Abstract
The methylotrophic yeast, Pichia pastoris, has been genetically engineered to produce many heterologous proteins for industrial and research purposes. In order to secrete proteins for easier purification from the extracellular medium, the coding sequence of recombinant proteins are initially fused to the Saccharomyces cerevisiae α-mating factor secretion signal leader. Extensive site-directed mutagenesis of the prepro region of the α-mating factor secretion signal sequence was performed in order to determine the effects of various deletions and substitutions on expression. Though some mutations clearly dampened protein expression, deletion of amino acids 57-70, corresponding to the predicted 3rd alpha helix of α-mating factor secretion signal, increased secretion of reporter proteins horseradish peroxidase and lipase at least 50% in small-scale cultures. These findings raise the possibility that the secretory efficiency of the leader can be further enhanced in the future.
Keywords: Pichia pastoris, α-Mating factor secretion signal, Recombinant protein expression
1. Introduction
In the past decade, the methylotrophic yeast, Pichia pastoris, has been genetically engineered to produce hundreds of recombinant proteins important to industrial, pharmaceutical, and basic research purposes. Many scientists favor the use of this particular yeast as an expression system for heterologous proteins because it can be easily manipulated on a genetic level, does not undergo fermentation like Saccharomyces cerevisiae, can add many eukaryotic posttranslational modifications, and has a number of strong promoters available to drive heterologous expression (Cereghino and Cregg, 2000; Macauley-Patrick et al., 2005).
Heterologous proteins expressed in P. pastoris can be produced either intracellularly or secreted extracellularly. Because P. pastoris secretes only small amounts of endogenous proteins, the secreted recombinant protein constitutes the vast majority of the total protein in the medium. Therefore, directing a heterologous protein to the culture supernatant can be a significant first step in purification (Lin-Cereghino et al., 2007).
In addition to native signal sequences, several signal peptides such as the P. pastoris acid phosphatase (PHOI) or PHA-E from Phaseolus vulgaris agglutinin (Bieszke et al., 1999; Raemaekers et al., 1999) have been used for secreted expression from P. pastoris. In fact, vector systems such as PichiaPink™ from Life Technologies (Carlsbad, CA) offer 8 different secretion signals with its Secretion Signal Set. However, the Saccharomyces cerevisiae α-mating factor prepro peptide is still the most commonly used signal sequence for recombinant cargo proteins.
The α-mating factor prepro peptide signal leader consists of a 19-amino acid signal (pre) sequence followed by a 67-residue (pro) sequence containing three consensus N-linked glycosylation sites and a dibasic Kex2 endopeptidase processing site (Kurjan and Herskowitz, 1982). Processing of the α-mating factor secretion signal occurs in three steps: first, the pre signal is removed by signal peptidases in the endoplasmic reticulum; second, Kex2 endopeptidase severs the pro leader sequence between the arginine and lysine; and finally Ste13 protein rapidly cleaves the Glu-Ala repeats in the Golgi (Brake et al., 1984).
The pre and pro regions play important roles in the secretion and processing of the mature protein. As is the case with almost all signal peptides, the pre-peptide consists of a basic amino-terminus, a hydrophobic core, and a carboxy terminal polar region ending with a cleavage site (Kaiser et al., 1987). The small pre-peptide is believed to be important for interaction with the signal recognition particle, subsequent translocation into the ER, and folding of the nascent protein (Hegde and Bernstein, 2006). The pre-peptide can also determine whether the protein will undergo co-translational or post-translational import into the ER in yeast. The role of the pro-peptide, a generally hydrophobic protein interrupted by short stretches of charged or polar amino acids, is less clear. It is thought to slow down and ensure proper folding of the entire protein prior to and after translocation into the ER (Chaudhuri et al., 1992; Kjeldsen et al., 1997). Furthermore, secretion of some proteins can only be achieved if the pro-peptide is included in the leader sequence. For these reasons, the pro-peptide can be considered a molecular chaperonin for the secreted cargo protein (Fabre et al., 1991).
For many reasons that are not yet understood, P. pastoris often cannot secrete some proteins even if they possess a proper secretion leader. Certain recombinant proteins fused to the α-mating factor prepro peptide are retained in the ER or Golgi, which greatly diminishes the export of the protein to the extracellular medium. One strategy to enhance secretion efficiency is to modify the secretion leader. A few studies have been done to improve the secretory potential of the α-mating factor secretion signal through directed evolution, codon optimization or the addition of spacer sequences in P. pastoris (Kjeldsen et al., 1996; Xiong et al., 2005; Rakestraw et al., 2009). However, even though some improvement has been seen with the adjustments, to date, no systematic studies have been done to determine the effect of mutagenesis on the functionality of α-mating factor prepro peptide. We wanted to determine if deletions of specific sets of amino acids associated with structural features would affect secretion levels of heterologous proteins. To achieve our goals, site-directed mutagenesis on the 86-amino acid α-mating factor secretion signal was performed. These mutations were fused to reporter proteins horseradish peroxidase and Candida antartica lipase B. Subsequently, enzyme activities and protein levels in the extracellular medium were assessed. Our findings indicate that deletions in several regions of the α-mating factor secretion signal can substantially influence the secretion of both reporter proteins. Taken together, the results of these different deletions mutants are used to formulate a model of the structure of α-mating factor secretion signal which will be subsequently utilized to further enhance its function in protein export
2. Materials and methods
2.1 Strains, media and reagents
Pichia pastoris yJC100 (wild type) is a derivative of the original wild type P. pastoris strain NRRL Y11430 (North Regional Research Laboratories, US Department of Agriculture, Peoria, IL) and has been described previously (Lin-Cereghino et al., 2006). Yeasts were cultured in either YPD medium (1% yeast extract, 2% peptone, 2% dextrose), Buffered Minimal Glycerol-complex Medium (BMGY: 1% yeast extract, 2% peptone, 100mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base, 4×10-5 % biotin and 1% glycerol), or Buffered Minimal Methanol-complex Medium (BMMY: 1% yeast extract, 2% peptone, 100mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base, 4×10-5 % biotin and 0.5% methanol). Media was supplemented with 100μg/ml Zeocin® (Life Technologies, Carlsbad, CA) or 0.5mg/ml G418 (Gold Biotechnology, Inc., St. Louis, MO), as necessary.
Recombinant DNA manipulations were carried out using the Escherichia coli strain TOP10 (Invitrogen Corp., Carlsbad, CA). TOP10 was cultured at 37°C in LB (0.5% yeast extract, 1% glucose, and 0.5% NaCl) supplemented with 25μg/ml Zeocin® or 30μg/ml kanamycin, as necessary for plasmid selection. Recombinant DNA methods, including bacterial transformation, were performed essentially as described (Sambrook et al., 1989). Plasmid DNA was purified from E. coli cultures using the Zyppy™ Plasmid Miniprep Kit (Zymo Research, Irvine, CA). PCR products were purified with the DNA Clean & Concentrator™-5 (Zymo Research, Irvine, CA) prior to restriction digestion. Restriction enzymes were purchased from Fermentas (Hanover, MD). DNA fragments digested with restriction enzymes were resolved on FlashGels™ (Lonza, Allendale, NJ) or TBE agarose gels. Fragments for cloning were purified from TBE agarose gels by using the Geneclean II Kit (Qbiogene, Carlsbad, CA). Chromosomal DNA from P. pastoris transformants was prepared using the OmniPrep™ kit from G-Biosciences (St. Louis, MO). Oligonucleotides were synthesized by Sigma Genosys (Plano, TX). All site-directed mutations and ligation junctions were confirmed by DNA sequencing (Quintara Biosciences, Albany, CA).
2.2 Construction of plasmids
In order to construct the plasmid pPICZαB-CalB the primers 5’CalB-EcoRI: GCAGGAATTCTCGCTCTTCCATCTGGTTC and 3’CalB-NotI: CCTCTTGAGCGGCCGCTTATGGGG were used to PCR the Candida antartica lipase B gene from the plasmid pPpT4_CalB (Krainer et al., 2012). The PCR fragment containing the lipase B gene was then restricted with EcoRI and NotI and cloned into the synonomous sites in pPICZαB to create pPICZαB-CalB. The ligation junctions of the construct were confirmed by sequencing. The plasmids pPICZαB-HRP (Morawski et al., 2000) and pPpT4_CalB were kind gifts from Anton Glieder (Technische Universität Graz, Austria).
2.3 Site-directed mutagenesis
Deletions of the MATα prepro region were generated using QuickChange II XL Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). Primers were designed to anneal to the parent plasmids and “loop” out an area of the MATalpha prepro sequence creating a deletion. The HPLC-purified primers used in this work are listed in Table 1. Mutagenesis was performed according to manufacturer’s directions. Briefly, primers were diluted to a concentration of 25ng/μl and the following reaction mixture was set up: 5μl of 10X reaction buffer, 1μl parent plasmid, 5μl of each primer, 1μl of 10mM dNTP, 3 μl of Quik-Solution, 1 μl of PfuTurbo DNA polymerase, and sterile water to a final volume of 50μl. Cycling parameters for the reaction were: 1 cycle of 95°C for 5 min; 18 cycles of 95°C for 30s, 55°C for 1min, 68°C for 7 min, followed by a single cycle of 68°C for 10 min. After the reaction was complete, 1 μl of DpnI was added directly to the reaction mix and incubated at 37°C for 1 h to destroy the methylated, parental DNA. Finally, 5μl of the digested reaction mix was transformed into One Shot® TOP10 chemically competent cells and transformants were plated on LB agar plates supplemented with Zeocin®.
Table 1.
Primers used to make α-mating factor secretion signal mutants
| Primer name | Sequence (5’-3’) Construct Name | |
|---|---|---|
| JCdelahelixtop | CGATGAGATTTCCTGCTGCTCCAGTCAACAC | |
| Δ4-18 | ||
| JCdelahelixbot | GTGTTGACTGGAGCAGCAGGAAATCTCATCG | |
| KOMNdelwholeMatalphaprotop | GCAGCATCCTCCGCATTAGCTAAAAGAGAGGCTGAAGCTCAG | |
| Δ20-83 | ||
| KOMNdelwholeMatalphaprobottom | CTGAGCTTCAGCCTCTCTTTTAGCTAATGCGGAGGATGCTGC | |
| JENMATSSDEL10TOP | CATTAGCTGCTCCAGTGCATTCCGGCTGAAGCTG | |
| Δ23-32 | ||
| JENMATSSDEL10BOTTOM | CAGCTTCAGCCGGAATGACTGGAGCAGCTAATG | |
| JENMATSSDEL3TOP | GTCAACACTACAACAACGGCACAAATTCCGG | |
| Δ27-29 | ||
| JENMATSSDEL3BOTTOM | CCGGAATTTGTGCCGTTGTTGTAGTGTTGAC | |
| MATalphaDK40delTOP | CTACAACAGAAGATGAATTAGAAGGGGATTTCG | |
| Δ30-43 | ||
| MATalphaDK40delBOTTOM | CGAAATCCCCTTCTAATTCATCTTCTGTTGTAG | |
| TopmatalphaDK40fillin | CAACAGAAGATGAAGCTGCTGCTGCTGCTGCTAAGCTTGCTGCTGCTGCTGCTGCTTTAGAAGGGGATTTC | |
| A:Δ30-43 | ||
| BottommatalphaDK40fillin | GAAATCCCCTTCTAAAGCAGCAGCAGCAGCAGCAAGCTTAGCAGCAGCAGCAGCAGCTTCATCTTCTGTTG | |
| DKdelAEAVIGYSDtop | GATGAAACGGCACAAATTCCGTTAGAAGGGGATTTC | |
| Δ35-37 | ||
| DKdelAEAVIGYSDbot | GAAATCCCCTTCGAACGGAATTTGTGCCGTTTCATC | |
| JENMATSSDEL7TOP | GCTGTCATCGGTTACTCAGTTGCTGTTTTGCCA | |
| Δ43-49 | ||
| JENMATSSDEL7BOTTOM | TGGCAAAACAGCAACTGAGTAACCGATGACAGC | |
| LLdelNSTXItop | GCTGTTTTGCCATTTTCCGCCAGCATTGCTGCTAAAG | |
| Δ57-70 | ||
| LLdelNSTXIbot | GATGAAACGGCACAAATTCCGTTAGAAGGGGATTTC | |
| TopMATalphaLL3fillin | GTTTTGCCATTTTCCGCTGCTGCTGCTGCTGCTAAGCTTGCTGCTGCTGCTGCTGCTGCCAGCATTGCTGCTAAAG | |
| A:Δ57-70 | ||
| BottomMATalphaLL3fillin | CTTTAGCAGCAATGCTGGCAGCAGCAGCAGCAGCAGCAAGCTTAGCAGCAGCAGCAGCAGCGGAAAATGGCAAAAC | |
| MATalpha del57-60 top | GTTTTGCCATTTTCCAACGGGTTATTGTTTATAAATAC | |
| Δ57-60 | ||
| MATalpha del57-60 bottom | GTATTTATAAACAATAACCCGTTGGAAAATGGCAAAAC | |
| Lauren5deltop | CCATTTTCCAACAGCACATTTATAAATACTACTATTGCC | |
| Δ61-65 | ||
| Lauren5delbottom | GGCAATAGTAGTATTTATAAATGTGCTGTTGGAAAATGG | |
| Lauren10deltop | CCATTTTCCAACAGCACAATTGCCAGCATTGCTGC | |
| Δ61-70 | ||
| Lauren10delbottom | GCAGCAATGCTGGCAATTGTGCTGTTGGAAAATGG | |
| MATalphaDK40delTOP | CTACAACAGAAGATGAATTAGAAGGGGATTTCG | |
| Δ30-43, Δ57-70 | ||
| MATalphaDK40delBOTTOM | CGAAATCCCCTTCTAATTCATCTTCTGTTGTAG | |
| DKdelDFDtop | CGGTTACTCAGATTTAGAAGGGGTTGCTGTTTTGCCATTTTCCAACAGC | |
| Δ47-49, Δ57-70 | ||
| DKdelDFDbottom | GCTGTTGGAAAATGGCAAAACAGCAACCCCTTCTAAATCTGAGTAACCG | |
2.4 P. pastoris transformation
Yeast transformations were done by electroporation methods as described (Lin-Cereghino et al., 2005; Lin-Cereghino et al., 2007). Plasmids were linearized within the AOX1 promoter with the restriction enzyme Mph1103I, purified with the DNA Clean & Concentrator™-5 (Zymo Research, Irvine, CA) after restriction digestion, and electroporated into competent yJC100 cells. Immediately after electroporation, cells were allowed to recover for 1 h at 30°C in 1ml of 50% 1M sorbitol/ 50% YPD and plated on selective media. Transformants were confirmed by colony PCR as described previously (Thor et al., 2005).
2.5 Real-Time PCR
Real-time PCR reactions were performed using the DyNAmo™ SYBR® green kit from New England BioLabs (Beverly, MA) to determine the copy number of the recombinant HRP genes in all strains. Reactions were composed of 10μl of 2X reaction buffer, 10ng of genomic DNA, and 0.3μM primers in a 20μl total reaction volume. PCR reactions were carried out in a BioRad MiniOpticon™ (Hercules, CA) with the following parameters: 95°C, 5 min (1 cycle); 95°C for 1 min, 59°C for 1 min, 72°C for 1 min (40 cycles). Primers used to amplify the HRP gene were: HRP_RT:Forward1 CAATTTCAGCAACACTGGGTTA and HRP_RT:Reverse1 AGGCCTTTCTGCTCCTCTAGAT. The primers for the MET2 (Thor et al., 2005) reference gene were: mets100 CGTTCTCGCAACTCTTTCGAA and metas100 CAATGGCATCAGTTATGACGGAAG. All samples were performed in duplicate and tested several times in different experiments as previously described (Lin-Cereghino et al., 2008). Q-gene software was used to analyze real-time PCR data (Muller et al., 2002).
2.6 Small scale induction of P. pastoris
Induction assays were performed in 50 mL conical tubes and deep well plates. Assays in conical tubes were done as previously described (Li et al., 2010). Yeast strains were grown in deep well plates (Bel-Art Scienceware, Wayne, NJ) essentially as described (Weis et al., 2004) with some modifications. Briefly, after filling perimeter wells with 500μl of sterile water, strains were inoculated into 250μl of BMD (buffered minimal dextrose: 1.34% yeast nitrogen base, 4×10-5% biotin, 200mM potassium phosphate buffer, pH, 6.0 and 1% dextrose). Plates were sealed with Parafilm® and grown at 29°C for 48hrs, 325 rpm. After 48 hrs, 250μl of 2XBMMY was added to the wells. Cells were allowed to continue growing and every 24 hrs, 50μl of BMM10 (buffered minimal methanol medium containing 5% methanol) was added to each well. After a total of 72 hours of methanol induction, optical density at 600nm was measured in a Spectronic™ Genesys 2 (Thermo Spectronic, Rochester, NY) and measurements were uniformly between 14 and 16 OD600. Cells were then centrifuged briefly and supernatants either used immediately for enzyme assays or frozen at -80°C for later use.
2.7 Horseradish peroxidase activity assay
Horseradish peroxidase activity was measured in yeast supernatants using the following procedure. Between 1 and 10μl of supernatant was added to 150μl of phosphate-buffered saline. At regular intervals, reactions were initiated with 50μl of TMB (3,3’,5,5’-Tetramethylbenzidine liquid substrate, Sigma-Aldrich, St. Louis, MO). After 45 seconds at room temperature, reactions were stopped by adding 200μl of 0.5M HCl. Samples were then read at 450nm in a Biotek ELx800 plate reader (Biotek Instruments, Winooski, VT). Activity was calculated using standard formulas from at least three separate isolates of each strain and divided by cell concentration (OD600) to arrive at activity/cell.
2.8 Horseradish peroxidase overlay plate assay
Yeast transformants grown on YNB-methanol plates were assayed for horseradish peroxidase activity using an overlay assay. A sterile mixture of 0.5% molecular biology grade agarose in phosphate buffered saline was liquefied in a microwave and held at 50°C. 2.5ml of TMB was mixed with an equal amount of melted agarose, vortexed gently, and poured over the plate of colonies. Samples were then observed for blue halos surrounding the colonies.
2.9 Lipase assay
Determination of lipase activity was performed essentially as described (Zhang et al., 2003). 50 μl of supernatant or 15μg of intracellular extracts were added to 350μl of assay solution (300mM Tris-HCl, pH 7; 1% DMSO, and 4mM p-nitrophenyl butyrate). Esterase activity was measured at room temperature every two minutes by following color development at 405nm in a Spectronic™ Genesys 2 (Thermo Spectronic, Rochester, NY) or Biotek ELx800 plate reader (Biotek Instruments, Winooski, VT). One unit is defined as the formation of 1μmol p-nitrophenol/min. Activity was calculated using standard formulas from at least three separate isolates of each strain and divided by cell concentration (OD600) to arrive at activity/cell.
2.10 Spot westerns
The spot western technique, though previously described (Lin-Cereghino et al., 2007), was performed with modifications. Supernatant volumes corresponding to equivalent numbers of cells were spotted onto nitrocellulose using a vacuum spot blot unit from Topac (Bristol, United Kingdom). Westerns were done with the SNAP i.d®. Protein Detection System from Millipore (Billerica, MA) following manufacturer’s instructions and using the following amounts of antibodies: 40μl rabbit anti-HRP (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA) and 2μl secondary goat anti-rabbit IgG (Applied Biosystems, Foster City, CA). Blots were developed with the femtoLUCENT™ PLUS-AP kit from G-Biosciences (St. Louis, MO). Blot images were captured and quantitated with a ChemiImager 5500 (Protein Simple, Santa Clara, CA).
2.11 Molecular modeling of α-mating factor prepro peptide
The secondary structure prediction for the protein sequence was performed by JPRED3 (Cole et al., 2008). The knob-socket model (Joo et al., 2012) based helix predictions were used to refine the boundaries of helices in the predicted secondary structure of the protein. Using amino acid preferences for the knobs, the packing interactions between helix 2 (35-43) and helix 3 (62-76) were predicted by socket patterns on each helix. Protein structure modeling program Modeller (Sali and Blundell, 1993) was used to generate the extended chain for the full-length sequence of the protein. The secondary structure constraints were specified to induce the designated regions as helices. An initial model was generated using the distance constraints from pairs of knob-socket packing at the interface between two helices. In the iterative process of modelling, additional distance constraints were specified to refine the packing interface. The loop region (44-61) between helix 2 and helix 3 was modeled using Modeller’s default loop building protocol and the final model was selected based on energy function ranking from Modeller. Visualization and manual packing iterations during model building process was performed using UCSF Chimera (Pettersen et al., 2004) visualization system.
2.12 Generation of cell-free extracts of Pichia pastoris
Intracellular extracts of Pichia pastoris were generated essentially as described (Thor et al., 2005) with the following modifications. Cell pellets, washed once in 500μl chilled phosphate buffered saline (PBS), were resuspended in 500μl PBS and Yeast/Fungal ProteaseArrest™ protease inhibitor cocktail from G-Biosciences (St. Louis, MO). To each sample tube, 150μl glass beads (500-600μm) were added. Tubes were then placed in a BioSpec Mini-Beadbeater-8 (Bartlesville, OK) and homogenized for a total of 3 minutes (1 minute pulses with 30 second pauses). Samples were then centrifuged at 13,200 rpm for 5 min. The supernatant was transferred to a clean tube and kept on ice. Protein levels were determined using the Pierce Bicinchoninic Acid (BCA) Protein Assay kit (Rockford, IL) with bovine serum albumin a standard. Assays were read at 562nm in a Biotek ELx800 plate reader (Biotek Instruments, Winooski, VT).
3. Results and discussion
3.1 Rationale for deletion mutagenesis
Addition of the α-mating factor secretion signal to heterologous proteins in Pichia pastoris usually results in the secretion of the intact heterologous protein to the extracellular medium. Because we desired to determine whether the absence of certain structural features would affect secretion, we generated several mutations in the leader peptide using site-directed mutagenesis based upon two strategies. First, because the residues composing pro-peptides are mostly hydrophobic, we originally hypothesized that the removal of regions rich in hydrophilic amino acids should significantly affect its ability to facilitate secretion of the HRP (horseradish peroxidase) cargo protein. Second, we predicted that deletions that disrupted secondary structures would also influence export of the reporter. Random mutagenesis resulting in single amino acid substitutions did not provide significant changes in secretion efficiency (data not shown). Therefore, site-directed mutagenesis was used either to remove regions of polar and charged amino acids or delete groups of residues that were essential to predicted helices or loop regions. A schematic of the α-mating factor secretion signal and its corresponding amino acid sequence are shown in Figure 1.
Figure 1.
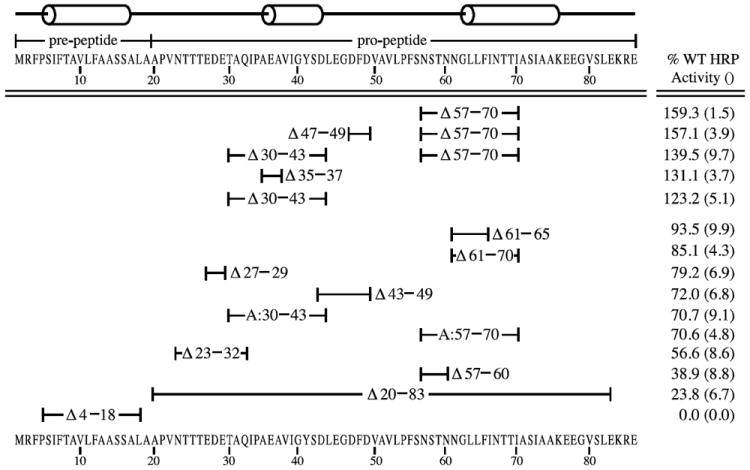
Horseradish peroxidase (HRP) activities of the α-mating factor secretion signal deletion mutants. The position of mutations are indicated by the amino acid numbers below the line drawing of α-mating factor secretion signal and the amino acid sequence of the pre-pro peptide. Amino acid deletions that have been substituted with alanines are indicated by an ‘A:’. P. pastoris strains expressing the mutant secretion signals fused to the HRP coding sequence were induced on methanol growth medium and the relative extracellular enzyme activities were determined. 100% corresponds to the specific activity of wild type α-mating factor secretion signal fused to HRP. The values represent averages of at least three trials on single copy strains. Standard deviations are indicated in parentheses.
The starting construct for our horseradish peroxidase (HRP) reporter assays was pPICZαB-HRP (Morawski et al., 2000) in which the α-mating factor prepro peptide is fused to HRP. In this construct, the methanol-inducible AOX1 promoter drives the expression of HRP, which is then directed to the extracellular medium by the α-mating factor secretion signal. We chose this particular reporter construct because there is a simple colorimetric enzyme assay using the liquid chemical substrate TMB (3,3’,5,5’-Tetramethylbenzidine) which can be added directly to agarose in a plate overlay assay or to diluted yeast supernatants to quantify HRP. In a positive reaction, the presence of HRP is indicated by a blue color that can be quantitated by the amount of absorbance at 450nm after acidification with 0.5M hydrochloric acid.
Using pPICZαB-HRP as a template for most deletions of single regions of amino acids, we used various oligonucleotide combinations to generate mutants. A list of oligonucleotides and resulting constructs are displayed in Table 1. Transformants of the deletion constructs into wild type P. pastoris strain yJC100 were selected on Zeocin®. To be certain that differences in HRP activity were not due to copy number differences, genomic DNAs from the various strains carrying modified alpha-mating factor constructs were isolated and the copy number of the HRP gene was found to be uniformly single copy (data not shown).
3.2 Deletion analysis
Single copy transformants were grown on medium to accumulate biomass, then induced in methanol for 72 hours and assayed for HRP activity. HRP activity was only found in supernatants and not in intracellular extracts, suggesting that HRP is only active after it has been secreted from the cell. A comparison of secreted enzyme activities is shown in Figure 1. All activities are shown relative to wild type activities.
As indicated in Figure 1, the α-mating factor signal peptide has two parts: a pre- and pro-region. To establish the necessity of both these regions deletion mutants of the pre- region (MATα:Δ4-18) and pro-region (MATα:Δ20-83) were constructed. As compared to wild type α-mating factor prepro peptide fused to HRP, a mutant deleted for the pre-region has no HRP activity. This is not surprising considering that this pre- region is thought to form an alpha helix which functions to bind the signal recognition particle required for entry into the endoplasmic reticulum (ER) (Stern et al., 2007). Deletion of the entire pro region (MATα:Δ20-83) severely affected HRP secretion, reducing export by about 75%, but was not entirely suppressed. This result confirmed that the pro region played a vital role in secretion efficiency and therefore deserved more detailed attention.
To investigate the contributions of hydrophilic amino acids to the function of the pro region, additional α-mating factor signal peptide deletions of MATα:Δ23-32, MATα:Δ27-29, and MATα:Δ43-49 were made. These deletions appear to dampen secretion approximately 20-40%. Such deletions may not change the overall essential structure of the signal peptide since they are in intervening loop structures (Figure 1). However, it is clear that increasing the overall hydrophobicity of the pro-peptide reduced the secretion efficiency of the HRP reporter, suggesting that these polar and charged residues play a positive role in secretion. These results are consistent with the findings of Xiong et al., 2006 who found that insertion of a 10-amino acid sequence rich in glutamic acid and alanine near the carboxy terminus of the α-mating factor leader peptide increased secretion of bacterial phytase over 5-fold in P. pastoris.
Based on structural predictions by the JPRED3 program (Cole et al., 2008) a model of the prepro leader was proposed using the knob-socket approach to verify helices and helical interactions (Joo et al., 2012). This model features: a) the pre region containing mainly an alpha helix and b) the pro region consisting of a large loop region framed by two interacting helices. These pro region helices sandwich a loop region that, according to our model shown in Figure 2, needs sufficient freedom in order to interact with the cargo protein or with trans-acting factors in the secretory organelles. Therefore, any mutations that increase this flexibility would favor interactions and thus secretion. In contrast, this model suggests that mutations constraining this loop restrains interactions and decreases secretion.
Figure 2.
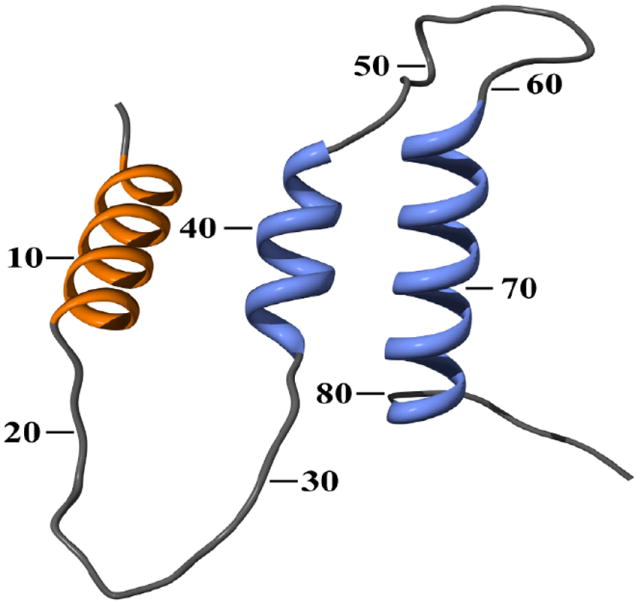
Predicted structure of the α-mating factor pre-pro secretion signal. The first helix is part of the pre-region, whereas the following two helices are part of the pro-region.
Deletions that influence secretion the most involve disrupting secondary structure motifs, specifically the removal of all or parts of either the 2nd or 3rd alpha helix located in the pro region. Deletion of part of the 2nd alpha helix (MATα:Δ35-37) or all of it (MATα:Δ30-43) have the same effect of raising secretion about 20-30%. Removal of the entire last helix and some residues N-terminal of it (MATα:Δ57-70) has a very large effect and appears to increase secretion by at least 50%. Because the amino acids that compose the pro-region are thought to slow down the rate of transport in order to provide ideal folding conditions for the protein to be secreted (Chaudhuri et al., 1992), it may be that the helical residues (30-43 and 57-70) of the pro-region constrain the interactions for proper secretion. Consequently, the removal of either one actually frees the loop (amino acids 44-56) such that cargo proteins fused to it move through the secretion pathway more efficiently. Shortening of the helix region by deletions MATα:Δ61-65 or MATα:Δ61-70 lowers secretion by at most 15%. The removal of these smaller stretches decreases the flexibility of the loop, according to our structural model, to interact with the secretion machinery, and therefore slightly decreases secretion efficiency. Interestingly, removal of 4 amino acids preceding the 3rd alpha helix (MATα:Δ57-60) dramatically decreases secretion of HRP. Based on our model, this mutation reduces the distance between the loop and the 3rd alpha helix which causes the 3rd helix to interfere with the loop’s ability to freely interact with the secretion pathway. However, further removal of residues into the 3rd helix (MATα:Δ61-70) disrupts this helix and thus removes the constraint on the loop’s interactions. In this way our model reconciles the result that deletion of residues 57-60 by itself greatly reduces secretion but that this deletion coupled with the removal of amino acids 61-70 increases the export of the HRP cargo protein.
3.3 Addition analysis and combination mutagenesis
Since it was found that deletion of either the 2nd or 3rd alpha helix of the α-mating factor signal peptide increased secretion of HRP by approximately 20-60% above wild type levels, we wanted to determine if the deletion of these specific amino acids or simply changing the length of the pro-region was responsible for the change. Therefore the mutants MATα:Δ30-43 and MATα:Δ57-70 were reconstructed by substituting alanines for the deleted amino acids. By doing this, the length of the pro region was replaced by alanines. In both cases, secretion dropped to approximately 70% of wild type, indicating that shortening of the regions, not just absence of the particular amino acids, was at the root of the secretion increases. As the stretches of polyalanine introduce large hydrophobic patches, the pro region has a higher likelihood of forming aggregate structures, and thus, indirectly interfering with loop flexibility. In light of our model, these results would indicate that the substitution of the normal amino acids with these polyalanine regions actually increased the level of constraint in the secretion leader, which caused decreased HRP export.
Additionally, a combination mutant of the deletion of both the 2nd and 3rd alpha helices was constructed with the thought that the two mutations combined might be additive or even synergistic. However, secretion of HRP in the mutant MATα:Δ30-43, Δ57-70 was actually at a level of approximately 140% of wild type, directly in between the secreted enzyme activity of each deletion by itself. Consistent with our model, the double deletion allows increased flexibility and interaction with the secretion machinery over the single MATα:Δ30-43, but not as much as with the MATα:Δ57-70. This result suggests that at least one of the two alpha helices is necessary for the secretion machinery to be most effective.
3.4 Western analysis
Because a secretion leader serves to assist in folding of a cargo protein, it could be argued that changes in the leader could cause the cargo protein to be folded into a more active conformation, which would lead to a higher activity in our HRP assays. Therefore, it is plausible that the increased HRP activities resulted not from increased amounts of exported HRP but instead on greater functional activity of the same amount of the secreted reporter. To confirm that the activity increases were a direct result of increase in heterologous protein secretion amounts, western spot analysis was performed on a select group of α-mating factor secretion signal mutants fused to horseradish peroxidase. As the results show in Figure 3, enzyme activity found in the culture medium can be roughly correlated with the amount of protein detected by anti-HRP antibody, demonstrating that the deletions change the number of HRP molecules being exported out of the yeast cell.
Figure 3.
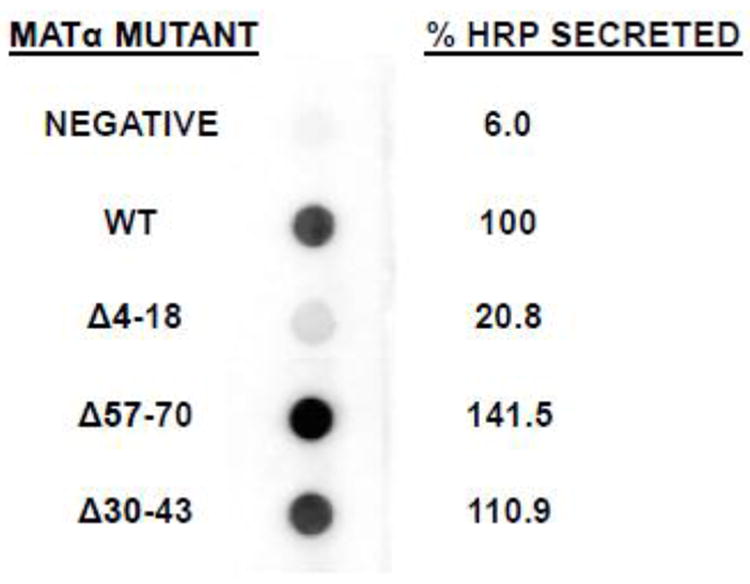
HRP spot western of selected α-mating factor secretion signal mutant constructs. Strains containing the indicated α-mating factor secretion signal mutants fused to HRP were induced on methanol medium and harvested after 72 h. Supernatant volumes corresponding to equivalent numbers of cells were spotted onto nitrocellulose and probed with anti-HRP antibodies. The blot was developed with the femtoLUCENT™ PLUS-AP detection system. Images captured with a ChemiImager 5500 were subjected to Integrated Densitometry Values (IDV) analysis and the numerical value of each spot was divided by that of the wild type strain in order to yield a relative percentage of HRP protein secreted. The negative sample is yJC100 alone.
3.6 Effect of deletions on an alternative reporter
For P. pastoris, the expression levels of different secreted recombinant proteins have ranged from 1mg/L to above 10g/L even though the same strains, expression vectors, and methodology were used (Cereghino and Cregg, 2000). Secretion efficiency is obviously not dictated solely by its signal leader peptide but rather depends in part on the structural nature of the entire protein (Smith et al., 1985; Verripsab et al., 2000). To determine whether the positive or negative effects of α-mating factor secretion signal deletions were transferable to another reporter protein, a selected group of α-mating factor prepro peptide deletions were fused to Candida antartica lipase B. Lipase B fused to a wild type α-mating factor secretion signal (pPICZαB-CalB) served as the template for site-directed deletion mutagenesis resulting in the generation of corresponding mutations Δ30-43, Δ43-49, Δ57-70, and Δ57-60 in the secretion signal. Although lipase activity seems to be more wide ranging (see Figure 4), the trend of the mutations follows that of the fusions to HRP, indicating that the α-mating factor secretion signal deletions have the same effect on the secretion of Lipase B as on HRP. Additionally, we wanted to confirm that the signal sequence modifications did not significantly alter the levels of the reporter intracellularly. When testing lipase activity in intracellular extracts, we found that mutant strains, such as Δ57-70, that secreted higher levels of lipase than wild type strains had intracellular levels of lipase comparable to or slightly lower than intracellular enzyme activity levels found in wild type (data not shown).
Figure 4.
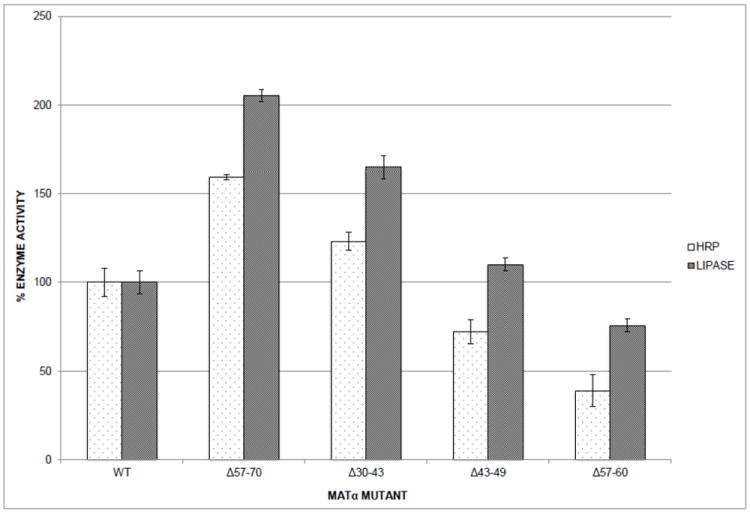
Relative enzyme activities of α-mating factor secretion signal mutant strains. Selected α-mating factor secretion signal deletion mutants were fused to reporter enzymes horseradish peroxidase (HRP) or Candida antartica lipase B (LIPASE). P. pastoris strains harboring the mutants were induced on methanol medium and harvested after 72 h. 100% corresponds to the specific activity of wild type α-mating factor secretion signal fused to HRP or lipase. The values represent averages of at least three trials on single copy strains and standard deviations are indicated by the error bars.
4. Conclusions
Using deletion mutagenesis of the Saccharomyces cerevisiae α-mating factor secretion signal, we have determined which stretches of amino acids can be removed to improve secretion and utilized these results to generate a model for its potential structure. Further studies to test this model will serve to elucidate the structure-function relationship it has to secretion in Pichia pastoris.
Highlights.
Mutations of the MATα pre-pro leader affected protein secretion in P. pastoris
Many deletions in α-mating factor pre-pro secretion signal reduced secretion
A deletion of amino acids 57-70 increased protein secretion at least 50%
Deletions had similar quantitative effects on two different reporters
Results were used to create a 3D model of the MATα leader
Acknowledgments
This work was supported by NIH-AREA grant GM65882-03 to J.L-C. and G.P.L-C. K.A. was a recipient of a Summer Undergraduate Research Fellowship from the University of the Pacific. N.S. and C.M.S. were both supported by the University of the Pacific Powell Scholars Program.
Abbreviations Gene-D-12-01318
- Ala
alanine
- AP
alkaline phosphatase
- BMGY
buffered minimal glycerol-complex medium
- BMMY
buffered minimal methanol-complex medium
- CalB
Candida antartica lipase B
- DMSO
dimethyl sulfoxide
- dNTP
deoxyribonucleoside triphosphate
- ER
endoplasmic reticulum
- Glu
glutamic acid
- HCl
hydrochloric acid
- HPLC
high-performance liquid chromatography
- HRP
horseradish peroxidase
- MATα
α-mating factor secretion signal
- P
Pichia
- PCR
polymerase chain reaction
- TBE
tris borate ethylenediaminetetraacetic acid
- TMB
3,3’,5,5’-tetramethylbenzidine
- YNB
yeast nitrogen base
- YPD
yeast peptone dextrose
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999;38:14138–45. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Brake AJ, Merryweather JP, Coit DG, Heberlein UA, Masiarz FR, Mullenbach GT, Urdea MS, Valenzuela P, Barr PJ. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984;81:4642–6. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Steube K, Stephan C. The pro-region of the yeast prepro-alpha-factor is essential for membrane translocation of human insulin-like growth factor 1 in vivo. Eur J Biochem. 1992;206:793–800. doi: 10.1111/j.1432-1033.1992.tb16986.x. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Nicaud JM, Lopez MC, Gaillardin C. Role of the proregion in the production and secretion of the Yarrowia lipolytica alkaline extracellular protease. J Biol Chem. 1991;266:3782–90. [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–71. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Joo H, Chavan AG, Phan J, Day R, Tsai J. An amino acid packing code for α-helical structure and protein design. J Mol Biol. 2012;419:234–54. doi: 10.1016/j.jmb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987;235:312–7. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Brandt J, Andersen AS, Egel-Mitani M, Hach M, Pettersson AF, Vad K. A removable spacer peptide in an alpha-factor-leader/insulin precursor fusion protein improves processing and concomitant yield of the insulin precursor in Saccharomyces cerevisiae. Gene. 1996;170:107–12. doi: 10.1016/0378-1119(95)00822-5. [DOI] [PubMed] [Google Scholar]
- Kjeldsen T, Pettersson AF, Hach M, Diers I, Havelund S, Hansen PH, Andersen AS. Synthetic leaders with potential BiP binding mediate high-yield secretion of correctly folded insulin precursors from Saccharomyces cerevisiae. Protein Expr Purif. 1997;9:331–6. doi: 10.1006/prep.1996.0695. [DOI] [PubMed] [Google Scholar]
- Krainer FW, Dietzsch C, Hajek T, Herwig C, Spadiut O, Glieder A. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb Cell Fact. 2012;11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurjan J, Herskowitz I. Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982;30:933–43. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Leung W, Yon A, Nguyen J, Perez VC, Vu J, Giang W, Luong LT, Phan T, Salazar KA, Gomez SR, Au C, Xiang F, Thomas DW, Franz AH, Lin-Cereghino J, Lin-Cereghino GP. Secretion and proteolysis of heterologous proteins fused to the Escherichia coli maltose binding protein in Pichia pastoris. Protein Expr Purif. 2010;72:113–24. doi: 10.1016/j.pep.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, de la Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–97. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Leung W, Lin-Cereghino J. Expression of protein in Pichia pastoris. In: Dyson M, Durocher Y, editors. Expression Systems: Methods Express. Scion Publishing Limited; Oxfordshire: 2007. pp. 123–145. [Google Scholar]
- Lin-Cereghino J, Hashimoto MD, Moy A, Castelo J, Orazem CC, Kuo P, Xiong S, Gandhi V, Hatae CT, Chan A, Lin-Cereghino GP. Direct selection of Pichia pastoris expression strains using new G418 resistance vectors. Yeast. 2008;25:293–9. doi: 10.1002/yea.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino J, Wong WW, Xiong S, Giang W, Luong LT, Vu J, Johnson SD, Lin-Cereghino GP. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques. 2005;38:44–48. doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–70. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- Morawski B, Lin Z, Cirino P, Joo H, Bandara G, Arnold FH. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 2000;13:377–84. doi: 10.1093/protein/13.5.377. [DOI] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–4. 1376, 1378–9. [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Raemaekers RJ, de Muro L, Gatehouse JA, Fordham-Skelton AP. Functional phytohemagglutinin (PHA) and Galanthus nivalis agglutinin (GNA) expressed in Pichia pastoris correct N-terminal processing and secretion of heterologous proteins expressed using the PHA-E signal peptide. Eur J Biochem. 1999;265:394–403. doi: 10.1046/j.1432-1327.1999.00749.x. [DOI] [PubMed] [Google Scholar]
- Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A Laboratory Handbook. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Smith RA, Duncan MJ, Moir DT. Heterologous protein secretion from yeast. Science. 1985;229:1219–1224. doi: 10.1126/science.3939723. [DOI] [PubMed] [Google Scholar]
- Stern B, Olsen L, Trosse C, Ravneberg H, Pryme I. Improving mammalian cell factories: The selection of signal peptide has major impact on recombinant protein synthesis and secretion in mammalian cells. Trends in Cell and Molecular Biology. 2007;2:1–17. [Google Scholar]
- Thor D, Xiong S, Orazem CC, Kwan AC, Cregg JM, Lin-Cereghino J, Lin-Cereghino GP. Cloning and characterization of the Pichia pastoris MET2 gene as a selectable marker. FEMS Yeast Res. 2005;5:935–42. doi: 10.1016/j.femsyr.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Verripsab T, Duboc P, Visser C, Sagt C. From gene to product in yeast: production of fungal cutinase. Enzyme Microb Technol. 2000;26:812–818. doi: 10.1016/s0141-0229(00)00176-9. [DOI] [PubMed] [Google Scholar]
- Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;2:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Xiong AS, Yao QH, Peng RH, Han PL, Cheng ZM, Li Y. High level expression of a recombinant acid phytase gene in Pichia pastoris. J Appl Microbiol. 2005;98:418–28. doi: 10.1111/j.1365-2672.2004.02476.x. [DOI] [PubMed] [Google Scholar]
- Zhang N, Suen WC, Windsor W, Xiao L, Madison V, Zaks A. Improving tolerance of Candida antarctica lipase B towards irreversible thermal inactivation through directed evolution. Protein Eng. 2003;16:599–605. doi: 10.1093/protein/gzg074. [DOI] [PubMed] [Google Scholar]


