GFP) and total intensities of LacI-GFP-HEC1. Data are from a total of ≥17 cells from two experiments. Ratios for LacI-GFP-HEC1WT–expressing cells are set to 1. Bars, 5 µm. WT, wild type; a.u., arbitrary unit.
These results showed that residues 1–207 of HEC1 (encompassing the CH domain and tail) were necessary for MPS1 localization and checkpoint activity. To determine whether HEC1 is also sufficient for MPS1 localization, we examined whether HEC1 can recruit MPS1 when targeted to a nonkinetochore location. To this end, GFP-HEC1 was targeted to an array of lac operator (lacO) sequences in an arm of chromosome 1, by fusion to LacI. Indeed, accumulation of HEC1 on the lacO array was followed by recruitment of endogenous MPS1 to those sites (Fig. 5, e and f). Importantly, the ectopic recruitment of MPS1 depended on the microtubule-binding domains of HEC1, as MPS1 did not localize to lacO arrays decorated with GFP-HEC1Δ207 (Fig. 5, e and f). Collectively, these data argue that the N-terminal, microtubule-binding region of HEC1 promotes efficient mitotic checkpoint activity by ensuring NTE-mediated localization of MPS1.
Control of MPS1 kinetochore localization by Aurora B is mediated by the TPR domain
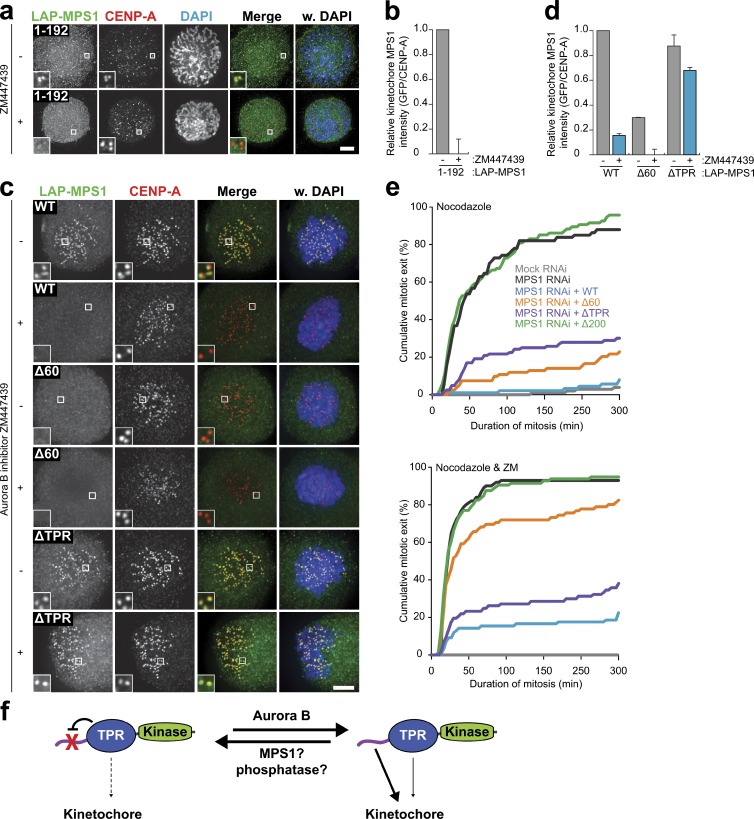
Inhibition of Aurora B prevents the accumulation of MPS1 on unattached kinetochores and delays establishment of the mitotic checkpoint in early mitosis (Saurin et al., 2011). Given the well-established regulation of the HEC1 N-terminal tail by Aurora B (Tooley and Stukenberg, 2011) and our finding that the HEC1 tail–CH region (1–207) is required to recruit MPS1, we hypothesized that Aurora B controls MPS1 localization by phosphorylating the HEC1 tail. To address this, MPS1 localization to kinetochores was assessed in HEC1-depleted cells reconstituted with GFP-HEC1Δ80 (which lacks the HEC1 tail), GFP-HEC19A (lacking the Aurora B phosphorylation sites in the tail), or GFP-HEC19D (in which the Aurora B sites were substituted to aspartate residues to mimic phosphorylation; Guimaraes et al., 2008; Miller et al., 2008; DeLuca et al., 2011). Surprisingly, all three HEC1 mutants were able to restore MPS1 kinetochore levels to the same extent as wild-type HEC1 (Fig. S5, a and b). We thus conclude that the regulation of MPS1 localization by Aurora B is not mediated by phosphorylation of the HEC1 tail.
We next asked whether Aurora B controls the MPS1 localization module. As shown in Fig. 6 (a and b), kinetochore binding of MPS11–192 was abolished by treatment with ZM447439, showing that Aurora B affects MPS1 localization by regulating binding of this minimal domain to kinetochores. Strikingly, although ZM447439 strongly reduced the amounts of MPS1WT at prometaphase kinetochores and abolished residual MPS1Δ60 levels, it had no effect on kinetochore binding of MPS1ΔTPR (Fig. 6, c and d). Consistently, although Aurora B inhibition weakened or abolished mitotic delays in nocodazole-treated cells expressing MPS1WT (20% exit after 5 h) or MPS1Δ60 (82% exit after 5 h), respectively, it left the (weakened) checkpoint in MPS1ΔTPR-expressing cells virtually unaffected (Fig. 6 e). In summary, removal of the TPR domain renders MPS1 localization independent of Aurora B activity. This suggests that the TPR domain normally prevents MPS1 localization, and this inhibitory effect is relieved by Aurora B (Fig. 6 f).
Figure 6.
Aurora B regulates MPS1 kinetochore localization by controlling function of the TPR domain. (a and b) Representative images (a) and quantification (b) of immunolocalization of LAP-MPS11–192 and centromeres (CENP-A) in prophase HeLaK FRT TetR cells depleted of MPS1 and treated with ZM447439, as indicated. DNA (DAPI) is shown in blue. Insets show magnifications of the boxed regions. Graph in b shows total kinetochore intensities (±SEM) of MPS1 relative to centromeres. Data are from ≥38 cells from two experiments. Ratios for mock-treated cells are set to 1. (c and d) Representative images (c) and quantification (d) of immunolocalization of the indicated LAP-MPS1 proteins and centromeres (CENP-A) in MPS1-depleted HeLaK FRT TetR cells treated with nocodazole and reversine, with or without ZM447439. DNA (DAPI) is shown in blue. Insets are magnifications of the boxed regions. Graph in d shows total kinetochore intensities (±SEM) of MPS1 relative to centromeres in DMSO-treated (gray bars) or ZM447439-treated (blue bars) cells. Data are from ≥32 cells from two experiments. Ratios for mock-treated, LAP-MPS1WT–expressing cells are set to 1. (e) Time-lapse analysis of the duration of mitotic arrest in HeLaK FRT TetR cells transfected with mock or MPS1 siRNA and expressing the indicated LAP-MPS1 proteins and treated with nocodazole and DMSO (top) or nocodazole and ZM447439 (ZM; bottom). Data indicate cumulative percentage of cells (from a total of ≥70 cells) that exit mitosis (scored as chromosomal decondensation) at the indicated times after NEB and are representative of at least two independent experiments. (F) Model of regulated MPS1 localization at unattached kinetochores. See Discussion for details. Bars, 5 µm. WT, wild type.
Discussion
Based on data presented in this study, we postulate that the mitotic checkpoint relies on the NTE-TPR module of MPS1 and that Aurora B–mediated control of the checkpoint impinges on this module. In our model (Fig. 6 f), MPS1 alternates between a localization-deficient and -proficient form, and the equilibrium can be driven to proficient by Aurora B activity. The TPR domain is important to maintain the deficient form, whereas the proficient form binds kinetochores predominantly through the NTE with some contribution from the TPR. Aurora B activity simultaneously inhibits the negative impact of the TPR domain on MPS1 localization and stimulates the contribution of the TPR domain to kinetochore binding. The model in Fig. 6 f is consistent with present and previously published data. The model predicts that (a) deletion of the TPR domain renders localization and function of MPS1 solely dependent on NTE and independent of Aurora B activity (Figs. 2 and 6), (b) deletion of the NTE allows weak, but Aurora B–dependent, MPS1 localization (Figs. 2 and 6), and (c) endogenous MPS1 can localize weakly in the absence of Aurora B activity. Indeed, we and others have shown that although MPS1 localization is potentiated by Aurora B activity (Santaguida et al., 2011; Saurin et al., 2011), it can weakly localize and eventually autoactivate without Aurora B (Saurin et al., 2011).
Important questions are how the TPR domain prevents the NTE from localizing MPS1 to kinetochores and how Aurora B alleviates this. The most straightforward mechanism that we envision is one in which the NTE interacts with the TPR domain, inhibiting both NTE- and TPR-mediated kinetochore binding. In this scenario, release of this interaction is promoted (directly or indirectly) by Aurora B activity, rendering both the NTE and TPR available as kinetochore binding sites. Because Aurora B affects TPR functionality, both the release of NTE and the kinetochore affinity of TPR will depend on Aurora B. Aurora B could directly phosphorylate the NTE, the TPR, HEC1, or even an unknown kinetochore protein that directly binds MPS1 and whose function relies on HEC1. The Aurora B sites in the tail of HEC1 are not involved, and we have not been able to find Aurora B–dependent phosphorylation in the N-terminal domains of MPS1 or in the CH domain of HEC1. Aurora B may thus indirectly control MPS1 localization, for instance by causing a conformational change in HEC1 or MPS1 that exposes potential interaction sites or by preventing PP1-dependent dephosphorylation of residues at the MPS1–kinetochore interface. Much work using cellular structure–function assays and in vitro interaction experiments is needed to uncover the mechanism behind the regulation of MPS1 localization.
Catalytically inactive MPS1 accumulates on kinetochores to higher levels than active MPS1 (Hewitt et al., 2010; Jelluma et al., 2010), suggesting that MPS1 kinase activity controls its own turnover at kinetochores. This accumulation can at least in part be explained by postulating that inactivated MPS1 has increased residence time at kinetochores (Jelluma et al., 2010). MPS1 may also promote its turnover at kinetochores by counteracting the effects of Aurora B on TPR function, affecting in a more direct manner its own localization domain. MPS1 is autophosphorylated on multiple sites in the NTE as well as in the TPR domain (Daub et al., 2008; Dephoure et al., 2008; Jelluma et al., 2008a; Oppermann et al., 2009; Xu et al., 2009; Dulla et al., 2010; Morin et al., 2012). This suggests that one or more of these phosphorylations either reduce the affinity of the NTE for its binding site at kinetochores or stimulate TPR-mediated inhibition of MPS1 localization. Detailing the mechanism by which MPS1 autoregulates its affinity for kinetochores will be an important future research effort.
The MPS1 localization module integrates the microtubule attachment site, tension-dependent signaling, and mitotic checkpoint activity
Production of the mitotic checkpoint complex from a single kinetochore is inhibited upon engagement of this kinetochore with spindle microtubules, as exemplified by absence of MAD1 and MAD2 on attached kinetochores (Kops and Shah, 2012). Removal of these proteins is at least in part mediated by dynein-dependent poleward transport, but other, dynein-independent, mechanisms have been proposed. These include microtubule binding to the N terminus of KNL1, attachment-dependent recruitment of phosphatases, and possibly an additional yet unresolved spindly-controlled pathway (Kops and Shah, 2012). Any of these could, in principle, impinge on MPS1 kinetochore binding or regulation thereof by Aurora B. Our finding that MPS1 localization is dependent on the microtubule-binding domain of HEC1 offers a tentative alternative model. Although it is unclear whether the molecular requirements of HEC1 to bind microtubules are the same as those that are required to promote MPS1 localization, the two functions of HEC1 could be mutually exclusive. In such a model, microtubule attachment would prevent MPS1 kinetochore binding, providing a direct mechanism of regulation. Absence of biorientation and the accompanying zone of Aurora B activity might continue to prime MPS1 kinetochore binding in case attachment is lost. This is consistent with asymmetric Mps1 localization on paired kinetochores during prometaphase and strongly reduced MPS1 levels on the attached sister kinetochore of a monotelic chromosome (Fig. S5, c and d), although dynein-dependent stripping could account for this behavior also. Further molecular insights into how the mitotic checkpoint machinery is integrated with the microtubule attachment site and the error correction machinery will be vital for understanding the coupling between attachment and tension and the cell cycle responses to the absence of either.
Materials and methods
Protein expression and purification of MPS162–239
MPS162–239 was transformed in Rosetta2 (DE3) cells (EMD Millipore). Cells were grown in lysogeny broth medium at 30°C until OD600nm = ∼0.6 and then cooled down to 18°C and induced at OD600nm = ∼0.8 for 16 h with 1 mM IPTG. For selenomethionine incorporation, the SelenoMet Medium (Molecular Dimensions Limited) was used according to the manufacturer’s instructions. Bacteria were harvested by centrifugation and resuspended in 100 ml buffer A (50 mM Tris, pH 7.5, 500 mM NaCl, 10 mM imidazole, pH 8.0, and 5 mM β-mercaptoethanol). Cells were lysed and cleared by high-speed centrifugation. The supernatant was treated with 2% streptomycin sulfate and further centrifuged. Finally, the soluble extract was loaded on a 1-ml affinity column (HiTrap; GE Healthcare) precharged with NiCl2. After extensive washing with buffer A, the protein was eluted with a linear gradient of imidazole to 250 mM. The eluate was diluted 1:1 to reduce salt concentration, loaded on a 1-ml HiTrap heparin column (GE Healthcare), and eluted with a linear gradient of NaCl to 2 M. The eluate was incubated with 3C protease for affinity tag cleavage, concentrated, and loaded on a Superdex G75 16/60 HiLoad (GE Healthcare) equilibrated in 25 mM Tris, pH 7.5, 150 mM NaCl, and 1 mM DTT. The protein eluted as a monomer and was concentrated to 10 mg/ml and flash frozen in liquid nitrogen until further use.
Crystallization, data collection, and structure solution
Crystals of MPS162–239 were grown in 0.1 M MIB (sodium malonate, imidazole, and boric acid buffer), pH 5.0, 25% Polyethylene Glycol 1500 (solution B2; pH, anion, and cation crystallization trial screen; QIAGEN; Newman et al., 2005) and were transferred into a cryoprotecting solution consisting of 25% glycerol before vitrification in liquid nitrogen. Data were collected at the European Synchrotron Radiation Facility on ID23-1 where the crystals diffracted to a 2.2-Å resolution in the space group P212121 with cell dimensions a = 79.9 Å, b = 80.1 Å, and c = 142.2 Å. Phases were obtained by single wavelength anomalous dispersion at the Swiss Light Source beamline PX1. All data were integrated by MOSFLM (Leslie, 2006) and scaled using SCALA (Evans, 2006). Because the a and b axes were very close to each other, many crystals appeared to belong to the primitive tetragonal rather than the primitive orthorhombic space group. Several datasets were collected, processed, and analyzed using POINTLESS (Evans, 2006) and PHENIX.XTRIAGE (Zwart et al., 2008). As the “more tetragonal” crystals appeared merohedrally twinned, we aimed to find orthorhombic crystals with minimal indications of twinning in the intensity distribution statistics. Such a dataset was identified with unit cell dimensions a = 79.77 Å, b = 79.81 Å, and c = 139.2 Å, and a highly complete and redundant dataset was collected to 3.2-Å resolution. That crystal was used for phasing, using autoSHARP (Vonrhein et al., 2007), based on the signal from the eight incorporated selenomethionine residues resulting from four molecules within the asymmetric unit, two in each molecule. As the expected signal was rather low even in theory (2–3% at that resolution), the initial phases had a rather low figure of merit (0.22), which was improved after solvent flattening and twofold noncrystallographic averaging to 0.86 (the four molecules were arranged in two pairs, as fourfold averaging was not useful in that case). These resulted in a good quality map that was used to build an initial model using BUCCANEER (Cowtan, 2006) and contained 582 residues (529 in sequence) dispersed in eight discrete chains; this model, however, contained quite a few wrongly or out of register–placed residues. A single molecule was manually isolated from that model using COOT (Emsley et al., 2010) and was used as a search model to find the four copies in the related high-resolution native dataset by molecular replacement using PHASER (McCoy et al., 2007). The map from PHASER was subsequently used for running APR/wARP (Langer et al., 2008) to yield a model with 504 residues in four chains (405 in sequence), which contained no errors. That model was manually completed in COOT, with alternate rounds of refinement using REFMAC (Murshudov et al., 2011) followed by refinement in autoBUSTER (Blanc et al., 2004) using autoncs and translation libration screw-motion refinement. The final model contained residues 62–199, 62–195, 62–195, and 62–199 in the A, B, C, and D molecules, respectively (preceded by residues GPG, which remained after protease cleavage), 229 water molecules, and several ordered components from the crystallization condition (11 glycerol, 5 polyethylene glycol, and 2 malonate molecules). Data collection and refinement statistics are shown in Table 1. The coordinates and structure factors have been deposited in the Protein Data Bank with accession no. 4B94.
Size-exclusion chromatography and multiangle laser light scattering analysis
For quaternary structure determination of MPS1 constructs, 100 µl of purified protein samples were injected (at 5 mg/ml for MPS11–196, MPS19–255, and MPS162–239 and at 15 mg/ml for MPS11–239) into a Superdex S75 10/30 column connected to a liquid chromatography system (ÄKTAFPLC; both obtained from GE Healthcare) and coupled to a light-scattering detector (MiniDawn; Wyatt Technology). The measurements were performed in 20 mM Hepes, pH 7.4, 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine, and the elution profiles were monitored at 280 nm. Data were recorded and analyzed with the Astra 5 software (Wyatt Technology) using a differential index of refraction value of 0.185.
Orthologue definition and phylogenetic analyses
MPS1 orthologues were defined as described previously (Vleugel et al., 2012). In short, we performed BLAST (Altschul et al., 1990) searches for hMPS1 against a local database comprised of genomes representative for all eukaryotic supergroups. The kinase domains of the resulting hits were aligned using MAFFT (Katoh and Standley, 2013) with option LINSI. Positions with too many gaps (>20%) were excluded from the alignment. Subsequently, an RAxML (Stamatakis et al., 2005) tree with 100 bootstraps was generated (option PROTGAMMAWAG). From the resulting tree, a subcluster corresponding to the orthologous group of which hMPS1 is a member was delineated. Potential TPR domains in these homologues were searched for by constructing a HMMER3 profile (Eddy, 2011) for the TPR domain of vertebrate MPS1 homologues. Significant sequences from additional MPS1 homologues were added to the profile in an iterative process until convergence. The domain topology (TPR and kinase) and resulting gene tree were visualized using iTOL (Interactive Tree Of Life; Letunic and Bork, 2007).
Cell culture and reagents
U2OS cells, HEK 293T cells, and HeLa cells were grown in DMEM supplemented with 9% FBS, 50 µg/ml penicillin/streptomycin, and 2 mMl-glutamine. All Flp recognition target (FRT) HeLa cells stably expressing H2B-mRED, a HA-tagged tetracycline repressor (TetR), and doxycycline-inducible MPS1 constructs were derived from the HeLa Kyoto (HeLaK) FRT TetR cell line (a gift from U. Kutay and P. Meraldi, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland; Zemp et al., 2009) by transfection with pCDNA5/FRT/TO vector (Invitrogen) and pOG44 (Invitrogen) and cultured in the same medium but containing 9% tetracycline-approved FBS (Takara Bio Inc.), 200 µg/ml hygromycin, and 1 µg/ml puromycin. All HeLa Flp-in cells stably expressing a TetR and doxycycline-inducible MPS1 or HEC1 constructs were derived from the HeLa Flp-In cell line (gift from S. Taylor, University of Manchester, Manchester, England, UK; Klebig et al., 2009) as in this paragraph and cultured in the same medium but containing 9% tetracycline-approved FBS, 200 µg/ml hygromycin, and 4 µg/ml blasticidin instead. The U2OS-LacO cell line, bearing an array of 256 lacO repeats on chromosome 1 (Janicki et al., 2004) was a gift from I. Cheeseman (Whitehead Institute, Cambridge, MA). To induce protein expression in the inducible cell lines, 1 µg/ml doxycycline was added for ≥8 h. 2 mM thymidine, 830 nM nocodazole, 10 µM MG132, 500 nM reversine, doxycycline, and 1 µg/ml puromycin were all obtained from Sigma-Aldrich. Hygromycin was purchased from Roche. 20 µM S-trityl-l-cysteine and 2 µM ZM447449 were both obtained from Tocris Bioscience. Blasticidin was obtained from PAA Laboratories.
Immunoprecipitation and immunoblotting
HEK 293T cells transfected with LAP-MPS1 (Fig. S4 e) or LAP-MPS1 and FLAG-MPS1 (Fig. S1 c) were treated with thymidine for 24 h and subsequently released into nocodazole for 16 h. Cells were lysed in lysis buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 1 mM β-glycerophosphate, 1 mM NaF, 1 mM Na3VO4, and protease inhibitor [Complete; Roche]). LAP-MPS1 was bound to GFP-Trap agarose beads (ChromoTek) for 1 h and washed four times in lysis buffer, and after removal of all buffer, sample buffer was added. Samples were separated by SDS-PAGE. Immunoblotting was performed using standard protocols; the signal was visualized and analyzed on a scanner (ImageQuant LAS 4000; GE Healthcare) using enhanced chemiluminescence (Figs. 2 b, S1 c, and S4 e) or analyzed on an scanner (Odyssey; LI-COR Biosciences) using fluorescently labeled secondary antibodies (Fig. S3, a and b).
Knockdown and reconstitution experiments with LAP-MPS1 and GFP-HEC1
For knockdown and reconstitution of MPS1 in HeLaK FRT TetR cell lines, cells were transfected with 10 nM MPS1 or mock siRNA for 16 h after which cells were arrested in early S phase for 24 h by addition of thymidine. Subsequently, cells were released from thymidine for 8–10 h and arrested in prometaphase by the addition of nocodazole and (in MPS1 immunolocalization experiments) treated with reversine to accumulate MPS1 at kinetochores and MG132 to prevent mitotic exit. LAP-MPS1 expression was induced by the addition of doxycycline at the release from thymidine. For knockdown and reconstitution of MPS1 in HeLa Flp-in cells, cells were transfected with 20 nM MPS1 or mock siRNA and, in some experiments, 20 nM HEC1, NUF2, or KNL1 siRNA and subsequently treated as the HeLaK FRT TetR cells. For knockdown and reconstitution of HEC1 in HeLa Flp-in cells, cells were transfected with 40 nM HEC1 or mock siRNA for 16 h, after which cells were arrested in S phase for 24 h by addition of 2 mM thymidine. Subsequently, cells were released from thymidine and were transfected again with 40 nM HEC1 or mock siRNA. 8–10 h after the release, cells were arrested for a second time in S phase for 14–16 h. Subsequently, cells were treated as the HeLaK FRT TetR cells. GFP-HEC1 expression was induced by the addition of doxycycline at the time of the second thymidine addition. To compensate for less efficient incorporation of GFP-HEC1Δ207 into kinetochores, its expression was induced at the time of the first thymidine addition. As a control, a cell line was used that inducibly expressed a full-length mRNA encoding for GFP-HEC1 in which a stop codon was introduced to replace the first amino acid of HEC1 (GFP-HEC1STOP), resulting in the expression of GFP.
Transfection and siRNA
For U2OS cells, plasmids were transfected using the calcium-phosphate method. Plasmids were transfected into HEK293T, HeLa, and U2OS-LacO cells using Fugene 6 (Roche) according to the manufacturer’s instructions. siRNAs used in this study were as follows: si-HEC1, 5′-CCCUGGGUCGUGUCAGGAA-3′ (custom; Thermo Fisher Scientific); si-MPS1, 5′-GACAGAUGAUUCAGUUGUA-3′ (custom; Thermo Fisher Scientific); si-mock (Luciferase GL2 duplex; D-001100-01-20; Thermo Fisher Scientific); si-NUF2, 5′-AAGCATGCCGTGAAACGTATA-3′ (custom; Thermo Fisher Scientific), and siKNL1, 5′-GCAUGUAUCUCUUAAGGAA-3′ (CASC5#5; J-015673-05; Thermo Fisher Scientific). All siRNAs were transfected using HiPerFect (QIAGEN) at 10, 20, or 40 nM (for HEC1 reconstitutions) according to the manufacturer’s instructions.
Antibodies
The following primary antibodies were used for immunofluorescence imaging and immunoblotting: MPS1–N terminal (EMD Millipore), α-tubulin (Sigma-Aldrich), CREST/anticentromere antibodies (Cortex Biochem), HEC1 (9G3; Abcam), GFP (custom rabbit polyclonal raised against full-length GFP as antigen; Jelluma et al., 2008b), GFP (mouse monoclonal; Roche), CENP-A (3–19; Abcam), KNL1 (ab70537; Abcam), MAD2 (custom rabbit polyclonal raised against full-length 6×His-tagged MAD2 as antigen; Sliedrecht et al., 2010), and pT676-MPS1 (custom rabbit polyclonal raised against the peptide CMQPDTpTSVVKDS coupled to keyhole limpet hemocyanin as antigen; Jelluma et al., 2008a). Secondary antibodies were high-crossed goat anti–human and anti–mouse Alexa Fluor 647 and goat anti–rabbit and anti–mouse Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes) for immunofluorescence experiments.
Live-cell imaging, immunofluorescence, and image quantification
For live-cell imaging, cells were plated in 24-well glass-bottom plates (MatTek Corporation), transfected, and imaged in a heated chamber (37°C and 5% CO2) using a 20×/0.5 NA UPLFLN objective (Olympus) on a microscope (IX-81; Olympus) controlled by Cell-M software (Olympus). Images were acquired using a camera (ORCA-ER; Hamamatsu Photonics) and processed using Cell-M software. For imaging of H2B-mRED, multiple z layers were acquired and projected to a single layer by maximum intensity projection.
For immunofluorescence, cells plated on 12-mm coverslips were preextracted with 0.1% Triton X-100 in PEM (100 mM Pipes, pH 6.8, 1 mM MgCl2, and 5 mM EGTA) for 45 s before fixation with 4% paraformaldehyde in PBS. Coverslips were washed with PBS and blocked with 3% BSA in PBS for 1 h, incubated with primary antibodies for 2–4 h at room temperature or 16 h at 4°C, washed with PBS, and incubated with secondary antibodies for an additional hour at room temperature. Coverslips were then incubated with DAPI for 2 min, washed, and mounted using antifade (ProLong; Molecular Probes). All images were acquired on a deconvolution system (DeltaVision RT; Applied Precision) with a 100×/1.40 NA U Plan S Apochromat objective (Olympus) using softWoRx software (Applied Precision). Images are maximum intensity projections of deconvolved stacks. For quantification of immunostainings, all images of similarly stained experiments were acquired with identical illumination settings; cells expressing comparable levels of exogenous protein were selected for analysis and analyzed using ImageJ (National Institutes of Health). An ImageJ macro was used to threshold and select all centromeres and all chromosome areas (excluding centromeres) using the DAPI and anticentromere antibodies channels as described previously (Saurin et al., 2011). This was used to calculate the relative mean kinetochore intensity of various proteins ([centromeres–chromosome arm intensity (test protein)]/[centromeres–chromosome arm intensity (CREST/CENP-A)]). Immunostainings on LacO arrays were quantified similarly, with the exception that the LacO dot was manually selected and that the relative mean LacO intensity of various proteins was calculated ([LacO–chromosome arm intensity (test protein)]/[LacO–chromosome arm intensity (GFP)]).
Fluorescence recovery after photobleaching
Flp-in HeLa cells were grown in 8-well glass-bottom dishes (LabTek Corporation), depleted of endogenous MPS1 by transfection with MPS1 siRNA, and induced to express MPS1WT or MPS1ΔTPR. The media were replaced with Leibovitz L-15 media (Invitrogen) supplemented with 10% FCS, 2 mM l-glutamine, and 100 U/ml penicillin/streptomycin. Cells were treated with 830 nM nocodazole, 10 µM MG132, and 500 nM reversine 30 min before imaging. Cells expressing similar levels of LAP-MPS1 were selected for imaging. Samples were imaged on a personal DeltaVision system equipped with a heated chamber and lens warmer (both set at 37°C), with a 100×/1.40 NA U Plan S Apochromat objective using softWoRx software. Images were acquired using a camera (CoolSNAP HQ2; Photometrics) and processed using softWoRx software and ImageJ. The EYFP-based LAP tag of LAP-MPS1 was bleached using the 488-nM laser line of an argon laser (max 20 mW) set to 100%. Areas centered on single kinetochore pairs were bleached once at 100% laser power for 200 ms. Fluorescence intensity of the entire cell was acquired for three prebleach iterations at a 500-ms interval and for 32 iterations after bleach at an adaptive time interval (∼600–800 ms). For each time point, the mean fluorescence intensity was measured in the area that encompassed kinetochore movement and in a similarly sized directly neighboring cytosolic area that was devoid of kinetochores throughout the experiment. Both areas were corrected for background, and the mean fluorescence of the cytosolic area was subtracted from the kinetochore area for each time point (area(KT − cyto)). For each measurement, the mean prebleach fluorescence intensity of the area(KT − cyto) was set to 100%, and the measured postbleach area(KT − cyto) signal was normalized to this value. Because a large volume of the cell was bleached, the total loss of YFP signal was calculated from the mean fluorescence recovery in the cytosol at the last three time points (mean fluorescence intensity postbleach/mean fluorescence intensity prebleach) and the postbleach area(KT − cyto) measurements were normalized for this loss in total fluorescence (area(KT −cyto)/[mean fluorescence intensity postbleach/mean fluorescence intensity prebleach]). Recovery half-times (ln(2)/rate constant) and signal recovery were determined by nonlinear curve fitting based on a one-phase association followed by a plateau using Prism software (GraphPad Software).
Fluorescence-assisted cell sorting
Cells were released from a 24-h thymidine-induced block into nocodazole for 16 h. All cells were harvested, washed once with PBS, and fixed in 70% ice-cold ethanol for 2 h. Cells were washed with PBST (PBS/0.1% Triton X-100), incubated with antiphospho-Ser/Thr-Pro antibody (MPM-2; EMD Millipore) in PBST for 1 h on ice, and washed again in PBST. Incubation with Cy3-conjugated donkey anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) was for 1 h on ice. After a final wash with PBST, DNA was stained with propidium iodide, and cells were treated with RNase A for 15 min and measured on a flow cytometer (FACS Calibur; BD). Flow cytometric analysis of transfected cells was based on Spectrin-GFP expression. As a control, a fraction of cells was lysed 48 h after transfection and analyzed by immunoblotting for expression of exogenous MPS1.
Plasmids and cloning
pOG44 (Invitrogen) encodes an Flp recombinase expression vector. The pSuper-based shRNA plasmids used in this study were mock, 5′-AGATTCTAGCTAACTGTTC-3′, and MPS1, 5′-GACAGATGATTCAGTTGTA-3′, as described previously (Jelluma et al., 2008b). pCDNA3-LAP-MPS1WT and pCDNA3-LAP-MPS1KD encode full-length, N-terminally LAP-tagged and shRNA-insensitive (modified codons 288 and 289) wild-type or kinase-dead (D664A) MPS1, respectively, and were described previously (Jelluma et al., 2008a). pCDNA3-YFP-MIS12-MPS1WT and pCDNA3-YFP-MIS12-MPS1KD were created by inserting the full MIS12 sequence into pCDNA3-LAP-MPS1 and were described previously (Jelluma et al., 2010). pEGFP-HEC1WT, a mammalian expression construct encoding N-terminally GFP-tagged full-length wild-type HEC1, and pEGFP-HEC19A and pEGFP-HEC19D (in which Ser4, Ser5, Ser8, Ser15, Ser55, Thr49, Ser55, Ser62, and Ser69 have been mutated to alanine or aspartic acid, respectively) have been described previously (Guimaraes et al., 2008). pCDNA3-LAP-MPS1Δ60 was created by introduction of an XhoI site at bases 174–179 of pCDNA3-LAP-MPS1WT and subsequent digestion with XhoI to excise bases 1–179 of MPS1. pCDNA3-LAP-MPS1Δ100 was created by introduction of an XhoI site at bases 294–299 of pCDNA3-LAP-MPS1WT and subsequent digestion with XhoI to excise bases 1–299 of MPS1. pCDNA3-LAP-MPS1Δ200 was created by introduction of an XhoI site at bases 594–599 of pCDNA3-LAP-MPS1WT and subsequent digestion with XhoI to excise bases 1–599 of MPS1. pCDNA3-LAP-MPS1ΔTPR was generated by PCR of the LAP tag and the first 186 bases of pCDNA3-LAP-MPS1WT using a reverse primer that contained a ClaI site and PCR of bases 577–1,995 of MPS1 with a forward primer that contained a NarI site and ligation of the ClaI site into the NarI site, creating a Ile-Ala linker. For generation of stable cell lines, MPS1 and HEC1 cassettes were subcloned into pCDNA5/FRT/TO vector. pCDNA5-FRT-TO-LAP-MPS1WT was created by ligation of the LAP-MPS1 module into the KpnI and ApaI sites of pCDNA5/FRT/TO. pCDNA5-FRT-TO-LAP-MPS11–192 was created by introduction of a stop codon at residue 193 of pCDNA3-LAP-MPS1WT and subsequent cloning of the MPS1 cassette into pCDNA5-LAP-MPS1WT with XhoI and ApaI. All other pCDNA5-FRT-TO-LAP-MPS1 constructs were created by ligation of the MPS1 cassette into the XhoI and ApaI restriction sites of pCDNA5-FRT-TO-LAP-MPS1WT. All pCDNA5-FRT-TO-FLAG-MPS1 constructs were created by ligation of a double FLAG tag into the BamHI and XhoI sites of pCDNA5/FRT/TO and subcloning of the MPS1 cassette into the XhoI and ApaI sites. All pCDNA3-YFP-MIS12-MPS1 constructs were created by ligation of the MPS1 cassette into the XhoI and ApaI restriction sites of pCDNA3-YFP-MIS12-MPS1WT. pCDNA5-FRT-TO-GFP-HEC1WT was created by digestion of pEGFP-HEC1WT, a gift of J. DeLuca (Colorado State University, Fort Collins, CO), with NheI and ApaI and ligation of the GFP-HEC1WT module into the XbaI and ApaI sites of pCDNA5/FRT/TO. pCDNA5-FRT-TO-GFP-HEC1STOP was generated by mutagenesis of the HEC1 ATG to TAG by site-directed mutagenesis. pCDNA5-FRT-TO-GFP-HEC1Δ80 was created by looping out bases 1–237 of pCDNA5-FRT-TO-GFP-HEC1WT by site-directed mutagenesis. pCDNA5-FRT-TO-GFP-HEC1Δ207 was created by looping out bases 1–618 of pCDNA5-FRT-TO-GFP-HEC1WT by site-directed mutagenesis. pCDNA5-FRT-TO-GFP-HEC19A was created by digestion of pEGFP-HEC19A, a gift of J. DeLuca, with NheI and ApaI and ligation of the GFP-HEC1WT module into the NheI and ApaI sites of pCDNA5-FRT-TO-GFP-HEC1WT. pCDNA5-FRT-TO-GFP-HEC19D was created by site-directed mutagenesis of pCDNA5-FRT-TO-GFP-HEC1WT using the full-length HEC19D gene, which was amplified by PCR from pEGFP-HEC19D-GFP, a gift of J. DeLuca, as a mutagenesis primer. pLacI-LAP was created by a LacI PCR from pKG194 (a gift from I. Cheeseman and K. Gascoigne, Whitehead Institute, Cambridge, MA) and subsequent cloning into the Nhe1 site of pLAP (pIC113). All pLacI-GFP-HEC1 constructs were created by subcloning of the GFP-HEC1 cassette from pCDNA5-GFP-HEC1 constructs into the SacII and AgeI sites of pLacI-LAP. The sequence encoding for residues 62–239 of MPS1 used for crystallographic experiments as well as all other protein expression constructs used in this study were cloned into the pETNKI-His-3C-LIC-kan vector by ligation independent cloning. The resulting construct was fused N terminally to the residues MAHHHHHHSAALEVLFQ-//-GPG, containing a human rhinovirus 3C protease cleavage site. All constructs were validated by sequencing of the full ORF.
Online supplemental material
Fig. S1 shows that MPS1 TPR lacks the characteristic KNL1-binding depression of BUB TPR domains and is monomeric in solution. Fig. S2 shows the phylogenetic analysis of the MPS1 TPR domain. Fig. S3 shows the expression of MPS1 and HEC1 in HeLa-FRT and HeLa Flp-in cell lines. Fig. S4 shows that N-terminal MPS1 mutants retain kinase activity and display normal residence time at unattached kinetochores. Fig. S5 shows that MPS1 localization is dependent on kinetochore–microtubule attachment status but independent of Aurora B phosphorylation of the HEC1 tail. A ZIP file is also provided that contains descriptions of background select, kinetochore select, and kinetochore measure macros used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201210033/DC1.
Supplementary Material
Acknowledgments
We thank Tatjana Heidebrecht for assisting in some cloning and purification experiments, Jonathan Grimes for help in autoSHARP phasing, Jennifer DeLuca for GFP-HEC1 constructs, Stephen Taylor for the HeLa Flp-In cell line, Ulrike Kutay and Patrick Meraldi for the HeLaK FRT TetR cells, Iain Cheeseman and Karen Gascoigne for the LacO/LacI system, and the Kops, Lens, Medema, and Rowland laboratories for insights and discussions.
This work was supported by an European Research Council starting grant (KINSIGN; to G.J.P.L. Kops), by the Dutch Cancer Society (KWF Kankerbestrijding; UU2012-5427; to G.J.P.L. Kops), by the European Molecular Biology Organization (long-term fellowship to E. von Castelmur), and by the Swiss National Science Foundation (PBBSP3-133408; fellowship to E. von Castelmur).
Footnotes
Abbreviations used in this paper:
- CH
- calponin homology
- FRT
- Flp recognition target
- HeLaK
- HeLa Kyoto
- lacO
- lac operator
- LAP
- localization and affinity purification
- NEB
- nuclear envelope breakdown
- NTE
- N-terminal extension
- TetR
- tetracycline repressor
- TPR
- tetratricopeptide repeat
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- Beaufils S., Grossmann J.G., Renault A., Bolanos-Garcia V.M. 2008. Characterization of the tetratricopeptide-containing domain of BUB1, BUBR1, and PP5 proves that domain amphiphilicity over amino acid sequence specificity governs protein adsorption and interfacial activity. J. Phys. Chem. B. 112:7984–7991 10.1021/jp711222s [DOI] [PubMed] [Google Scholar]
- Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., Bricogne G. 2004. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60:2210–2221 10.1107/S0907444904016427 [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Beaufils S., Renault A., Grossmann J.G., Brewerton S., Lee M., Venkitaraman A., Blundell T.L. 2005. The conserved N-terminal region of the mitotic checkpoint protein BUBR1: a putative TPR motif of high surface activity. Biophys. J. 89:2640–2649 10.1529/biophysj.105.063511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Kiyomitsu T., D’Arcy S., Chirgadze D.Y., Grossmann J.G., Matak-Vinkovic D., Venkitaraman A.R., Yanagida M., Robinson C.V., Blundell T.L. 2009. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure. 17:105–116 10.1016/j.str.2008.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Lischetti T., Matak-Vinković D., Cota E., Simpson P.J., Chirgadze D.Y., Spring D.R., Robinson C.V., Nilsson J., Blundell T.L. 2011. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure. 19:1691–1700 10.1016/j.str.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62:1002–1011 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- D’Arcy S., Davies O.R., Blundell T.L., Bolanos-Garcia V.M. 2010. Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J. Biol. Chem. 285:14764–14776 10.1074/jbc.M109.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H., Olsen J.V., Bairlein M., Gnad F., Oppermann F.S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. 2008. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 31:438–448 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., DeLuca J.G. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105:10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulla K., Daub H., Hornberger R., Nigg E.A., Körner R. 2010. Quantitative site-specific phosphorylation dynamics of human protein kinases during mitotic progression. Mol. Cell. Proteomics. 9:1167–1181 10.1074/mcp.M900335-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. 2011. Accelerated Profile HMM Searches. PLOS Comput. Biol. 7:e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hached K., Xie S.Z., Buffin E., Cladière D., Rachez C., Sacras M., Sorger P.K., Wassmann K. 2011. Mps1 at kinetochores is essential for female mouse meiosis I. Development. 138:2261–2271 10.1242/dev.061317 [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Johnston R.C., Smith D.L., Murray A.W. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871–882 10.1083/jcb.148.5.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt L., Tighe A., Santaguida S., White A.M., Jones C.D., Musacchio A., Green S., Taylor S.S. 2010. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1–C-Mad2 core complex. J. Cell Biol. 190:25–34 10.1083/jcb.201002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38(Suppl. 2):W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., van der Burg M., Szuhai K., Kops G.J., Medema R.H. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 333:1895–1898 10.1126/science.1210214 [DOI] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., McLeod I., Yates J.R., III, Cleveland D.W., Medema R.H., Kops G.J. 2008a. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 3:e2415 10.1371/journal.pone.0002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N., Brenkman A.B., van den Broek N.J., Cruijsen C.W., van Osch M.H., Lens S.M., Medema R.H., Kops G.J. 2008b. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 132:233–246 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Jelluma N., Dansen T.B., Sliedrecht T., Kwiatkowski N.P., Kops G.J. 2010. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J. Cell Biol. 191:281–290 10.1083/jcb.201003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. In press 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S., Stach M., Knapp M., Ortiz J., Pfannstiel J., Ruppert T., Lechner J. 2009. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28:1099–1110 10.1038/emboj.2009.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J.P.L., Shah J.V. 2012. Connecting up and clearing out: how kinetochore attachment silences the spindle assembly checkpoint. Chromosoma. 121:509–525 10.1007/s00412-012-0378-5 [DOI] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1–kinetochore subunit Knl1 interaction. J. Cell Biol. 196:451–467 10.1083/jcb.201110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Cleveland D.W. 2010. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J. Cell Biol. 190:21–24 10.1083/jcb.201006080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G., Cohen S.X., Lamzin V.S., Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3:1171–1179 10.1038/nprot.2008.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Thebault P., Freschi L., Beaufils S., Blundell T.L., Landry C.R., Bolanos-Garcia V.M., Elowe S. 2012. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J. Biol. Chem. 287:5988–6001 10.1074/jbc.M111.307355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.G. 2006. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62:48–57 10.1107/S0907444905039107 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23:127–128 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Liu S.T., Chan G.K., Hittle J.C., Fujii G., Lees E., Yen T.J. 2003. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 14:1638–1651 10.1091/mbc.02-05-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J., George K.A., Terret M.E., Zhang C., Shokat K.M., Jallepalli P.V. 2010. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190:89–100 10.1083/jcb.201001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., Nigg E.A. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Miller S.A., Johnson M.L., Stukenberg P.T. 2008. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr. Biol. 18:1785–1791 10.1016/j.cub.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin V., Prieto S., Melines S., Hem S., Rossignol M., Lorca T., Espeut J., Morin N., Abrieu A. 2012. CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr. Biol. 22:289–295 10.1016/j.cub.2011.12.048 [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67:355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Newman J., Egan D., Walter T.S., Meged R., Berry I., Ben Jelloul M., Sussman J.L., Stuart D.I., Perrakis A. 2005. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr. D Biol. Crystallogr. 61:1426–1431 10.1107/S0907444905024984 [DOI] [PubMed] [Google Scholar]
- Oppermann F.S., Gnad F., Olsen J.V., Hornberger R., Greff Z., Kéri G., Mann M., Daub H. 2009. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics. 8:1751–1764 10.1074/mcp.M800588-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Tighe A., D’Alise A.M., Taylor S.S., Musacchio A. 2010. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190:73–87 10.1083/jcb.201001036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., Musacchio A. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M., Kops G.J. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2:316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliedrecht T., Zhang C., Shokat K.M., Kops G.J. 2010. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PLoS ONE. 5:e10251 10.1371/journal.pone.0010251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 21:456–463 10.1093/bioinformatics/bti191 [DOI] [PubMed] [Google Scholar]
- Stucke V.M., Silljé H.H., Arnaud L., Nigg E.A. 2002. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 21:1723–1732 10.1093/emboj/21.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke V.M., Baumann C., Nigg E.A. 2004. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 113:1–15 10.1007/s00412-004-0288-2 [DOI] [PubMed] [Google Scholar]
- Sudakin V., Chan G.K., Yen T.J. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936 10.1083/jcb.200102093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., van Dam T.J., Karagöz G.E., von Castelmur E., Hubner N.C., Duarte A.M., Vleugel M., Perrakis A., Rüdiger S.G., Snel B., Kops G.J. 2012. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev. Cell. 22:1321–1329 10.1016/j.devcel.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181:893–901 10.1083/jcb.200712028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley J., Stukenberg P.T. 2011. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome Res. 19:377–391 10.1007/s10577-010-9180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink M., Simonetta M., Transidico P., Ferrari K., Mapelli M., De Antoni A., Massimiliano L., Ciliberto A., Faretta M., Salmon E.D., Musacchio A. 2006. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr. Biol. 16:755–766 10.1016/j.cub.2006.03.057 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Hoogendoorn E., Snel B., Kops G.J. 2012. Evolution and function of the mitotic checkpoint. Dev. Cell. 23:239–250 10.1016/j.devcel.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Vonrhein C., Blanc E., Roversi P., Bricogne G. 2007. Automated structure solution with autoSHARP. Methods Mol. Biol. 364:215–230 [DOI] [PubMed] [Google Scholar]
- Xu Q., Zhu S., Wang W., Zhang X., Old W., Ahn N., Liu X. 2009. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol. Biol. Cell. 20:10–20 10.1091/mbc.E08-03-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Wild T., O’Donohue M.F., Wandrey F., Widmann B., Gleizes P.E., Kutay U. 2009. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol. 185:1167–1180 10.1083/jcb.200904048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart P.H., Grosse-Kunstleve R.W., Lebedev A.A., Murshudov G.N., Adams P.D. 2008. Surprises and pitfalls arising from (pseudo)symmetry. Acta Crystallogr. D Biol. Crystallogr. 64:99–107 10.1107/S090744490705531X [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.