Abstract
Cells in the cardiovascular system are constantly exposed to complex mechanical stimulation due to the pulsatile nature of blood flow and the haemodynamic forces that are key to the regulation of vascular development, remodeling and pathophysiology. Mechanical stretch can also modulate the differentiation of stem cells toward vascular cell lineages (i.e., vascular smooth muscle cells), and represent a critical factor in vascular tissue engineering. Here we report on the development of a microchip platform that can emulate several key aspects of the vascular mechanical environment, such as cyclic stimulation and circumferential strain. This chip consists of an array of microfluidic channels with widths ranging from 20 to 500 micrometers. These channels are covered by suspended deformable membranes, on which cells are cultured and stimulated by cyclic circumferential strain of up to 20% via hydrodynamic actuation of the fluid in the microfluidic channels, thereby mimicking the biomechanical conditions of small blood vessels. We show that human mesenchymal stem cells (MSCs) can be cultured and continuously stimulated by cyclic stretch over a period of 7 days with no evidence of device fatigue or performance degradation. We observed localization and alignment of MSCs when mechanical stretch is larger than 10%, indicating the importance of mechanical stimulation in modulating cellular behavior. We further demonstrated simultaneous detection of proteins in multiple signaling pathways, including SMAD1/SMAD2 and canonical Wnt/β-catenin. This microchip represents a generic and versatile platform for high-throughput and rapid screening of cellular responses, including signal transduction cascades, in response to mechanical cues. The system emulates the physiological conditions of blood vessels and other tissues that are subject to cyclic strain, and may have a wide range of applications in the fields of stem cell mechanobiology, vascular tissue engineering, and other areas of regenerative medicine.
Introduction
Mechanical stimulation is an important modulator of cell fate and function, and plays a crucial role in tissue development and disease. In the vascular system, cells including endothelial cells and smooth muscle cells experience complex mechanical stimuli such as shear, tensile stresses, and compressive strains. While endothelial cells constantly experience shear stress exerted by pulsatile blood flow, smooth muscle cells that constitute the majority of vascular tissue in the medial layer are mainly under cyclic circumferential stretch and compressive pressure. Significant evidence has shown that these mechanical stimuli strongly influence vasculature development, remodeling and pathogenesis, implying that in vitro study of vascular cell biology should be performed in conditions that emulate the in vivo mechanical environment of the vascular system.
Mesenchymal stem cells (MSCs) are self-renewing, multipotent adult stem cells. They can be differentiated into osteoblasts, chondrocytes, adipocytes, and other connective tissue cell types1. They have been reported as one of the most promising cell sources for cardiovascular regenerative medicine2-7. Several groups have shown directed differentiation of MSCs to vascular smooth muscle cells8-10, yet the efficiency is still low. Recent studies indicate that mechanical forces significantly influence the differentiation of human stem cells including MSCs. For example, cyclic uniaxial stretch was found to be a key factor driving the differentiation of bone marrow derived MSCs toward smooth muscle cell lineages in both 2D and 3D cultures8,11-13. In a study that exploited a novel bioreactor with pulsatile mechanical actuation to growth blood vessels, cyclic strain alters the production of collagen molecules and induces extracellular matrix remodeling, which contributes to enhanced mechanical properties14. Due to the complexity of tissue microenvironments and the interplay of mechanical stimuli with a variety of biochemical cues, a platform that can execute high throughput examination of cellular responses to mechanical and biochemical cues in a combinatorial manner could have broad applicability.
Recently, substrate deformation-based mechanical stimulation platforms have been reported, in which cells are stretched in either a uniaxial or bi-axial manner. Several parameters can be tuned independently using these platforms. The Braille display-actuated system allows for rapid adjustment of stimulation frequency, but is unable to probe cellular response over a wide range of strain magnitudes15. In addition, this type of device does not generate a uniform strain field. Microdevices developed by Tan and colleagues are capable of detecting cellular responses to different strains, and cells cultured on this device were found to differentially respond to distinct strain fields and magnitudes16. In this system, the strain field is not uniform; instead, it continuously changes across the whole culture surface with no apparent boundary, and thus it is difficult to quantitatively compare cellular behavior and the strain magnitude. The FlexCell system (FlexCell International Cooperation, Hillsborough, NC, USA) is the most versatile technology that allows for the application of either tensile or compressive strains to cells cultured in six testing wells in parallel17. But the Flexcell system requires a large number of cells (on the order of 1 million or more) for a single experiment, and cannot test different mechanical environments in a high-throughput manner, which was improved by a miniaturized FlexCell-type microdevice recently reported by Simmon et al.18 These technologies have successfully incorporated mechanical cues into cell culture systems, but none of them was designed to emulate the mechanical conditions in the vascular system, especially small blood vessels (<1 mm). Most prior systems are incapable of conducting combinatorial experiments using mechanical and/or biochemical cues, and cannot examine an array of signaling pathways for comprehensive investigation of the molecular mechanisms underlying vascular biomechanics and development.
Herein we report the development of a microfluidics platform that can emulate several key aspects of vascular mechanical conditions, such as cyclic stimulation and circumferential strain, on a hemi-cylindrical substrate. The system is comprised of an array of microchannels (20, 50, 100, 200 and 500 micrometers in width) spanning the range of sizes from capillaries to small arteries or arterioles. The microchannels are covered by suspended, deformable membranes that act as the “vessel” wall, and cells cultured on these membranes can be stimulated by cyclic quasi-circumferential strain of up to 20%, via hydrodynamic actuation of fluid in the channels. A multi-well cell culture system was built on top of the membrane to allow for combinatorial examination of cellular responses. We demonstrate that human MSCs can be cultured on this device for 7 days and remain viable under continuous cyclic stretch over 6 days using this platform, and that the device shows no evidence of fatigue or performance degradation over this time frame. Six days is by far the longest testing time reported for cell cultures on a microchip that provides cyclic mechanical stimulation. To show the ability to investigate the response of cell signal transduction to dynamic mechanical loading, we managed to perform immunofluorescence (IF) assay simultaneously on multiple signaling pathways including, SMAD1/SMAD2 and the canonical Wnt/β-catenin pathway. The microchip may have a wide range of applications in the fields of stem cell mechanobiology, vascular tissue engineering, and other areas of regenerative medicine.
Results
Microchip design and operation
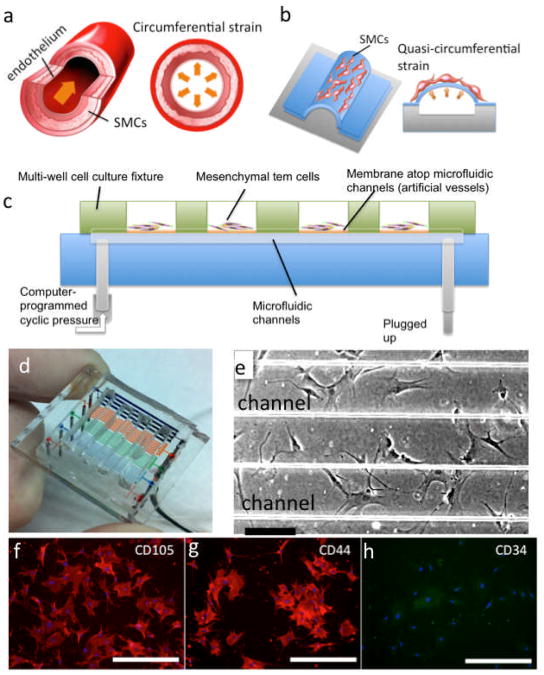
Our microfluidic device was designed to simulate vasculature deformation under hydrodynamic pulsation. Native arteries are circumferentially stretched during each cardiac cycle (Fig. 1a). The microfluidic vessel chip we designed has an array of thin membranes bonded atop microchannels to form closed artificial “vessels”. When a hydrostatic pressure is applied to the liquid-filled microchannels, the membrane inflates outward (Fig. 1b). When the pressure is released, the membrane returns to its original position. Repeating this process results in a cyclic actuation of the membrane that recapitulates the deformation of native arteries under hemodynamic pressure. In our experiments, the microfluidic artificial vessels were pressurized and stretched by using an external compressed nitrogen gas line. The frequency of cyclic strain is controlled using a LabVIEW program that operates a set of solenoid valves (see Materials & Methods).
Figure 1.
Design and operation of the microfluidics artificial “vessel” chip. a. Mechanical condition of native arteries. b. Schematic depiction of microfluidic artificial vessels for generating quasi-circumferential strain resembling the mechanical strains in blood vessels. c. Schematic depiction of fully packaged microfluidic culture system. d. Photograph of a packaged microchip. e. Human mesenchymal stem cells cultured on two microfluidic vessels. (Scale bar = 200 μm) f-h. Immunofluorescence staining of cell surface markers CD105, CD44, and CD34. (Scale bar = 400 μm)
Our device is comprised of five sets of parallel microchannels made of polydimethylsiloxane (PDMS) with widths of 20, 50, 100, 200, and 500 μm to mimic blood vessels ranging from capillaries to small arteries (Fig. 1d). These microchannels are sealed with a layer of PDMS membrane that can be actuated by the hydrodynamic pressure underneath the suspended membrane. On top of this device, a multi-well PDMS slab is assembled, which serves as the cell culture wells for multiplexed analysis of cellular responses. The microchip is fabricated using a two-step molding and bonding soft lithography approach (see more details in Fig. S1 and Materials & Methods). The cells cultured atop the microchannels would experience a mechanical condition similar to that for smooth muscle cells in the blood vessel wall. On a single chip, a set of microchannels with different widths are operated in parallel, ideally giving rise to the test of a range of mechanical strains in the same run; the channels of smaller diameters produce smaller strains and the larger channels result in higher strains under the same pressure. Given the versatility and high-throughput of this device for cell mechanical stimulation, we would like to point out two issues that are associated with the current device design. First, the edges of the PDMS membrane are rigidly fixed to the underlying thick PDMS substrate. Upon the application of an external pressure, the region at the center of the membrane undergoes largest strain whereas the region close to the microchannel edges that is anchored to the bulk PDMS substrate exhibits minimal deformation. This edge effect is more significant at the smaller channels (<100 μm). The strain distribution in the middle of the large channel membranes (>200 μm) can be nearly uniform if the membrane is infinitely thin. Second, the suspended membrane in the current device is, however, relative thick (∼35 μm) such that the ratio of membrane thickness t to the channel width w become another factor strongly influencing the membrane deformation and the strain field. In our device, the t/w ratio varies from 1.75 to 0.07 with the channel width increasing from 20 μm to 500 μm. When t/w is large (∼1), the suspended “membrane” is essentially a thick block of PDMS and appears very rigid. Only when t/w ≪ 1, the suspended membrane is sufficiently compliant and can be viewed as a free-standing thin membrane. Both the edge effect and the membrane thickness effect contribute to reduced deformability of the membrane. These issues can be partly resolved by decreasing the membrane thickness and consequently increase the t/w ratio, which is being under investigation in our laboratory. With respect to the current microchip described in this paper, the circumferential strain of the membranes atop the small channels (<100 μm) is nearly equal to zero.
In order to successfully culture human mesenchymal stem cells on the microchip and improve the adhesion of cells to the suspended membrane, the membrane was treated by coating a thin layer of extracellular matrix molecule (e.g., gelatin, collagen I, fibronectin, etc.) (see Materials & Methods). With a multi-well cell culture fixture, human MSCs were seeded on the membrane within the wells (Fig. 1c). It was observed that MSCs adhered to and spread on the PDMS surface within 6 hours and maintained the shape, morphology and dimensions characteristic of MSCs cultured in standard 2-dimensional culture. During long-term culture in the device, the microchip was taken out of the incubator every one or two days to replace the culture medium and allow for periodic imaging assessment. Figure 1e shows the MSCs cultured on the chip for 3 days - all cells appear viable and healthy. IF staining was performed for three MSC markers to ensure the cells remain MSC phenotype. As shown in Figures 1f, g and h, cells were positive for both CD105 and CD44, which are key MSC markers, but negative for CD34, a hematopoietic surface marker19. The results confirmed that the cells maintain the MSC phenotype when cultured on gelatin-coated PDMS membranes of the microchip.
Characterization of strain and applied pressure
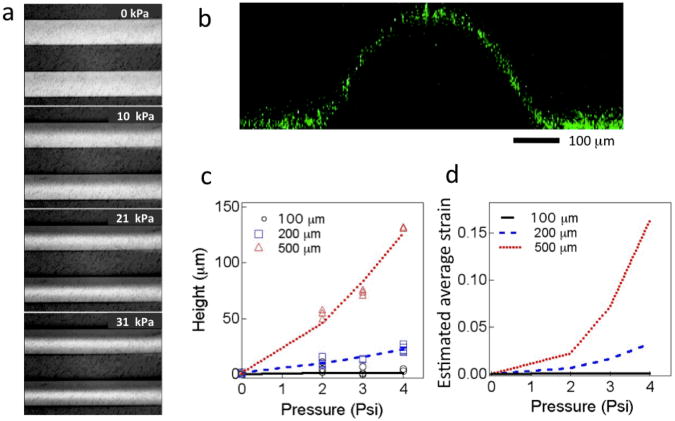
Pressure applied to the fluid-filled microchannels dictates the magnitude of the strain. First, we directly visualized the membrane deformation in 200 μm microchannels with pressures ranging from 0 to 10, 21, and 31 kPa (Fig. 2a). When the pressure was released, the membrane fully recovered to the initial position with no hysteresis, indicating the feasibility of actuating these membranes at a frequency similar to the human cardiac cycle (i.e., 1 Hz). We also used fluorescent nanoparticles (fluorescein-loaded PLGA nanoparticles, ∼100 nm in diameter) casted onto the suspended membrane to perform 3D confocal imaging to confirm membrane deformation. Figure 2b shows the cross-section of a 500 μm-wide membrane with the application of ∼21 kPa inner pressure. To quickly obtain the magnitude of membrane deformation and perform calibration of each device, we exploited a precision micrometer equipped on a high-resolution light microscope. Membrane deformation was measured by measuring the change of the focal plane in response to the pressure applied using gold nanoparticles as the tracer. Gold nanoparticles (diameter ∼300 nm) were drop-casted onto the membrane to enable accurate focusing in the z (height) direction. We measured the maximum height at the center of the deformed membrane as a function of the pressure applied to all the five sets of microchannels (20, 50, 100, 200 and 500 μm) (Fig. 2c). As anticipated, the membranes on the small channels (20 and 50 μm) do not show detectable deformation by measuring height change. To estimate the strain, we assume the membrane deformation adapts a cylindrical profile and then compute the average circumferential strain (Fig. 2d) using a simple mathematic model described in Supplementary Materials: Method. The wide channels allow for a significantly large range of strain magnitude (e.g., in the current device, we can obtain an estimated average strain up to 20% for 500 μm channels and 7% for 200 μm channels through the application of an external pressure of 28 kPa). Reducing the thickness of the PDMS membranes and applying higher pressures may further increase the strain magnitude, even for the membrane atop the small channels.
Figure 2.
Calibration of mechanical strain. a. Optical micrographs showing the deformation of suspended membranes upon the application of hydrostatic pressure (0-31.0 kPa) to the fluid in the 200 μm-wide microchannels. b. Confocal fluorescence image showing the cross-section of a deformed membrane on top of a 500 μm microchannel. Fluorescein-loaded nanoparticles were casted onto the membrane surface as the tracer. c. Membrane deformation (height) measured under varying pressures applied. d. Computed average circumferential strain as a function of the external pressure applied.
Long-term culture of MSCs under dynamic mechanical stimulation
To evaluate the durability and robustness of the device for long-term cell stimulation, we cultured human MSCs on the chip for up to 7 days, among which 6 days subject to continuous cyclic stretch (pressure = 21 kPa, close to the normal blood pressure region) for totally 520,000 cycles. During the experiments, the human MSCs were cultured statically during the first day and then further cultured under dynamic stimulation for 2 or 6 days. Cells continued to display the typical spindle and polygonal MSC morphology after cyclic mechanical stimulation (Fig. S2). The cells located on the suspended membranes adhered well, with no distinguishable difference as compared to the cells located between microchannels, which were not subjected to direct mechanical stimulation. All of the devices remained functional after 6 days of cyclic action, and the membranes were intact with no cracks or delamination. To our best knowledge, this result represents the longest culture of cells to date on a microchip that provides cyclic mechanical stimulation.
Cell alignment and localization induced by cyclic mechanical stimulation
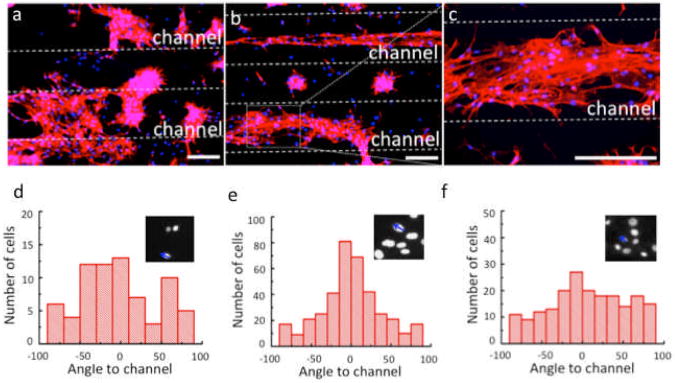
Mechanical cues can significantly influence the behavior of cells including cell adhesion, spreading, morphology, and migration. After the MSCs were cultured on chip under cyclic stretch, the cells were fixed, stained with Alexa Fluor 647 conjugated phalloidin for actin filaments and DAPI for nuclei, and then visualized by fluorescence microscopy. First, we performed a static control experiment in which MSCs were cultured on the microfluidic chip without the application of cyclic stretch. These cells showed minimal alignment on the membrane surface including on the widest channels (Fig. 3d). The degree of cell alignment is defined based on the angle between the longitudinal direction of the nucleus and the direction of the microchannels (Fig. 3d, inserts). It was suspected that the geometrical effect of the one-dimensional microchannels may induce preferential cell alignment along the channels, but this was not observed in our current device presumably because the cells were cultured on top of the membrane and could not sense the topological variation in the underneath microchannels. Second, in a separate experiment, MSCs were cultured statically on the microchip for 1 day to allow the cells to adhere and spread on the surface (Fig. 3a). Then these cells were stimulated with cyclic stretching (∼10% circumferential strain) for 6 more days (totally 7 days). The cells on the 500 μm channels unambiguously show preferential alignment along the direction of the microchannels (Fig. 3b and 3c). Quantitative analysis of cell alignment was performed by measuring nuclear orientation (Fig. 3e before cyclic stimulation, Fig. 3f after cyclic stimulation for 7 days). The degree of cell alignment is defined based on the angle between the longitudinal direction of the nucleus and the direction of the microchannels (Fig. 3e&f, inserts). When the absolute angle is less than 30°, this cell is considered as being aligned parallel to the channels. The results showed that ∼70% of the MSCs cultured with cyclic stretching were aligned, while the cells cultured without cyclic stretching showed relatively random angle distribution within the whole range from -90° to 90°. Interestingly, we found that cells in the vicinity of these channels appear to migrate toward the channel center. This could be attributed to the edge effect – the strain field is the highest in the center of the membrane and decreases in the region at the edges. Third, on the same microfluidic chip, neither cell alignment nor localization was observed for the cells cultured on the smaller microfluidic vessels (≤ 200 μm), on which the circumferential strain is very small. Finally, another experiment that performed cyclic stretch in a similar manner, but at lower circumferential strains, i.e., 5% or 7.5% circumferential strain on 500 μm channels, were not able to induce cell alignment or localization, either. All these results indicate that preferential cell alignment was caused by dynamic mechanical stimulation and there exists a possible threshold strain (∼10%) for the induction of MSC alignment. This result is in agreement with a prior study, which showed that MSCs were aligned perpendicularly to the axis of cyclic uniaxial strain, i.e., 10%, to reduce the effective stress in their cytoskeleton11,13.
Figure 3.
Cyclic strain-induced cell alignment and localization. a. Immunofluorescence image showing nuclear and cytoskeleton staining of the cells statically cultured for 1 day. b. Immunofluorescence image showing nuclear and cytoskeleton staining of the cells cultured under cyclic stretching for 6 more days (totally 7 days). c. Enlarged view of a selected area in b. nuclei (blue), actin filaments (red). Quantify the cell alignment according to the angle between the longitudinal direction of nuclei and the direction of microchannels (insert in d, e, and f) for 7-day static cell culture (d), 1-day static cell culture (e) and 7-day dynamic cell culture (f). (Scale bar = 400 μm)
Signaling pathways in response to dynamic mechanical stimuli
In the next three sections, we show the applicability of our microfluidic chip to the study of cell signaling in response to dynamic mechanical stimulation. We used a multi-well PDMS slab to perform parallel IF studies of proteins involved in cell signaling. In a typical setup, a four micro-well fixture was assembled on top of the microfluidic chip. In each well, there are five sets of microchannels (20, 50, 100, 200 and 500 μm). This in principle constitutes 20 testing conditions per chip. We chose to use two microwells for signaling protein staining, and two other microwells for MSC phenotype and morphology (nuclear/cytoskeleton) staining, respectively. A dual color detection scheme was employed to study signaling pathways, and thus we chose to examine four signaling proteins associated with TGF-β/SMAD and canonical Wnt/β-catenin signaling cascades (Fig. S3). SMAD signaling molecules are intracellular intermediates that are activated by the TGF-β superfamily20, and are involved in either maintaining the undifferentiated state of MSCs, or in guiding the differentiation of MSCs along a defined lineage21,22. TGF-β superfamily ligands bind to their receptors, leading to the phosphorylation of the receptor-regulated SMADs, which further bind with a coSMAD, SMAD4, to form a complex, together translocating into the nucleus to express transcription factors23. SMAD1 is activated by bone morphogenesis proteins (BMPs), i.e., BMP2, BMP4, and BMP7, and might play an essential role in maintaining the multipotency of MSCs. SMAD2 is activated by activins and TGF-βs, e.g., TGF-β1. TGF-β1 has been demonstrated to enhance the differentiation from MSCs into vascular smooth muscle cells (SMCs)8,24,25. Canonical Wnt signaling also plays a critical role in MSC proliferation and in maintaining MSC in an undifferentiated state26. β-catenin is a key component in canonical Wnt signaling (Fig. S3). In the absence of Wnts, β-catenin in the cytoplasm goes through proteolysis via a “destruction complex” formed by GSK-3β. However, when Wnt signaling is on, GSK-3β is recruited toward the cell membrane, and β-catenin is stabilized in cytoplasm. β-catenin then translocates into nucleus to alter gene expression.
Herein, we performed IF measurements of four signaling proteins, i.e., Phospho-SMAD1, SMAD2/Phospho-SMAD2, β-catenin/Phospho-β-catenin, and GSK-3β. We evaluated MSCs that were statically cultured on the chip for 1 day, and then dynamically cultured with the application of cyclic mechanical stimulation for 2 more days (total 3 days) and 6 more days (total 7 days) (Supplementary Fig. S4), respectively. The cells at day 1 were used as the reference. It has been reported that mechanical strain can enhance the expression of SMC markers in differentiating MSCs at a stage as early as in three days11. Both early stage (α-actin) and intermediate stage (calponin) SMC markers could be observed in MSCs under uniaxial dynamic mechanical stretch after 7 days13. Thus, we examined the signaling proteins of MSCs cultured for 1 day, 3 days, and 7 days for comparative study.
TGF-β/SMAD signaling
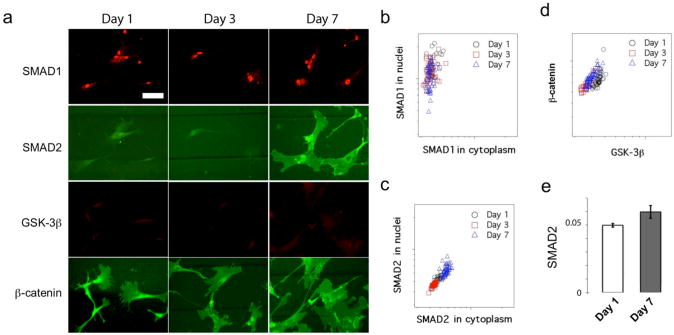
The data collected on 200 μm microfluidic vessels were selected as representative results of the TGF-β/SMAD signaling (Fig. 4a). The average circumferential strain on these channels was 1.6%. We observed that phospho-SMAD1 is highly accumulated in nucleus from the beginning of cell culture to the end of the experiment after 7 days, suggesting a possible constitutive activation of the SMAD1 pathway in these cells. While it has been reported that BMP/SMAD1 signaling helps to maintain the embryonic stem cell phenotype in mouse27, the reported role of SMAD1 in MSCs is complex. In our experiments, SMAD1 was constantly activated throughout culture, as assessed by IF. We further conducted a single cell quantitative fluorescence analysis. The results are summarized as a scatter plot (Fig. 4b) for the cells cultured on chip for 1, 3 and 7 days. This plot shows the SMAD1 fluorescence intensity in the nucleus vs. in the cytoplasm quantified on all single cells and each dot represents a single cell. Quantitative analysis also shows that the phospho-SMAD1 signal is localized in nucleus (all the dots are closer to the y-axis), which indicates the activation of the SMAD1 signaling pathway regardless of the time of cell culture on the microchip. Persistent activation of SMAD1 signaling we observed is likely due to the conditioned cell culture medium, which contains 10% fetal bovine serum (FBS) and might have proteins that function as the BMP-like ligand to induce SMAD1 signaling pathway activation.
Figure 4.
Cell signaling pathways in response to cyclic mechanical stimulation. a. Immunofluorescence images showing SMAD1, SMAD2, β-catenin, and GSK-3β expression in human mesenchymal stem cells statically cultured for 1 day, dynamically stimulated for 2 additional days (totally 3 days) and for 6 days (totally 7 days) on 200 μm channels, respectively. (Scale bar = 100 μm). b,c, and d are scatter plots showing single cell SMAD1 intensity in nuclei vs. in cytoplasm (b), single cell SMAD2 intensity in nuclei vs. in cytoplasm (c), and the intensity of β-catenin vs. GSK-3β in cell (d). The circle, square and triangle symbols correspond to cells examined at different time points (1, 3, and 7 days). e. Comparison of SMAD2 between Day 1 and Day 7 samples shown in (c). It shows the average and the variation of SMAD2 intensity from all single cells in each sample. These two samples show significant difference (p<0.01).
In three of the five devices we tested for 7 days, the IF intensity of total SMAD2 became significantly elevated in the MSCs upon constant cyclic mechanical stimulation. Only modest or little elevation of the IF intensity of total SMAD2 was observed on the other two devices, especially when cells were at high density (>200 cells/mm2) and exhibited strong alignment. These results suggest that the expression of total SMAD2 is likely to be responsive to mechanical stimulation, but the whole system is complex and other factors such as cell-cell contact and preferential alignment may also contribute to SMAD2 signaling. It is well known that the activation of SMAD2 signaling pathway requires translocation of phosphorylated SMAD2 from cytoplasm to nucleus to elicit gene expression, including the genes for specification and differentiation of MSCs into SMCs. We further performed quantitative single-cell fluorescence analysis and the results (scatter plot of nuclear SMAD2 vs. cytoplasm SMAD2, Fig. 4c) did not show apparent nuclear accumulation of total SMAD2 although the overall IF intensity of total SMAD2 slightly changes with increasing mechanical strain. We also co-stained phospho-SMAD2 with total SMAD2. Their signals overlapped with each other although the phospho-SMAD2 intensity was relatively weaker as compared to total SMAD2 (Supplementary Fig. S8). There is a slight increase of IF intensity of total SMAD2 in cytoplasm in the Day 7 sample as compared to the Day 1 and Day 3 samples, which is also confirmed by statistic analysis of all single cells imaged on the 200-μm microchannels (Fig. 4e). To further assess the effect of cyclic stretch on SMAD2, we conducted quantitative analysis of SMAD2 signal on the cells cultured on all the five sets of microchannels (Supplementary Fig. S5). The IF intensity of total SMAD2 in cytoplasm remained undetectable in the cells cultured on microchannels smaller than 100 μm, where the circumferential strain is less than 0.5%. The wider channels that generate greater mechanical strain gave rise to higher SMAD2 signals (Supplementary Fig. S6). For example, the normalized fluorescence intensity of SMAD2 slightly increased from the base line 0.0047 (20 μm microchannels, set as reference, strain = 0%) to 0.0059 (200 μm microchannels, strain = 1.6%, P < 0.01) and 0.0072 (500 μm microchannels, strain = 7.2%, P < 0.01), suggesting a possible link between the SMAD2 expression and dynamic mechanical stimulation.
Wnt/β-catenin signaling
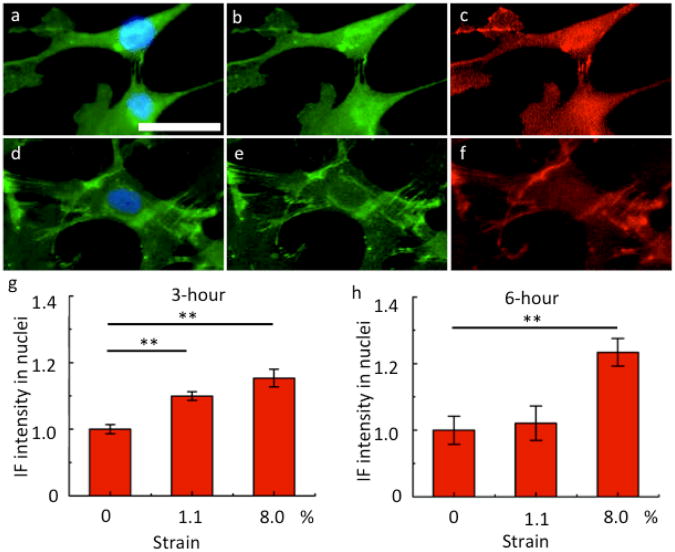
We measured the IF intensity of β-catenin and GSK-3β using a dual-color IF analysis of the cells cultured on five different microfluidic vessels on Day 1, 3 and 7. We observed that the IF intensity of β-catenin in whole cell remains high during the entire course of mechanical stimulation. However, we only observed negligible nuclear accumulation of β-catenin. We also examined the IF intensity of β-catenin in the cells cultured on different microchannels and observed little dependence of β-catenin on the magnitude of mechanical strain (Supplementary Fig. S7). The IF signal of GSK-3β remained low in all the MSC samples throughout the entire course of mechanical stimulation. The scatter plot of β-catenin vs. GSK-3β from single cell quantitative fluorescence analysis (Fig. 4d) also confirmed this observation, which is as expected because GSK-3β acts as an inhibitor to the Wnt/β-catenin signaling pathway through the ubiquitination of cytoplasmic β-catenin proteins. Simmons and his colleagues reported a slight increase of nuclear accumulation of β-catenin as an early response to biaxial cyclic strains on MSCs within the first several hours (3 to 6 h), which was noticeable only when the strain was larger than 8%18. But the nuclear accumulation of β-catenin returned to its basal level at 6 h for strain > 15%. We suspect this could be the reason why we did not observe nuclear enrichment of β-catenin in our long time experiment. Then we performed a set of short period experiments (i.e., 3 hours and 6 hours) on MSCs in response to dynamical mechanical stimulation. Indeed, we observed slight nuclear enrichment of β-catenin/Phospho-β-catenin with the application of dynamic stimulation as shown in the immunofluorescence images, although the Phospho-β-catenin signal was weaker than the β-catenin signal (Fig. 5a-f). At 3 hours, notable nuclear accumulation of β-catenin was observed for the average strain applied at both 1.1% and 8% (p < 0.01) (Fig. 5g). But at 6 hours, significant nuclear accumulation was only observed when strain is as large as 8% (p < 0.01) (Fig. 5h). Our results are in agreement with the Simmon's results18 despite of the difference in cell type, culture condition and device design.
Figure 5.
β-catenin signaling in response to short-time cyclic mechanical stimulation. a. Immunofluoscence image showing β-catenin (green) in the cells cultured with cyclic stretching for 3 hours. d. Immunofluoscence image showing β-catenin (green) in the cells cultured with cyclic stretching for 6 hours. Nuclei are stained with DAPI (blue). b and e are single channel (green) images showing β-catenin only. c and f are single channel (red) images showing Phospho-β-catenin only. (Scale bar = 50 μm) g and h are quantitative analyses of normalized fluorescence intensity (with respect to the fluorescent intensity at zero strain) in nuclei under different average circumferential strains at 3 hours (g) and at 6 hours (h). ** p<0.01, n>100. The error bars represent the standard error of the mean.
Discussion
In this study, we report the development of an integrated microfluidic vessel chip, which is versatile, durable and high-throughput platform for the assessment of cellular responses to mechanical stimulation, and which emulates the physiological conditions of the vascular system. This platform in principle allows for simultaneous operation of five different artificial vessel channels with the sizes ranging from 20 μm to 500 μm. The conventional FlexCell device is a macroscopic mechanical stimulation system, while a miniaturized testing array chip developed by Simmons and colleagues contains cyclic loading units that are 0.8 mm and above. It was estimated that the application of a modulated external pressure at 21 kPa can generate mechanical strain of up to 10%, which is comparable to the physiologically-relevant value in the vascular system, on the large channels (500 μm). Higher strain magnitude can be achieved by reducing the membrane thickness. Hence, this system can simulate the mechanical conditions of small blood vessels or even capillaries.
A multi-well PDMS slab was designed to enable culture of multiple samples at the same time. The current design allows for 20 testing conditions to be incorporated onto a single chip the size of half of a microscope slide. In each condition there are four to eight microfluidic vessels for parallel measurements, giving rise to high throughput and high content as compared to conventional cell mechanical stimulation systems. If a microfluidics-based cell culture chip (channels 200 μm in width) were designed and placed on top of the artificial “vessel” chip, the multiplexing capability could be further increased to as much as 200 conditions per chip, making the platform even more appealing for drug screening and signaling pathway profiling. In addition, the quantity of cells required to perform a complete test using this microchip platform is much less than using conventional technologies. In the typical experiments, <200 cells were used for each testing condition, which may represent a unique advantage when the sample size is limited (e.g. rare cells from primary tissues or stem cell isolates). It is challenging to conduct conventional protein or gene expression analysis of very small numbers of cells retrieved from our microchip using western blotting or PCR. However immunofluorescence assay and high-content imaging still allow for quantitative cell signaling analysis on chip.
This multi-channel platform is robust and durable for long-term mechanical stimulation of cultured cells. We demonstrated successful culture of human MSCs on the microchip under continuous cyclic stretching for 6 days and observed no fatigue or performance degradation. The successful long-term culture of MSCs on this microchip indicates the feasibility of applying this platform to investigate the entire course of MSC differentiation into vascular SMCs or other cell types under dynamic mechanical stimulation. Using this technology, we observed that mechanical cues do influence the behavior of cells, including cell morphology and orientation. MSCs cultured under >10% dynamical mechanical stretch showed preferential alignment along the longitudinal axis of the microchannels. On a single chip, multiple mechanical conditions can be created in parallel for high throughput assessment of cellular responses to mechanical stimulation.
Though a 2D chip design provides advantages for handling and imaging cells, the suspended membranes on microchannels do not completely replicate the mechanical characteristics of blood vessel walls in terms of the strain field distribution. The cells cultured in the center of the membrane experience greater strains than those located at the edges. The non-uniformity of the strain field is less significant for wider microchannels. This issue may be reduced by decreasing the membrane thickness and selectively seeding cells at the center of membranes using advanced patterning techniques such as micro-contact printing or inkjet printing.
We further demonstrated the ability of performing cell signal transduction analysis on chip in response to dynamic mechanical stimulation. SMAD1 was constitutively activated regardless of mechanical stimulation, which was presumably due to the culture medium condition required for MSC self-renewal and growth. It is known that TGF-β-induced SMAD2 activation is one of the key mechanisms that drive the differentiation of MSCs into vascular SMCs. In some cases, we observed a slight increase of IF intensity of SMAD2 in cytoplasm, but did not find significant SMAD2 accumulation in the nuclei. This observation may suggest a very early event in the SMAD2 signaling process, prior to the nuclear translocation and full activation of SMAD2 signaling pathway. The canonical Wnt/β-catenin signaling pathway was also studied. MSCs cultured on the chip showed constant expression of β-catenin while the expression of GSK-3β, a β-catenin signaling inhibitor, remained low. This observation appeared to be independent of the extent of mechanical strain applied to the cells. We did not observe apparent accumulation of β-catenin in nuclei in the long-time culture experiments. A slight increase of nuclear accumulation of β-catenin was observed in the cells treated for very short periods such as several hours. Overall, these results demonstrate that the artificial microfluidic vessel chip is a versatile platform for high throughput investigation of cell signaling pathways. However the cellular biology in the condition of dynamic mechanical stimulation is very complex and further studies are required to make definitive conclusions about the associated cell signaling pathways.
Materials and Methods
Device fabrication
This device consists of two layers of poly(dimethylsiloxane) (PDMS). The lower layer is parallel microgrooves fabricated by soft lithography and the top layer is a free-standing thin membrane. The microgrooves were replicated from a 1 × 1 in2 SU-8 negative photoresist pattern on a silicone wafer. The thin membrane (∼35 μm in thickness) was obtained by spin-casting PDMS on a flat silicone wafer at 2000 rpm for 60 s. In order to achieve a strong bonding between the membrane and the microgrooves, these two layers were prepared by PDMS with cure agent to base solution ratio of 1:20 (membrane) and 1:5 (microgrooves), respectively28. These two layers were separately cured at 80 °C for 30 min (membrane) and 50 min (microgrooves), and then were bonded together to further cure at 80 °C overnight to achieve extra crosslinking between the interface of the two layers and strengthened adhesion.
Computer interface and microfluidics control system for programmable cyclic loading
In order to obtain accurate and programmable device control, both a pressure control system and a computer control system were applied. First, the microchannels were filled with water with one end closed and the other end connected to a pressure system controlled by solenoid valves. The inner pressure of each set of microchannels could be adjusted separately to achieve different mechanical strain of the suspended membranes. Inside one set of microchannels, the inner pressure was equal everywhere due to the quasi-static conditions. The accuracy of the adjustable inner pressure was 3.5 kPa. The solenoid valves were controlled by a computer system through a Data Acquisition Card (National Instruments Corporation, Austin, TX, USA). The on/off frequency and duration of the solenoid valves were programmed by LabVIEW 8.6 (National Instruments Corporation, Austin, TX, USA), which determined the frequency and duration of the dynamic mechanical pulsing applied to the microfluidic devices. A constant inner pressure was applied to all channels to obtain different strain of suspended membranes. The inflation and deflation duty cycles were the same, and lasted for 50% of the entire cycle. The pulsing frequency was 1 Hz, roughly mimicking the human heart rate.
Human mesenchymal stem cell culture
Human Mesenchymal Stem Cells (hMSCs) were purchased from Lonza and cultured in Poietics™ MSCGM (Lonza Group Ltd., Switzerland) to maintain their undifferentiated status. The hMSCs used in the experiments were at passage 3-7. These cells were seeded onto PDMS device at a density of ∼25000 cells/cm2 and placed in an incubator under 5% CO2.
Device surface functionalization
The PDMS surfaces were modified by ECM molecules, e.g., collagen I, fibronectin, and gelatin, before cell seeding in order to improve cell adhesion. A 20 μg/ml Collagen I (rat tail, Invitrogen) coating solution was prepared by dissolving in 0.02 N acetic acid. The human fibronectin (BD Bioscience) coating solution of the same concentration was prepared by dissolving it in 1X phosphate buffered saline (PBS) (Invitrogen). A 0.2% gelatin (Sigma-Aldrich) coating solution was obtained by dissolving gelatin powder in DI water. Then the ECM solutions were used to treat the PDMS surface for 1 hour at room temperature. The amount of solution needed for a microchip is determined by the surface area with the loading level at ∼5 μg/cm2. After coating, the extra solution was aspirated, and then PBS or DI water (for fribronectin coating) was used to rinse the PDMS surface three times. The coated surfaces were immunofluorescence stained with anti-collage I (Abcam) and anti-fibronectin (Abcam) antibodies and visualized using fluorescent secondary antibodies to verify the successful coating of ECM molecules onto PDMS. In addition, we also tested the culture of A549 lung epithelial cells (ATCC) on the PDMS surfaces coated with collagen I, fibronectin, gelatin, laminin, and control. Within a day, we observed apparent difference of cell adhesion between coated and uncoated PDMS surfaces, indicating the functional validity of our ECM coating method.
Dynamic mechanical stimulation on chip
During experiments, a multi-well cell culture fixture was attached to the top of the membrane. Gelatin 0.2% was used to coat the PDMS membranes for 1 h at room temperature before seeding cells. For the first 24 h, a static pressure of less than 14 kPa was applied to the device. This low pressure only introduced less than 2% strain to all channels but was high enough to prevent the free suspended membrane on 500 μm microchannels from retracting into the microgrooves. A continuously dynamic pulsing was introduced to the device after 24 h and lasted for 144 h. The total cell culture time was 7 days, one day under static pressure and 6 days under dynamic pressure. At the end of 1 day, 3 days, and 7 days, the devices were retrieved and cells were fixed for further characterization. The cells at day 1 were used as a reference. It has been reported that mechanical strain can regulate the expression of SMC markers in MSCs even as early as within 3 days11. Both early stage (α-actin) and intermediate stage (calponin) SMC markers could be observed in the MSCs treated by uniaxial dynamic mechanical stretch for 7 days13. Medium was changed every one to two days.
Immunofluorescence staining
To detect different signaling pathways, a standard immunofluorescence staining procedure was performed. Briefly, the MSCs on the PDMS membranes were fixed with 4% paraformaldehyde for 15 minutes and then rinsed 3 times with 1X phosphate buffered saline (PBS) (Invitrogen). The cells were blocked in blocking buffer (5% bovine serum albumin (BSA) (Sigma-Aldrich) and 0.3% Triton X-100 (Sigma-Aldrich) in 1X PBS) for 60 minutes. After rinsed with 1X PBS three times for 5 minutes each, the cells were incubated with primary antibodies in dilution buffer (1% BSA and 0.3% Triton X-100 in 1X PBS) for 60 minutes at room temperature. Following this, the cells were rinsed again and incubated with secondary antibodies for 60 minutes at room temperature. The primary antibodies used in this experiment were β-catenin (Cell Signaling), phospho-β-catenin (Cell Signaling), GSK-3β (Cell Signaling), phospho-SMAD1 (Invitrogen), SMAD2 (Invitrogen), and phospho-SMAD2 (Cell Signaling). The secondary antibodies are FITC conjugated goat anti-mouse IgG (Immunotech) and Alexa Fluor 555 conjugated anti-rabbit IgG (Cell Signaling). Before fluorescent imaging, the actin filaments were stained with Alexa Fluor 647 conjugated phalloidin (Invitrogen) for 20 minutes and the nuclei were stained with DAPI (Cell Signaling) for 5 minutes.
Fluorescence microscopy and quantitative image analysis
Images were collected under a fluorescent microscope with a CCD camera (EVOS fl, Advanced Microscopy Group, Bothell, WA, USA). All images in a given group were collected with the same hardware and software settings. CellProfiler 2.0 (Broad Institute, Cambridge, MA, USA), cell image analysis software, was used to analyze the cell number, cell shape, nucleus shape, and fluorescence intensity in cells. The nuclear staining was used to count the cell number and define the shape of the nuclei. The actin filament staining was applied to identify the cell shape and cytoplasmic area. The mean fluorescence intensity of each signaling antibody in cells was independently measured from GFP or RFP images based on the nucleus and cell shapes indentified from nuclear and actin filament staining. The intensity of GSK-3β and β-catenin were measured by the mean intensity in the whole cell. The nucleus and cytoplasm intensity of SMAD1 and SMAD2 were the mean intensity collected from nuclei and cytoplasm separately. Each point shown in the intensity plot represents one single cell.
Statistics
ANOVA analysis was used to analyze the fluorescence intensity between each group. p < 0.05 was defined as significant difference with * mark in plot, and p < 0.01 was defined as very significant difference with ** mark in plot. Statistic analysis of single-cell fluorescence intensity was performed on all the cells imaged on every microchannels with the number of cells analyzed >100. Seven independent experiments were conducted to examine cell alignment under long term dynamic stretching (7 days), including five SMAD1/2 measurements and four β-catenin/GSK3β measurements.
Supplementary Material
Insight, innovation, integration.
Mechanical forces play a crucial role in vascular development, remodeling and pathophysiology. A system that can emulate the complex mechanical conditions in a high throughput manner has tremendous value for the study of vascular mechanobiology and tissue engineering. Here, we developed such a system using integrated microfluidics technology. Cells cultured on chip can be stimulated by cyclic quasi-circumferential strain similar to the mechanical conditions of small blood vessels. It demonstrates that human mesenchymal stem cells can be continuously stimulated by cyclic stretch for up to 6 days. We further assess the feasibility to probe multiple signaling pathways including SMAD1/SMAD2 and canonical Wnt/β-catenin in response to dynamic mechanical stimulation on chip via a combinatorial approach. This microfluidic artificial vessel chip represents a generic and versatile platform for high-throughput and rapid screening of cellular responses to mechanical cues that emulate the physiological conditions of blood vessels, and may have a wide range of applications in the fields of stem cell mechanobiology, vascular tissue engineering, and other areas of regenerative medicine.
Acknowledgments
This research was supported by the National Institutes of Health Award (NIH R01 EB008366 and EB008366-03S1, PI: L.E.N.). J.Z. thanks Dr. Yu Wu for technical help on using cell profiler programs and valuable discussions. J.Z. also thanks Dr. Yao Lu for assistance in device fabrication. J.Z. also thanks Dr. Wenwen Tang for help with confocal fluorescent microscopy.
Footnotes
Competing financial interest: L.E.N. has a financial interest in Humacyte, Inc, a regenerative medicine company. Humacyte did not fund these studies, and Humacyte did not affect the design, interpretation, or reporting of any of the experiments herein.
References
- 1.Caplan AI, Bruder SP. Mesenchymal stem cells: building blocks for molecular medicine in the 21st century. Trends Mol Med. 2001;7:259–264. doi: 10.1016/s1471-4914(01)02016-0. [DOI] [PubMed] [Google Scholar]
- 2.Nesselmann C, et al. Mesenchymal stem cells and cardiac repair. Journal of Cellular and Molecular Medicine. 2008;12:1795–1810. doi: 10.1111/j.1582-4934.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrmann JL, et al. Cell-Based Therapy for Ischemic Heart Disease: A Clinical Update. Annals of Thoracic Surgery. 2009;88:1714–1722. doi: 10.1016/j.athoracsur.2009.05.079. [DOI] [PubMed] [Google Scholar]
- 4.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circulation Research. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 5.Mangi AA, et al. Mesenchymal stem cells modified with Akt prevent remodeling and restore performance of infarcted hearts. Nature Medicine. 2003;9:1195–1201. doi: 10.1038/nm912. [DOI] [PubMed] [Google Scholar]
- 6.Tomita S, et al. Autologous transplantation of bone marrow cells improves damaged heart function. Circulation. 1999;100:247–256. doi: 10.1161/01.cir.100.suppl_2.ii-247. [DOI] [PubMed] [Google Scholar]
- 7.Shake JG, et al. Mesenchymal stem cell implantation in a swine myocardial infarct model: Engraftment and functional effects. Annals of Thoracic Surgery. 2002;73:1919–1925. doi: 10.1016/s0003-4975(02)03517-8. [DOI] [PubMed] [Google Scholar]
- 8.Gong Z, Niklason LE. Small-diameter human vessel wall engineered from bone marrow-derived mesenchymal stem cells (hMSCs) Faseb Journal. 2008;22:1635–1648. doi: 10.1096/fj.07-087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang DJ, et al. Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor beta 1 stimulation. Journal of Biological Chemistry. 2004;279:43725–43734. doi: 10.1074/jbc.M407368200. [DOI] [PubMed] [Google Scholar]
- 10.Gojo S, et al. In vivo cardiovasculogenesis by direct injection of isolated adult mesenchymal stem cells. Experimental Cell Research. 2003;288:51–59. doi: 10.1016/s0014-4827(03)00132-0. [DOI] [PubMed] [Google Scholar]
- 11.Park JS, et al. Differential effects of equiaxial and uniaxial strain on mesenchymal stem cells. Biotechnology and Bioengineering. 2004;88:359–368. doi: 10.1002/bit.20250. [DOI] [PubMed] [Google Scholar]
- 12.Kurpinski k, Park Jennifer, Thakar Rahul G, Li Song. Regulation of vascular smooth muscle cells and mesenchymal stem cells by mechanical strain. Molecular & Cellular Biomechanics. 2006;3:21–34. [PubMed] [Google Scholar]
- 13.Hamilton DW, Maul TM, Vorp DA. Characterization of the response of bone marrow-derived progenitor cells to cyclic strain: implications for vascular tissue-engineering applications. Tissue Eng. 2004;10:361–369. doi: 10.1089/107632704323061726. [DOI] [PubMed] [Google Scholar]
- 14.Niklason LE, et al. Functional arteries grown in vitro. Science. 1999;284:489–493. doi: 10.1126/science.284.5413.489. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Zhu XY, Futai N, Cho BS, Takayama S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan W, Scott D, Belchenko D, Qi HJ, Xiao L. Development and evaluation of microdevices for studying anisotropic biaxial cyclic stretch on cells. Biomedical Microdevices. 2008;10:869–882. doi: 10.1007/s10544-008-9201-8. [DOI] [PubMed] [Google Scholar]
- 17.Maul TM, Chew DW, Nieponice A, Vorp DA. Mechanical stimuli differentially control stem cell behavior: morphology, proliferation, and differentiation. Biomech Model Mechanobiol. 2011 doi: 10.1007/s10237-010-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moraes C, Chen JH, Sun Y, Simmons CA. Microfabricated arrays for high-throughput screening of cellular response to cyclic substrate deformation. Lab on a Chip. 2010;10:227–234. doi: 10.1039/b914460a. [DOI] [PubMed] [Google Scholar]
- 19.Haynesworth SE, Baber MA, Caplan AI. Cell-Surface Antigens on Human Marrow-Derived Mesenchymal Cells are Detected by Monoclonal-Antibodies. Bone. 1992;13:69–80. doi: 10.1016/8756-3282(92)90363-2. [DOI] [PubMed] [Google Scholar]
- 20.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 21.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nature Cell Biology. 2007;9:1000–1004. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 22.Dijke P, Heldin Carl-Henrik. Smad signal transduction: smads in proliferation, differentiation and disease. Springer; 2006. [Google Scholar]
- 23.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 24.Kurpinski K, et al. Transforming Growth Factor-beta and Notch Signaling Mediate Stem Cell Differentiation into Smooth Muscle Cells. Stem Cells. 2010;28:734–742. doi: 10.1002/stem.319. [DOI] [PubMed] [Google Scholar]
- 25.Kinner B, Zaleskas JM, Spector M. Regulation of smooth muscle actin expression and contraction in adult human mesenchymal stem cells. Experimental Cell Research. 2002;278:72–83. doi: 10.1006/excr.2002.5561. [DOI] [PubMed] [Google Scholar]
- 26.Ling L, Nurcombe V, Cool SM. Wnt signaling controls the fate of mesenchymal stem cells. Gene. 2009;433:1–7. doi: 10.1016/j.gene.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 28.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.